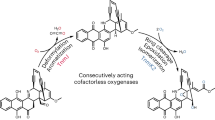
Abstract
Oxepinamides are derivatives of anthranilyl-containing tripeptides and share an oxepin ring and a fused pyrimidinone moiety. To the best of our knowledge, no studies have been reported on the elucidation of an oxepinamide biosynthetic pathway and conversion of a quinazolinone to a pyrimidinone-fused 1H-oxepin framework by a cytochrome P450 enzyme in fungal natural product biosynthesis. Here we report the isolation of oxepinamide F from Aspergillus ustus and identification of its biosynthetic pathway by gene deletion, heterologous expression, feeding experiments, and enzyme assays. The nonribosomal peptide synthase (NRPS) OpaA assembles the quinazolinone core with d-Phe incorporation. The cytochrome P450 enzyme OpaB catalyzes alone the oxepin ring formation. The flavoenzyme OpaC installs subsequently one hydroxyl group at the oxepin ring, accompanied by double bond migration. The epimerase OpaE changes the d-Phe residue back to l-form, which is essential for the final methylation by OpaF.
Similar content being viewed by others
Introduction
Oxepinamides are a class of oxepins with a fused pyrimidinone ring and were mainly found in fungi. For example, oxepinamides F, G, and H were isolated from Aspergillus puniceus1,2, varioloid A and varioxepine A from Paecilomyces variotii3,4, dihydrocinereain from Aspergillus carneus5, circumdatin A/B from Aspergillus ochraceus6, and versicoloid A/B from Aspergillus versicolor7 (Fig. 1a). Some oxepinamides show high affinity to liver X receptors (LXRs) and are potential agents for the treatment of Alzheimer’s disease, atherosclerosis, diabetes, and inflammation1,2. Oxepinamides are usually derived from quinazolinones with an anthranilyl (Ant) residue in common. They differ from each other by incorporation of two other varying amino acids and additional modifications. Until now, four trimodular nonribosomal peptide synthases (NRPSs) for assembling quinazolinones containing Trp, Ala, and Gly have been functionally characterized (Fig. 1b). In comparison to AldpA8 and CtqA8, TqaA9 and FmqA10 have an additional epimerization (E) domain in the Trp module, which is responsible for the conversion of l- to d- tryptophan in fumiquinazoline (FQF). As shown in Fig. 1a, phenylalanine, valine, leucine, and isoleucine residues are often found in the oxepinamide structures. To the best of our knowledge, genes responsible for quinazolinones with these amino acids have not been reported prior to this study.
In comparison to 1H-oxepins with three C=C in the oxepin ring, varioxepine A, varioloid A, oxepinamide F and G feature a 3H-oxepin structure with two C=C in the ring and one exo C=N bond (Fig. 1a). To the best of our knowledge, despite the intriguing structural features and biological activities, studies on the biosynthesis of oxepinamides, especially the formation of the 3H-oxepin ring, have not been reported yet.
We identify in this study an oxepinamide (opa) biosynthetic gene cluster (BGC) in Aspergillus ustus by bioinformatic analysis. Gene deletion, heterologous expression, feeding experiments, and in vitro assays with purified enzymes prove the biosynthetic steps and the 3H-oxepin formation by consecutive ring expansion and regio- and stereospecific hydroxylation. Furthermore, the d-phenylalanyl epimerase OpaE converts the d-Phe residue back to l-form for the last methylation step to form oxepinamide F.
Results
Identification of the opa BGC
Oxepinamide F (1) was isolated, together with its nonmethylated congener oxepinamide E (2) from a rice culture of A. ustus 3.3904. Their structures were confirmed by mass spectrometry (MS), nuclear magnetic resonance (NMR), optical rotation, and circular dichroism (CD) analyses (NMR data are listed in Supplementary Tables 5–8 and spectra in Supplementary Figs 7–30. CD spectra are given in Supplementary Fig. 3)1. Typical NMR signals for 3H-oxepins were observed at δH 6.7 (d, 7.3 Hz), δH 5.5 (t, 7.1 Hz), δH 6.2 (dd, 10.0, 7.0 Hz), and δH 5.8 ppm (d, 10.1 Hz), as well as δC 144, 104, 130, and 128 ppm in their spectra.
To find the biosynthetic genes of 1, we sequenced the A. ustus 3.3904 genome, carried out prediction for putative BGCs by using AntiSMASH11, and compared them with the published data12. The core structure of 1 is proposed to be assembled from Ant, Phe, and Ile by a NRPS containing at least three A-T-C (A: adenylation, T: thiolation, C: condensation) modules13. 1 differs from 2 merely in the methyl group at OH-12, implying that an O-methyltransferase (O-MeT) should be involved in its biosynthesis. Comprehensive sequence analysis revealed the presence of at least 11 genes for putative NRPSs. Two of them, KIA75458 and KIA75688, contain three A-T-C modules. Furthermore, a gene coding for a putative O-MeT (KIA75453) was only found neighboring to the gene for KIA75458. Thus, the KIA75458-related BGC was the best candidate for the 1 and 2 biosynthesis.
Inspection of the genomic neighborhood of these candidate genes in A. ustus revealed the presence of a putative BGC containing six genes opaA–opaF, coding for the putative proteins KIA75458–KIA75453 in the database (Fig. 2a). Sequence analysis and comparison suggested their functions as putative cytochrome P450 enzyme (OpaB, KIA75457), FAD-dependent oxygenase (OpaC, KIA75456), transporter (OpaD, KIA75455), epimerase (OpaE, KIA75454), and O-MeT (OpaF, KIA75453) (Supplementary Table 1).
a The oxepinamide biosynthetic gene cluster (opa) from A. ustus 3.3904. b HPLC analysis at 254 nm of the extracts after 7 day cultivation. c Structures of oxepinamide F (1), E (2), and 15-epi-oxepinamide E (5), as well as the CD spectra of 2 and 5 (200–400 nm). Blue double-headed arrows represent NOESY interactions.
Further sequence analysis revealed that KIA75458, named OpaA in this study, has a domain architecture of A-T-C-A-T-E-C-A-T-C (Fig. 2a). To prove its function, opaA in A. ustus was replaced with a hygromycin B resistance cassette by using a split marker gene replacement protocol14. The generated mutants were verified by PCR (Supplementary Fig. 1) and cultivated in rice media. High-performance liquid chromatography (HPLC) analysis of the culture extract of the ΔopaA mutant revealed the abolishment of both 1 and 2 production (Fig. 2b), proving its involvement in their biosynthesis. Deletion of opaF abolished 1, but not 2 production, indicating that OpaF acts as a methyltransferase for the conversion of 2 to 1 (Fig. 2b). Based on above results, the opa gene cluster is indeed responsible for biosynthesis of 1 and 2.
The oxepin ring in 1 and 2 was proposed to be formed by oxidative benzene ring expansion1,15. However, it is unknown whether the P450 enzyme OpaB, the oxidase OpaC or both are responsible for this conversion. To figure out their functions, we firstly deleted opaB from A. ustus genome. Deletion of opaB led to the abolishment of 1 and 2, together with the accumulation of a new peak 3 at 16.2 min with a [M + H]+ ion at m/z 362.189 (Fig. 2b). Typical signals for oxepins were not observed in the NMR spectra of 3. Extensive interpretation of the spectroscopic data including NMR and CD spectra and comparison with known compounds16 proved 3 to be protuboxepin K, a quinazolinone derivative of Ant, d-Phe, and l-Ile (Fig. 3)17.
a HPLC analysis of the conversion of 3 to 4 in the opaB expression strain of A. nidulans at 254 nm. b A sequence logo for the conserved EXXR motif in OpaB using sequences of 96 P450 enzymes (Supplementary Table 9). c Proposed mechanism for oxepin formation catalyzed by OpaB (SH: substrate).
Bioinformatic analysis and comparison with known proteins revealed that OpaA consists of a deduced AAnt-T1-C1-APhe-T2-E-C2-AIle-T3-CT domain structures, similar to CtqA, AldpA, FmqA, and TqaA. OpaA has the same AAnt domain for anthranilic acid activation (Fig. 1b). As aforementioned, the E domain in FmqA and TqaA is responsible for incorporation of a d-Trp residue in FQF. In analogy, the presence of an E domain in the second module of OpaA and the d-Phe residue in 3 imply that the second A domain is for activating of l-Phe, which is then converted to d-form by the E domain. The third A domain is responsible for l-Ile activation and the terminal CT domain for cyclization and releasing the NRPS product. Thus, OpaA is a quinazolinone synthase using different amino acids from those listed in Fig. 1b, which expands clearly quinazolinone structure diversity.
OpaB functions as an oxepinase
The results from deletion experiments provide unambiguous evidence that OpaB is responsible for the expansion of the benzene to the oxepin ring. For further understanding of its function, opaB was amplified from genomic DNA and cloned into pYH-gpdA-afpyrG18 via homologous recombination in yeast19 for heterologous expression in Aspergillus nidulans. The obtained plasmid pLZ61 was linearized by SwaI and integrated into the genome of A. nidulans LO803020 (Supplementary Fig. 2). Feeding 3 in the obtained overexpression strain A. nidulans LZ61 led to the detection of compound 4, which was not observed in a negative control. 4 was confirmed to be protuboxepin A by NMR analysis and by comparison of its optical rotation value with that reported in literature21. This confirmed that OpaB alone is responsible for the oxepin ring formation (Fig. 3a). Sequence alignment indicated that the conserved P450 motif ExxR is present as ETMR in OpaB (Fig. 3b and Supplementary Table 9)22,23,24. In analogy to other P450 catalyzed alkane hydroxylations and alkene epoxidations25,26, we postulated the electrophilic oxoferryliron (FeIV=O) as the active oxygen intermediate in the OpaB reaction. Attacking of the oxoferryliron species by the nucleophilic benzene double bond in 3 would result in the formation of an arene oxide, which is in rapid spontaneous equilibrium with the oxepin 4 through 6π electrocyclic ring opening (Fig. 3c)15,27. The oxepin form is expected to be more stable at room temperature than its arene oxide27,28. Only the 1H-oxepin 4 was detected as OpaB poduct in this study, which differs from an oxepin intermediate important for both phenylacetate degradation and tropone biosynthesis. In those cases, the isomerase PaaG forms a stable 3H-oxepin from a labile 1H-oxepin29. On the other hand, both 1H- and 3H-oxepin derivatives listed in Fig. 1 were isolated as stable fungal metabolites. Phylogenetic analysis of OpaB with 51 known P450 enzymes from bacteria and fungi indicates clearly the presence of different clades (Supplementary Fig. 4). The bacterial P450 enzymes catalyzing diverse reactions, including the two hydroxylases P450cin and P450cam with trace oxepin formation activity28, build a distinct clade from fungal enzymes. The majority of the fungal P450s in the phylogenetic tree catalyzes hydroxylations of diverse substrates. OpaB is located near to the epoxidase AtaY, but far away from the oxepinase AtaF, both involved in the biosynthesis of acetylaranotin in Aspergillus terreus30.
OpaC catalyzes hydroxylation accompanied by double bond migration
Comparing the planar structures, 2 differs from 4 in the OH-12 and two different double bonds. Sequence analysis suggested OpaC to be a FAD-containing monooxygenase and could be a good candidate for a consecutive hydroxylation at C12 of the oxepin ring. The double bonds could be shifted during the hydroxylation reaction. To verify its function biochemically, opaC was amplified from complementary DNA (cDNA) and cloned into pET28a (+) for overexpression in Escherichia coli. The recombinant N-terminally His6-tagged protein was purified to near homogeneity with a yield of 3.5 mg per liter culture (Fig. 4b). To our surprise, one product peak 5 at 13.3 min, instead of 2 at 12.8 min, was detected in the incubation mixture of 4 with the purified OpaC in the presence of NADPH (Fig. 4a). 5 shares the same UV visible light absorption and mass spectral features with 2. NMR data and CD data supported that 5 and 2 are diastereomers and differ from each other merely in the configuration at C15. 5 was, therefore, named 15-epi-oxepinamide E (Fig. 2c). Deletion of opaC from the A. ustus genome led indeed to the accumulation of 5 (Fig. 2b). Biochemical characterization revealed that OpaC also used NADH as a cofactor, but with a significantly reduced activity (Fig. 4a). No substrate consumption was observed in the incubation mixture of 3 and OpaC (Supplementary Fig. 5), proving the prerequisite of the oxepin ring for an acceptance by OpaC. The kinetic data of the OpaC reaction with 4 in the presence of NADPH fit well to a typical velocity equation with substrate inhibition31,32. An apparent Km value at 0.43 ± 0.04 mM, a turnover number (kcat) at 0.16 ± 0.01 s−1, the catalytic efficiency (kcat/Km) at 0.37 mM−1 s−1 and a substrate inhibition constant (Ki) at 0.39 ± 0.03 mM (Fig. 4c) were calculated by using the software GraphPad Prism 6.0.
a HPLC analysis of in vitro assays of 4 with OpaC at 254 nm. b SDS-PAGE analysis of the purified OpaC. The experiments were repeated twice. c Determination of the kinetic parameters of the OpaC reaction. The error bars represent standard errors of velocity and the standard errors of mean (SEMs) are given as ±values (n = six independent experiments). d Proposed mechanism for the OpaC reaction with a C4a-peroxyflavin as a key intermediate.
Based on these results, a mechanism with a C4a-hydroperoxyflavin intermediate33 was postulated for the OpaC reaction (Fig. 4d). The oxidized flavin Flox is converted to its reduced form Flred by external electron donor NADPH. Subsequent reaction of Flred with O2 produces the electrophilic reagent C4a-hydroperoxyflavin. The elimination of the proton at C3 in 4 results in the double bond migration and attack on the C4a-hydroperoxyflavin, leading to the formation of 5 and C4a-hydroxyflavin. The latter one is then regenerated to Flox by removal of one water molecule for the next reaction cycle.
Sequence analysis and biochemical investigation revealed that OpaC belongs to the well-studied class A flavin-dependent monooxygenases34. Phylogenetic analysis of representatives from this group (Supplementary Fig. 6) indicates the presence of at least three clades. OpaC is located closely together with AspB in the biosynthesis of asperlicins and PhqK in that of paraherquamides. AspB catalyzes the hydroxylation at C3 of an indole moiety, resulting in the formation of a hexahydropyrrolo[2, 3-b]indole framework35. PhqK converts an indole ring to a 2-keto indoline ring via a postulated arene oxide intermediate36. In both cases, the C4a-hydroperoxyflavin species was proposed to serve as oxygen transferring agent, consistent with other class A flavin monooxygenases and our mechanistic proposal.
d-Phenylalanyl epimerization catalyzed by OpaE
As aforementioned, 2 differs from 5 only in the configuration at C15. The S-configuration at this position in 2 corresponds to that of l-Phe, which was epimerized to R-configuration by the E domain of OpaA in 3. Conversion of 5 to 2 would need an epimerase like OpaE. Deletion of opaE led indeed to the abolishment of 1 and 2 production and main accumulation of 5 with a trace amount of a methylated product 6 (15-epi-oxepinamide F, Fig. 2b). To prove OpaE function in vitro and to understand the epimerization mechanism, the coding region of opaE was cloned and overexpressed as described for OpaC. HPLC analysis of the reaction mixture of 5 with the recombinant OpaE revealed 2 as the mere product peak (Fig. 5), proving unequivocally its function as an epimerase. Configuration change at C15 is a prerequisite for further methylation to the final product 1, because only trace amounts of 5 were converted to its methylated product 6 in ΔopaE mutant (Fig. 2b). Determination of the kinetic parameters of the OpaE reaction with 5 via Michaelis–Menten equation gave a Km value of 1.41 ± 0.05 mM, a turnover number (kcat) of 0.28 ± 0.01 s−1 and the catalytic efficiency (kcat/Km) at 0.20 mM−1 s−1 (Fig. 5c).
a HPLC analysis of in vitro assays with OpaE at 254 nm. b SDS-PAGE analysis of the purified OpaE. The experiments were repeated twice. c Determination of the kinetic parameters for OpaE with 5.The error bars represent standard errors of velocity and SEMs are given as ± values (n = six independent experiments). d LC–MS monitoring of the OpaE reactions in D2O/H2O and negative controls. e Proposed mechanism for the OpaE reaction.
Incubation of 5 and OpaE in Tris-HCl buffer containing D2O/H2O (9:1) and subsequent analysis on liquid chromatography–mass spectrometry (LC–MS) led to detection of the shifted [M + H]+ isotopic pattern of 2 ([M + H]+ 395.184), which is 1 Da larger than that in H2O ([M + H]+ 394.177) (Fig. 5d). Incubation of 2 in Tris-HCl buffer containing D2O/H2O (9:1) did not change the isotopic pattern. These results proved the involvement of an enol intermediate in the OpaE-catalyzed epimerization, as proposed in Fig. 5e37.
Discussion
Taken together, we identified the opa cluster for the oxepinamide F biosynthesis. OpaA was proven to activate Ant, l-Ile, and l-Phe, change the configuration of l-Phe to d-Phe by an epimerase domain, and assemble the quinazolinone 3. The benzene ring in the Ant residue of 3 was expanded to an oxepin ring in 4 by the P450 enzyme OpaB alone. The regio- and stereospecific hydroxylation of 4 catalyzed by OpaC was accompanied by double bond migration from C4-N5 and C6-C12 in 4 to C3-C4 and N5-C6 in 5, leading to the conversion of 1H-oxepin to 3H-oxepin system. The R-configuration in 5 was changed to S-configuration by the single epimerase OpaE for the final methylation of the OH-12 by the O-methyltransferase OpaF, to the end product 1 (Fig. 6).
Ant, l-Phe and l-Ile are assembled to protuboxepin K (3) by the NRPS OpaA with epimerization at the phenylalanyl residue. The oxepinase OpaB catalyzes the oxepin ring formation in protuboxepin A (4). The regio- and stereospecific hydroxylation of 4 by OpaC is accompanied by a double bond migration, leading to the conversion of a 1H-oxepin to a 3H-oxepin system in 5. The d-Phe configuration in 5 is changed to l-Phe in 2 by the single epimerase OpaE. The O-methyltransferase OpaF catalyzes the conversion of 2 to the end product 1 by methylation of the hydroxyl group at C-12.
Oxepin rings also play important roles in the bacterial degradation of phenylacetic acid and the biosynthesis of tropone natural products that both depend on the multicomponent epoxidase PaaABCE and oxepin isomerase PaaG29,38. Stok et al.28 observed the formation of a simple oxepin derivative as a minor side product in the hydroxylation of tert-butyl benzene by two bacterial P450 enzymes, which usually use the monoterpenes camphor and 1, 8-cineole as natural substrates, respectively. Wang and coworkers reported the formation of a dihydrooxepin ring in the biosynthesis of an epipolythiodioxopiperazine by three enzymes, the epoxidase AtaF, the acetyltransferase AtaH, and the cyctochrome P450 enzyme AtaY30. The oxepin ring was formed from an acetoxylated cyclohexan-diene structure.
In peptide biosynthesis, conversion of l- to d-form amino acids during the peptide biosynthesis is usually catalyzed by epimerase domains of NRPSs or by radical S-adenosylmethionine-dependent enzymes in microoganisms39,40. No single enzyme was reported to catalyze the conversion of d- to l-form of an amino acid residue. To the best of our knowledge, an oxepinamide biosynthetic gene cluster that includes the characterization of involved biosynthetic enzymes and reactions steps has not been reported before. Furthermore, conversion of an amino acid residue from l- to d-configuration and then back to the l-configuration by two different epimerases for structural modification has not been described in literature.
Methods
Genome sequencing and sequence analysis
The genome of A. ustus 3.3904 was sequenced by Genewiz (Suzhou, China) using Nova-seq6000/X-ten (Illumina). Initial prediction and analysis of biosynthetic gene clusters were carried out by using AntiSMASH41. Prediction of the enzyme function was performed with the online BLAST programmer (http://blast.ncbi.nlm.nih.gov). The genomic DNA sequence of the opa cluster (Supplementary Table 1) reported in this study corresponds well to that depicted at GenBank under accession number: JOMC01000153.1.
The phylogenetic trees were created by MEGA version 7.0 (http://www.megasoftware.net). Protein sequence alignments were performed with the program ClustalW (https://www.genome.jp/tools-bin/clustalw) to identify strictly conserved amino acid residues.
Strains, media, and growth conditions
Escherichia coli DH5α and BL21(DE3) cells were grown in LB medium (1% NaCl, 1% tryptone, and 0.5% yeast extract) at 37 °C. In all, 50 μg mL−1 ampicillin or 50 μg mL−1 kanamycin were supplemented for cultivation of recombinant strains.
Saccharomyces cerevisiae HOD114-2B cells were grown in YPD medium (1% yeast extract, 2% peptone and 2% glucose, 1.5% agarose was used for plate). The SC-uracil medium (6.7 g L−1 yeast nitrogen base with ammonium sulfate, 650 mg L−1 CSM-His-Leu-Ura (MP Biomedicals), 20 mg L−1 His and 60 mg L−1 Leu, pH 6.2 − 6.3, 1.5% agarose was used for plate) with 2.0% glucose was used for selection.
Fungal strains used in this study are listed in Supplementary Table 2. Aspergillus ustus 3.3904 was purchased from China General Microbiological Culture Collection Center (Beijing, China) and cultivated in rice medium (20 g Alnatura long-grain rice with 30 mL H2O in 250 mL flask) at 25 °C for of secondary metabolite production.
Aspergillus nidulans LO8030 and derivatives were grown at 37 °C on GMM medium (1.0% glucose, 50 mL L−1 salt solution, 1 mL L−1 trace element solution, and 1.6% agar) for sporulation and transformation with appropriate nutrition as required. The salt solution comprises (w/v) 12% NaNO3, 1.04% KCl, 1.04% MgSO4 ∙ 7H2O, and 3.04% KH2PO4. The trace element solution contains (w/v) 2.2% ZnSO4 ∙ 7H2O, 1.1% H3BO3, 0.5% MnCl2 ∙ 4H2O, 0.16% FeSO4 ∙ 7H2O, 0.16% CoCl2 ∙ 5H2O, 0.16% CuSO4 ∙ 5H2O, 0.11% (NH4)6Mo7O24 ∙ 4H2O, and 5% Na4EDTA.
Genomic DNA isolation
The A. ustus 3.3904 and A. nidulans mycelia were dried on filter paper and transferred into 2 mL Eppendorf tubes. After addition of 400 μL LETS buffer (10 mM Tris-HCl pH 8.0, 20 mM EDTA pH 8.0, 0.5% SDS, and 0.1 M LiCl) and four glass beads (2.85 mm in diameter), the tubes were vigorously mixed for 4 min. After addition of another 300 μL LETS buffer, the mixtures were treated with 700 μL phenol: chloroform: isoamyl alcohol (25: 24: 1). The genomic DNA in the aqueous phase was precipitated by addition of 900 μL absolute EtOH, followed by centrifugation at 17,000 x g for 30 min and washing with 70% EtOH. The obtained DNA as pellet was dried at 55 °C for 15 min and dissolved in 50–100 μL distilled H2O.
RNA isolation from A. ustus 3.3904 and cDNA synthesis
For this purpose, A. ustus 3.3904 was cultivated in rice medium at 25 °C for 7 days. The mycelia were collected by washing with 50 mL H2O and subsequent centrifugation. Fungal RNA Mini kit (VWR OMEGA bio-tek E.Z.N.A) was used for RNA extraction following the standard protocol provided by the manufacturer. For cDNA synthesis, the ProtoScript II First Strand cDNA Synthesis kit (BioLabs) was used with Oligo-dT as primers.
The deduced polypeptide from the coding region of opaC obtained from cDNA comprises 457 amino acids, lacking the five residues from 248 to 252 in KIA75456:
PCR amplification, gene cloning, and plasmid construction
Primers and plasmids used in this study are listed in Supplementary Tables 3 and 4, respectively. Seqlab GmbH (Göttingen, Germany) synthesized the PCR primers. Phusion® High-Fidelity DNA polymerase from New England Biolabs (NEB) were used for PCR amplification, which was carried out on a T100TM Thermal cycler from Bio-Rad by following the manufacturer´s suggestion for temperature profiles.
Genetic manipulation in A. ustus 3.3904 and cultivation of deletion mutants
To get germlings for protoplast preparation, fresh spores of A. ustus 3.3904 were inoculated in a 250 mL flask containing 50 mL LMM medium and incubated at 230 rpm and 30 °C. After 11 h, the germlings were harvested by centrifugation at 2280 x g in 4 °C for 15 min and washed with distilled H2O. The pellets were suspended in 10 mL osmotic buffer (1.2 M MgSO4 in 10 mM sodium phosphate, pH 5.8) containing 40 mg lysing enzyme from Trichoderma harzianum (Sigma) and 30 mg yatalase from Corynebacterium sp. OZ-21 (OZEKI Co., Ltd.) and incubated at 100 rpm and 30 °C for 10 h. The formation of protoplasts was monitored by controlling under microscope. The mixture was then transferred into a 50 mL falcon tube and overlaid gently with 10 mL of trapping buffer (0.6 M sorbitol in 0.1 M Tris-HCI, pH 7.0). The protoplasts appeared as an interface between the two buffer systems after centrifugation at 2280 x g and 4 °C for 15 min. This interface was transferred carefully into a 15 mL falcon tube. The protoplasts in this phase were collected by centrifugation at 2280 x g, and resuspended in 100 μL of STC buffer (1.2 M sorbitol, 10 mM CaCl2 in 10 mM Tris-HCI, pH 7.5).
For transformation, the obtained protoplasts were incubated with DNA samples (2–3 μg in 8–10 μL) on ice for 50 min. 1.25 mL of PEG solution (60% PEG 4000, 50 mM CaCl2, 50 mM Tris-HCI, pH 7.5) was then added, gently mixed and incubated at room temperature for 30 min. In all, 5 mL STC buffer were added into the mixture and spread on plates with SMM bottom medium (1.0% glucose, 50 mL L−1 salt solution, 1 mL L−1 trace element solution, 1.2 M sorbitol and 1.6% agar) containing 100 µg mL−1hygromycin B. The plates were overlaid softly by SMM top medium (1.0% glucose, 50 mL L−1 salt solution, 1 mL L−1 trace element solution, 1.2 M sorbitol, and 0.8% agar) containing 50 µg mL−1hygromycin B. After cultivating at 30 °C for 3–4 days, the fungal colonies were transferred on fresh PDA plates (PD medium with 3% agar) containing 100 µg mL−1 hygromycin B for selection. The potential transformants were verified via PCR amplification (Supplementary Fig. 1), cultivated in rice medium at 25 °C for 7 days and extracted with EtOAc. After evaporation, the extracts were dissolved in dimethyl sulfoxide (DMSO) and subjected to LC–MS for analysis.
Heterologous expression in A. nidulans
In this study, we used A. nidulans LO803042 as expression host. The protoplast preparation procedure was similar to that for A. ustus 3.3904 as mentioned above. The germlings were shaken with 40 mg lysing enzyme from Trichoderma harzianum (Sigma) and 20 mg yatalase from Corynebacterium sp. OZ-21 (OZEKI Co., Ltd.) at 100 rpm and 37 °C for 3 h. pLZ61 containing the cytochrome P450 gene opaB was transformed into A. nidulans LO8030 to create the expression strain LZ61. The transformants were verified by PCR (Supplementary Fig. 2) and cultivated for biotransformation with protuboxepin K (3).
Overproduction and purification of OpaC and OpaE
The opaC and opaE expression plasmids pLZ62 and pLZ63 were transferred separately into E. coli BL21(DE3).Terrific Broth (TB) medium (2.4% yeast extract, 2.0% tryptone, 0.4% glycerol, 0.1 M phosphate buffer, pH 7.4) was used for cultivation. OpaC overproduction was induced with 0.5 mM IPTG at 16 °C for 16 h, and OpaE with 1 mM IPTG at 20 °C for 20 h. The recombinant His6-tagged proteins were purified by Ni-NTA affinity chromatography (Qiagen, Hilden). The protein concentration was determined on a Nanodrop 2000c spectrophotometer (Thermo Scientific, Braunschweig, Germany) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Figs. 4b and 5b). Protein yields of 3.5 and 20 mg per liter bacterial culture were calculated for OpaC and OpaE, respectively.
In vitro assays of OpaC and OpaE
To test the enzyme activity of OpaC, the reaction mixtures (50 μL) containing 50 mM Tris-HCl (pH 7.5), 5 mM NADPH, 1 mM protuboxepin A (4), 10 μg (3.9 µM) of the purified recombinant OpaC were incubated at 30 °C for 30 min. The reactions were terminated by addition of 50 μL methanol.
For OpaE, the reaction mixtures (50 μL) containing 50 mM Tris-HCl (pH 7.5), 1 mM 15-epi-oxepinamide E (5), 5 μg (3.2 μM) of the purified recombinant OpaE were incubated at 37 °C for 30 min. The reactions were terminated by addition of 50 μL methanol.
For deuterium labeling experiment with OpaE, the stock solutions of the assay components in H2O were diluted with D2O to a final D2O/H2O ratio of 9:1 in 50 mM Tris-HCl. The reaction mixtures were incubated and analyzed on LC–MS as described for standard assays.
Determination of kinetic parameters
Enzyme assays for determination of the kinetic parameters for OpaC (50 μL) contained 50 mM Tris-HCl (pH 7.5), 5 mM NADPH, 4 μg (1.6 μM) of the purified recombinant OpaC and protuboxepin A (4) at final concentrations from 0.025 to 2.5 mM. The incubations were carried out at 30 °C for 30 min. After extraction of the reaction mixtures with EtOAC and evaporation of the solvent to dryness, the residues were dissolved in DMSO and analyzed on HPLC.
For OpaE, 50 μL reaction mixtures contained 50 mM Tris-HCl (pH 7.5), 2 μg (1.3 μM) of the purified recombinant OpaE and compound 15-epi-oxepinamide E (5) at final concentrations from 0.02 to 2 mM. The incubations were carried out at 37 °C for 30 min. After addition of MeOH and centrifugation, the supernatants were analyzed on HPLC. The data presented in Figs. 4c and 5c were obtained from six independent experiments. SEMs are given as ± values (n = six independent experiments).
Kinetic parameters were determined by nonlinear regression using the software GraphPad Prism 6.0 via substrate inhibition velocity equation for OpaC and Michaelis–Menten equation for OpaE.
Large-scale fermentation, extraction, and isolation of secondary metabolites
The strains were cultivated in 0.75 kg rice (1.75 L) for isolation. The rice culture was extracted with equal volume of EtOAc for three times. The EtOAc extracts were concentrated under reduced pressure to afford the crude extracts for further purification.
To isolate oxepinamide F (1) and E (2), the crude extract of A. ustus wildtype was subjected to silica gel (230‒400 mesh) column chromatography, by using a gradient of CH2Cl2/CH3OH (100:0‒0:100) to give eight fractions (Fr.1‒Fr.8). Fr.4 was purified on a semi-preparative HPLC (ACN/H2O), leading to 38.42 mg of oxepinamide F (1), 57.24 mg of oxepinamide E (2). Similarly, 77.68 mg protuboxepin K (3) was obtained from a culture of the ΔOpaB mutant, 79.24 mg protuboxepin A (4) from a culture of the ΔOpaC mutant, 23.88 mg 15-epi- oxepinamide E (5) 16.88 mg 15-epi-oxepinamide F (6) from a culture of the ΔOpaE mutant and 15.78 mg oxepinamide E (2) from a culture of the ΔOpaF mutant.
Feeding experiments in the opaB expression strain A. nidulans LZ61
To figure out the function of the cytochrome P450 enzyme OpaB, opaB was overexpressed in A. nidulans with the gpdA promoter from A. fumigatus18. Ten milliliter PDB with 75 μL 0.5 mg mL−1 riboflavin and 10 μL 0.5 mg mL−1pyridoxine in a 25 mL erlenmeyer flask were inoculated at 230 rpm and 30 °C. One milligram of protuboxepin K (3) (8 mg mL−1DMSO) was added into 3 days old culture of the transformant A. nidulans LZ61. 16 h later, 1 mL culture was extracted with 1 mL EtOAc. The extracts were dried and dissolved in 100 μL DMSO for LC–MS analysis. Strain LZ61 without feeding protuboxepin K (3) and A. nidulans assembled empty vector with or without feeding protuboxepin K (3) were used as negative controls.
HPLC analysis and metabolite isolation
Fungal extracts were analyzed on an Agilent HPLC series 1200 (Agilent Technologies) by using an Eclipse XDB-C18 column (Agilent Technologies, 5 μm, 4.6 × 150 mm) and ACN/H2O as elution solvents. A linear gradient from 10 to 90% ACN in H2O containing 0.1% (v/v) HCOOH in 20 min was used. After washing with 100% ACN for 5 min, the column was equilibrated with 10% ACN for another 5 min. A photodiode array detector was used for detection and the absorptions at 254 nm are illustrated in this study.
For product isolation, a Multospher 120 RP-18 column (5 μm, 10 × 250 mm) was used on the same HPLC system, with the same elution solvents at a flow rate of 2 mL min−1. Separation was performed by isocratic elution with 45–70% ACN in H2O containing 0.1% (v/v) HCOOH for 10–20 min, and.
LC–MS and MS analysis
Extracts were also analyzed on an Agilent HPLC 1260 series system equipped with a Bruker microTOF QIII mass spectrometer by using a Multospher 120 RP-18-5μ column (5 μm, 250 × 2 mm). Separation was accomplished in a 40 min linear gradient from 5 to 100% ACN in H2O, both containing 0.1% (v/v) HCOOH at a flow rate of 0.25 mL min−1. The column was then washed with 100% ACN for 5 min followed by equilibration with 5 % ACN for 10 min. Positive ions were scanned in the range of m/z 100–1500 under the following conditions: capillary voltage with 4.5 kV, collision energy with 8.0 eV and electrospray ionization. Mass calibration was achieved by using sodium formate in each run. Data collection and analysis were carried out with the Compass DataAnalysis 4.2 software (Bruker Daltonik, Bremen, Germany).
NMR analysis
Samples in high purity were dissolved in DMSO-d6 or CDCl3 and subjected to JEOL ECA-500 or ECA-400 (JEOL, Akishima, Tokyo, Japan) for taking NMR spectra. MestReNov.9.0.0 (Mestrelab Research, Santiago de Compostella, Spain) was used for spectral processing.
CD spectroscopic analysis
The samples were dissolved in CH3OH and measured in the range of 200–400 nm by using a 1 mm path length quartz cuvette (Hellma Analytics, Müllheim, Germany) on a J-815 CD spectrometer (Jasco Deutschland GmbH, Pfungstadt, Germany). The CD spectra are shown in Supplementary Fig. 3.
Measurement of optical rotations
Jasco DIP-370 at 25 °C equipped with the D-line of the sodium lamp at λ = 589.3 nm was utilized to measure the optical rotations of the isolated compounds in CHCl3 or CH3OH. The polarimeter was calibrated with the respective solvent before the measurement.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The information of opa gene cluster can be obtained from JOMC01000153.1 in the NCBI database. The authors declare that all relevant data supporting the finding of this study are available within the paper and its Supplementary Information files. All data are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Lu, X. et al. Oxepinamides: Novel liver X receptor agonists from Aspergillus puniceus. Eur. J. Org. Chem. 2011, 802–807 (2011).
Liang, X. et al. Diketopiperazine-type alkaloids from a deep-sea-derived Aspergillus puniceus fungus and their effects on liver X receptor α. J. Nat. Prod. 82, 1558–1564 (2019).
Zhang, P., Li, X., Wang, J. & Wang, B. Oxepine-containing diketopiperazine alkaloids from the algal-derived endophytic fungus Paecilomyces variotii EN-291. Helv. Chim. Acta 98, 800–804 (2015).
Zhang, P. et al. Varioxepine A, a 3H-oxepine-containing alkaloid with a new oxa-cage from the marine algal-derived endophytic fungus Paecilomyces variotii. Org. Lett. 16, 4834–4837 (2014).
Zhuravleva, O. I. et al. New metabolites from the algal associated marine-derived fungus Aspergillus carneus. Nat. Prod. Commun. 8, 1071–1074 (2013).
González-Jartín, J. M., Alfonso, A., Sainz, M. J., Vieytes, M. R. & Botana, L. M. Identification of circumdatins produced by Aspergillus ochraceus. J. Agr. Food Chem. 65, 4843–4852 (2017).
Wang, J. et al. Antifungal new oxepine-containing alkaloids and xanthones from the deep-sea-derived fungus Aspergillus versicolor SCSIO 05879. J. Agric. Food Chem. 64, 2910–2916 (2016).
Yan, D. et al. Complexity and diversity generation in the biosynthesis of fumiquinazoline-related peptidyl alkaloids. Org. Lett. 21, 1475–1479 (2019).
Gao, X. et al. Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat. Chem. Biol. 8, 823–830 (2012).
Wang, P. M. et al. TrpE feedback mutants reveal roadblocks and conduits toward increasing secondary metabolism in Aspergillus fumigatus. Fungal Genet. Biol. 89, 102–113 (2016).
Blin, K. et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019).
Pi, B. et al. A genomics based discovery of secondary metabolite biosynthetic gene clusters in Aspergillus ustus. PLoS ONE 10, e0116089 (2015).
Walsh, C. T., Haynes, S. W., Ames, B. D., Gao, X. & Tang, Y. Short pathways to complexity generation: fungal peptidyl alkaloid multicyclic scaffolds from anthranilate building blocks. ACS Chem. Biol. 8, 1366–1382 (2013).
Fan, J. et al. Peniphenone and penilactone formation in Penicillium crustosum via 1, 4-Michael additions of ortho-quinone methide from hydroxyclavatol to γ-butyrolactones from crustosic acid. J. Am. Chem. Soc. 141, 4225–4229 (2019).
Beaudry, C. M., Malerich, J. P. & Trauner, D. Biosynthetic and biomimetic electrocyclizations. Chem. Rev. 105, 4757–4778 (2005).
Fan, Y. et al. Alkaloids with cardiovascular effects from the marine-derived fungus Penicillium expansum Y32. Mar. Drugs 13, 6849–6504 (2015).
Ohte, S. et al. A new diketopiperazine-like inhibitor of bone morphogenetic protein-induced osteoblastic differentiation produced by marine-derived Aspergillus sp. BFM-0085. J. Antibiot. 73, 554–558 (2020).
Yin, W. B. et al. Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synth. Biol. 2, 629–634 (2013).
van Leeuwen, J., Andrews, B., Boone, C. & Tan, G. Rapid and efficient plasmid construction by homologous recombination in yeast. Cold Spring Harb. Protoc. 2015, 853–861 (2015).
Chiang, Y. M. et al. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J. Am. Chem. Soc. 135, 7720–7731 (2013).
Lee, S. U. et al. Protuboxepins A and B and protubonines A and B from the marine-derived fungus Aspergillus sp. SF-5044. J. Nat. Prod. 74, 1284–1287 (2011).
Syed, K. & Mashele, S. S. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE 9, e95616-1–e95616-14 (2014).
Schneider, T. D. & Stephens, R. M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Jin, S., Bryson, T. A. & Dawson, J. H. Hydroperoxoferric heme intermediate as a second electrophilic oxidant in cytochrome P450-catalyzed reactions. J. Biol. Inorg. Chem. 9, 644–653 (2004).
Sono, M., Roach, M. P., Coulter, E. D. & Dawson, J. H. Heme-containing oxygenases. Chem. Rev. 96, 2841–2888 (1996).
Vogel, E. & Günther, H. Benzene oxide-oxepin valence tautomerism. Angew. Chem. Intern. Ed. 6, 385–401 (1967).
Stok, J. E. et al. Direct observation of an oxepin from a bacterial cytochrome P450-catalyzed oxidation. Chem. Eur. J. 22, 4408–4412 (2016).
Spieker, M., Saleem-Batcha, R. & Teufel, R. Structural and mechanistic basis of an oxepin-CoA forming isomerase in bacterial primary and secondary Metabolism. ACS Chem. Biol. 14, 2876–2886 (2019).
Guo, C. J. et al. Biosynthetic pathway for the epipolythiodioxopiperazine acetylaranotin in Aspergillus terreus revealed by genome-based deletion analysis. J. Am. Chem. Soc. 135, 7205–7213 (2013).
Valentino, H. et al. Structure and function of a flavin-dependent S-monooxygenase from garlic (Allium sativum). J. Biol. Chem. 295, 11042–11055 (2020).
Copeland, R. A. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. (Wiley, New York, 2000).
Teufel, R. Flavin-catalyzed redox tailoring reactions in natural product biosynthesis. Arch. Biochem. Biophys. 632, 20–27 (2017).
Huijbers, M. M. E., Montersino, S., Westphal, A. H., Tischler, D. & van Berkel, W. J. Flavin dependent monooxygenases. Arch. Biochem. Biophys. 544, 2–17 (2014).
Haynes, S. W., Gao, X., Tang, Y. & Walsh, C. T. Assembly of asperlicin peptidyl alkaloids from anthranilate and tryptophan: a two-enzyme pathway generates heptacyclic scaffold complexity in asperlicin E. J. Am. Chem. Soc. 134, 17444–17447 (2012).
Fraley, A. et al. Molecular basis for spirocycle formation in the paraherquamide biosynthetic pathway. J. Am. Chem. Soc. 142, 2244–2252 (2020).
Wiese, A., Pietzsch, M., Syldatk, C., Mattes, R. & Altenbuchner, J. Hydantoin racemase from Arthrobacter aurescens DSM 3747: heterologous expression, purification and characterization. J. Biotechnol. 80, 217–230 (2000).
Duan, Y., Petzold, M., Saleem-Batcha, R. & Teufel, R. Bacterial tropone natural products and derivatives: Overview of their biosynthesis, bioactivities, ecological role and biotechnological potential. Chembiochem 21, 2384–2407 (2020).
Hur, G. H., Vickery, C. R. & Burkart, M. D. Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat. Prod. Rep. 29, 1074–1098 (2012).
Ogasawara, Y. & Dairi, T. Peptide epimerization machineries found in microorganisms. Front. Microbiol. 9, 1–8 (2018).
Weber, T. et al. Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol. 33, 15–26 (2015).
Chiang, Y. M. et al. Development of genetic dereplication strains in Aspergillus nidulans results in the discovery of aspercryptin. Angew. Chem. Int. Ed. Engl. 55, 1662–1665 (2016).
Acknowledgements
We thank Rixa Kraut, Lena Ludwig-Radtke, and Stefan Newel for taking MS and NMR spectra, Lena Ludwig-Radtke and Yiling Yang for technical assistance. This project was funded in part by the Deutsche Forschungsgemeinschaft (DFG, INST 160/620-1). Liujuan Zheng (201604910536) is a scholarship recipient from the China Scholarship Council. The CD spectrometer was provided by the core facility for protein biochemistry and spectroscopy in the Institute of Cytobiology.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
S-M.L. directed the research. L.Z. conducted genetic manipulation and biochemical studies. H.W. performed compound isolation and structure elucidation. L.Z., H.W., A.F., and S-M.L. designed the experiments, analyzed the data, and wrote the manuscript. L.Z. and H.W. contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zheng, L., Wang, H., Fan, A. et al. Oxepinamide F biosynthesis involves enzymatic d-aminoacyl epimerization, 3H-oxepin formation, and hydroxylation induced double bond migration. Nat Commun 11, 4914 (2020). https://doi.org/10.1038/s41467-020-18713-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-020-18713-0
This article is cited by
-
Design and synthesis of novel quinazolinyl-bisspirooxindoles as potent anti-tubercular agents: an ultrasound-promoted methodology
Molecular Diversity (2023)
-
A basidomycetous hydroxynaphthalene-prenylating enzyme exhibits promiscuity toward prenyl donors
Applied Microbiology and Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.