Abstract
Ghrelin is a gastric peptide hormone with important physiological functions. The unique feature of ghrelin is its Serine 3 acyl-modification, which is essential for ghrelin’s activity. However, it remains to be elucidated why the acyl-modification of ghrelin is necessary for activity. To address these questions, we solved the crystal structure of the ghrelin receptor bound to antagonist. The ligand-binding pocket of the ghrelin receptor is bifurcated by a salt bridge between E124 and R283. A striking feature of the ligand-binding pocket of the ghrelin receptor is a wide gap (crevasse) between the TM6 and TM7 bundles that is rich in hydrophobic amino acids, including a cluster of phenylalanine residues. Mutagenesis analyses suggest that the interaction between the gap structure and the acyl acid moiety of ghrelin may participate in transforming the ghrelin receptor into an active conformation.
Similar content being viewed by others
Introduction
Ghrelin, a peptide hormone consisting of 28 amino acids, was originally discovered in the stomach as an endogenous ligand for GHSR (growth hormone secretagogue receptor, now called the ghrelin receptor), a member of the β-branch in the class A GPCRs1,2. Ghrelin is the only peptide hormone that induces appetite stimulation following peripheral injection, and is thus functionally opposite to leptin3. Moreover, ghrelin has a wide range of physiological functions, playing roles in growth hormone secretion, adiposity, energy homeostasis, memory formation, and hippocampal neurogenesis4.
A salient feature of ghrelin is the O-acyl-modification at Ser32,5, which is essential for its activity; des-acyl ghrelin (i.e., ghrelin lacking the acyl-modification) is inactive. No other peptide hormone is known to require such an acyl-modification for its activity. The acyl-modification of ghrelin is catalyzed by ghrelin O-acyltransferase (GOAT)6,7,8. Although GOAT can transfer fatty acids ranging in size from acetic to palmitic acid onto des-acyl ghrelin, GOAT prefers medium-chain fatty acids as its substrates9. The main acyl-modified and most potent active form of ghrelin is n-octanoyl ghrelin, but other medium-chain fatty acids are also detected as endogenous forms of ghrelin. For example, n-decanoyl ghrelin has been purified from the stomachs of frog and bird at levels comparable to those of n-octanoyl ghrelin10,11. Moreover, calcium mobility assays using synthesized ghrelin with fatty acids of various lengths revealed that ghrelin is less active when modified with a shorter-chain fatty acid12. In addition, several reports indicated that the ghrelin receptor exhibits strong ligand-independent activity13,14,15. The structural basis for its high constitutive activity is thought to be an aromatic amino acid cluster on transmembrane helixes 6 and 715. However, because of the lack of structural information about the ghrelin receptor, the precise mechanisms of its high constitutive activity and activation by acyl-modified ghrelin remained to be elucidated.
Here, we report the first high-resolution crystal structure of the ghrelin receptor bound to an antagonist. We found that the ghrelin receptor has a wide gap (crevasse) between the TM6 and TM7 bundles, which is rich in hydrophobic amino acids including a cluster of phenylalanine residues. Thus, the interaction between the gap structure and the acyl acid moiety of ghrelin may participate in transforming the ghrelin receptor into an active conformation.
Results
Crystal structure of the ghrelin receptor
To facilitate the determination of the structure, we designed an engineered ghrelin receptor construct (Supplementary Fig. 1). For crystallization, 28 and 20 residues were removed from the N and C termini of the receptor, respectively, and the thermostabilized apocytochrome b562RIL (bRIL) protein from Escherichia coli was fused to the deleted N terminus. Deletion of the N-terminal 28 amino acids resulted in loss of cell-surface expression and receptor activity, whereas both expression and activity were retained when the C-terminal 20-amino acids were deleted (Supplementary Fig. 2a–d). In addition, two mutations were introduced: T1303.39K (superscripts indicate residue numbering using the Ballesteros–Weinstein nomenclature) to improve thermostability and expression16, and N188Q in extracellular loop 2 (ECL2) to avoid glycosylation. Moreover, both a Fab fragment antibody (Fab 7881) specific for the ghrelin receptor and an antagonist, Compound 21, were added to bRIL-conjugated truncated ghrelin receptor to increase thermostability (Supplementary Fig. 3a, b). Compound 21 was synthesized from YIL-781, another ghrelin receptor antagonist, to improve antagonistic activity and lipophilic efficiency17. Compound 21 had no effect on the constitutive activity of the ghrelin receptor, indicating that it is a neutral antagonist (Supplementary Fig. 3c). Binding assays using [3H]-Compound 21 confirmed that Compound 21 binds to the ghrelin receptor construct, although the binding affinity of the construct (dissociation constant, Kd = 17.2 ± 7.1 nM) was lower than that of the WT ghrelin receptor (Kd = 4.42 ± 0.76 nM) (Supplementary Fig. 3d). Fab 7881 binds to intracellular loop 3 (ICL3); the flexibility of ICL3 prevents the crystallization of the receptor. Thus, Fab 7881 seems to stabilize ICL3 and improve the thermostability of the ghrelin receptor, allowing it to form crystals. The ghrelin receptor was crystallized in lipidic cubic phase and solved at 3.3 Å resolution (Fig. 1, Supplementary Fig. 3e–g, Supplementary Table 1).
Overall structure of the bRIL–hGHSR–Fab 7881–Compound 21 complex. The ghrelin receptor is shown in cartoon representation and colored in orange. The bRIL fusion protein is shown in pink cartoon representation. For Fab 7881, the crimson and green-yellow cartoons represent the heavy and light chains, respectively. Compound 21 is shown as spheres and sticks with carbon atoms in yellow, oxygen in red, and nitrogen in blue.
The ghrelin receptor structure, as a class A GPCR, shares a canonical seven-transmembrane helical architecture and an intracellular amphipathic helix 8 (Fig. 1)18. Similar to other peptide hormone receptors, ECL2 forms anti-parallel β-strands with a short hairpin and is stabilized by the highly conserved disulfide bond between C1163.28 and C198ECL2. ECL3 between TM6 and TM7 has not been observed, probably due to its unstable structure. The structure superimposed well with that of the inactive OX2 orexin receptor (PDB ID: 4S0V)19, with a root-mean-squared deviation of 0.819 Å. Thus, our structure of the ghrelin receptor probably represents the inactive state.
Bifurcated ligand-binding pocket and the gap structure
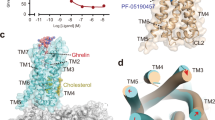
The ghrelin receptor has two main differences relative to other peptide hormone GPCRs. It is the bifurcated ligand-binding pocket and the hydrophobic wide gap structure of TM6 and TM7. The binding pocket of ghrelin receptor is bifurcated into two cavities by a salt bridge between E1243.33 and R2836.55 (Fig. 2a–c, Supplementary Fig. 4). We call the larger one cavity I, and the smaller one cavity II (Fig. 2b, c). In addition, the top of cavity II is covered with ECL2. Note that the A204E mutation in ECL2 causes loss of constitutive activity of the ghrelin receptor and is associated with familial short stature20. In fact, we observed loss of basal receptor activity and a decrease in ghrelin-induced receptor function (Supplementary Fig. 5a, b). Thus, mutation of A204 may block ghrelin’s access to the cavities.
a The cutaway surface for the ghrelin receptor in complex with the antagonist Compound 21 reveals a bifurcated ligand-binding pocket. b The ligand-binding pocket of the ghrelin receptor. c Schematic model of Fig. 2b. d The phenylalanine cluster forms a crevasse in the ghrelin receptor. The five phenylalanines are shown as sticks in dark orange. The crevasse is shown as a red dashed line. G282 is shown as spheres with carbon atoms in orange, nitrogen in blue, and oxygen in red.
No such bifurcated pocket has been observed in the class A peptide hormone GPCRs in the β-branch family whose structures have been determined thus far (e.g., the neurotensin receptor NTSR121, the OX1 and OX2 orexin receptors19,22,23, the endothelin type B receptor24, and the NPY Y1 receptor25) (Supplementary Fig. 6a–e).
Another characteristic feature of ghrelin receptor is the gap structure between TM6 and TM7. The extracellular side of the TM7 bundle curves outwardly to a greater degree than the corresponding structures of other class A GPCRs. Thus, the TM6 and TM7 bundles kink each other. Furthermore, the G2826.54 and the bulky side chains of the five phenylalanines (F2796.51, F2866.58, F2906.62, F3097.39, and F3127.42) in TM6 and TM7 are positioned to open the space between the TM6 and TM7 bundles. These structural features enable the ghrelin receptor to form a wide gap between the TM6 and TM7 bundles. We refer to this gap as the crevasse (Fig. 2b, d, Supplementary Fig. 7a). However, within the β-branch of class A GPCRs, with the exception of the ghrelin receptor, the TM6–TM7 interface is occupied by bulky side chains that completely separate the ligand binding cavity from the lipid bilayer; consequently, ligands cannot be observed from outside the receptors (Supplementary Fig. 7b–f). In contrast to the peptide hormone GPCRs, several GPCRs with lipid ligands have gaps between TM1 and TM7 (S1P1, EP4, and TP)26,27,28, TM3 and TM4 (GPR40)29, and TM4 and TM5 (PAFR and LPA6)30,31 (Supplementary Fig. 7g–l). These gaps have been proposed to be the access routes of the lipid ligands from outside the lipid bilayers into the pockets where they bind.
Binding mode of Compound 21
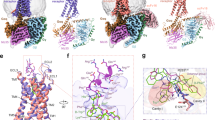
Compound 21 covers the salt bridge and fills the two cavities (Fig. 3a, b). The crystal structure of ghrelin receptor bound to Compound 21 revealed that the 1-isopropylpiperidine moiety of the Compound 21 sits in cavity I, whereas the 4-bromo-2-fluorobenzene moiety sits in cavity II, and the 5-azaquinazolinone moiety (5-aza core) links between cavities I and II. Moreover, the 5-nitrogen of the 5-aza core forms a hydrogen bond with R2836.55, which connects with E1243.33 through a salt bridge and forms the boundary between the cavities I and II (Fig. 2b, Supplementary Fig. 4). In addition, the 5-aza core makes a π–cation interaction with R1022.63 (Fig. 3a, b). The nitrogen of the 1-isopropylpiperidine moiety of Compound 21 forms hydrophobic interactions with the F2796.51, F3097.39, and F3127.42 (Fig. 3a, b). In particular, the F2796.51A and F3127.42A mutants decreased the specific binding of [3H]-Compound 21 and basal receptor activity (Supplementary Fig. 8f, i, j, Supplementary Fig. 9k), although cell-surface expression of the mutant ghrelin receptors was retained (Supplementary Fig. 9f, j). The 4-bromo-2-fluorobenzene moiety makes three hydrophobic contacts with residues I1784.60, L1814.63, and M2135.39 in cavity II. The fact that Compound 21 covers the two cavities and interacts with eight residues may explain the high antagonistic affinity of Compound 21 for the ghrelin receptor. However, the results of the binding assay using [3H]-Compound 21 and site-directed mutants of the ghrelin receptor suggest that the amino acids in cavity I are more important than those in the cavity II for the interaction between Compound 21 and the receptor (Supplementary Fig. 8a–j, Supplementary Fig. 9, Supplementary Fig. 10).
a Side chain interactions within 4.0 Å residues are shown in stick representation. Hydrogen bonds are shown as black dashed lines. b Schematic representation of the interactions between the ghrelin receptor and Compound 21, analyzed using the Discovery Studio 2016. The black dot line indicates a hydrogen bond.
Amino acid residues in the ligand-binding pocket
Mutations in the salt bridge, E1243.33A and R2836.55A, completely abolished ghrelin-induced receptor function, whereas replacement with the cognate amino acid mutants E1243.33D and R2836.55K retained function, although their ghrelin-induced receptor activities were significantly reduced (Fig. 4c, d). Note that the cell-surface expression of E1243.33A mutant was retained at 111.5% of the WT level, while that of R2836.55A mutant was significantly decreased (66.3% of the WT level) (Supplementary Fig. 9a, d, e). Recent NMR data and structural modeling suggest that the N-terminus of ghrelin extends down into the bottom of the ligand-binding pocket of the receptor, where it interacts with E1243.33 32. Furthermore, alanine mutation of two other polar amino acids in the ligand-binding pocket, D992.60 and R1022.63, abolishes the receptor activity (Fig. 4a, b). Polar amino acids in the ligand-binding pocket are also important in other peptide hormone GPCRs, such as R3276.54 in NTSR1 (PDB ID: 4GRV)21, R3436.55 and R3577.24 in ETB (PDB ID: 5GLH)24, and K2155.42, D2796.58, and D2977.32 in AT2R (PDB ID: 5XJM)33. These polar amino acids interact with the peptide core and side chains to fix the ligand in the binding pocket.
In addition, our crystal structure of the ghrelin receptor suggests a possible mechanism for receptor activation by acyl-modified ghrelin, and in particular the role of the acyl acid. We propose that the interaction between the acyl acid of ghrelin and the hydrophobic environment in the crevasse induces the transformation of the ghrelin receptor into an active form. Mutagenesis analyses of the phenylalanine cluster in the crevasse suggested that the cluster is important for the receptor activity. When F2796.51, F3097.39, and F3127.42 were individually mutated to alanine, basal and ghrelin-induced receptor activities were significantly reduced (Fig. 4e, h, i, Supplementary Fig. 9k), although the cell-surface expression of the mutant receptors was retained at 78.9 − 112.1% of the WT level. By contrast, the basal and ghrelin-induced receptor activities of the F2866.58A and F2906.62A mutants were only slightly lower than that of the WT ghrelin receptor (Fig. 4f, g, Supplementary Fig. 9k). Moreover, when the phenylalanines were each mutated to tyrosine, the receptor activities were remained at WT levels, with the exception of F3127.42Y (Fig. 4e–i). It should be noted that both F2866.58 and F2906.62 are located near the extracellular surface of the receptor, whereas F2796.51, F3097.39, and F3127.42 are located in the bottom of the ligand-binding pocket (Fig. 2d). These observations suggest that both F2866.58 and F2906.62 play a role in the entry of ghrelin into the receptor core, whereas F2796.51, F3097.39, and F3127.42 contribute to recognition of the acyl-modification of ghrelin. A recent study combining NMR with molecular modeling supports the concept that the n-octanoyl moiety of ghrelin is essential for access to the ligand-binding pocket34.
Meanwhile, we found that I1784.60, L1814.63, and M2135.39 in cavity II are not important for the interaction with ghrelin, as their alanine mutations caused no decrease in ghrelin-induced activity, although the basal receptor activities were suppressed (Supplementary Fig. 10a–c). In fact, we found that des-acyl ghrelin not only was unable to activate the ghrelin receptor, but also did not bind to the ghrelin receptor at all (Supplementary Fig. 11), suggesting that the acyl-modification is necessary for ghrelin to enter the binding pocket of the ghrelin receptor. An in silico docking study of inverse agonists to a homology model of ghrelin receptor also suggested that F2796.51, F3097.39, and F3127.42 are key residues for receptor activation/inactivation35. For other lipid receptors, the equivalent cavities contain multiple phenylalanine residues and accommodate hydrophobic moieties of their lipo-ligands. For example, the acyl tail of ML056, an antagonist of the S1P1 receptor, and both the tricyclic tetrahydrocannabinol ring and alkyl chain of AM11542, an antagonist of the CB1 receptor, are placed in the cavity (Fig. 5a, b)26,36. These facts suggest that the acyl moiety of ghrelin could be located in the cavity, where it interacts with phenylalanine residues essential for receptor activation.
The ghrelin receptor surface and its cross-section were colored according to electrostatic potential from red (negative) to blue (positive) using APBS tools. a The S1P1 receptor (PDB ID: 3V2Y) and (b) the CB1 receptor (PDB ID: 5XRA) are superposed onto the ghrelin receptor. The 2796.51, F3097.39, and F3127.42 of the ghrelin receptor and Compound 21 are depicted as orange and yellow sticks, respectively. The sphingolipid mimic S1P1 antagonist (ML056) and the CB1 antagonist (AM11542) are depicted as pink and cyan sticks, respectively.
Discussion
This study provides insights into the interactions between the ghrelin receptor and ghrelin, and our findings may explain why the acyl-modification of the ghrelin peptide is necessary for ghrelin receptor activation. Several ghrelin mimetics are under development for treatment of cancer cachexia or metabolism-linked disorders, and our results will promote the design of more potent and effective ghrelin mimetics.
The ghrelin receptor may have properties of both peptide hormone GPCRs and lipid GPCRs. As described above, the binding pocket of the ghrelin receptor is bifurcated into two cavities, in which cavity I is more important for the binding of ghrelin, as determined by the results of mutagenesis analyses. In the peptide hormone GPCRs, such as NTSR1, ETBR, and AT2 receptor, the polar amino acids in the ligand binding pockets are important for recognition of the peptide cores of the endogenous ligands. In the case of the ghrelin receptor, the polar amino acids in cavity I is the core for recognition of the peptide part of ghrelin, because the alanine mutations of the polar amino acids in the cavity I abolish receptor activity (Fig. 4a–d).
On the other hand, in many lipid GPCRs, a wide gap between transmembrane bundles is proposed to serve as the access route for lipid ligands. However, in the case of the ghrelin receptor, the gap structure (crevasse) is probably not the access route for ghrelin into the ligand binding pocket, but instead seems to be the recognition site for the acyl-modification of ghrelin. This idea is supported by the results of hydrophobic amino acid mutations in the gap region, in particular mutations of the phenylalanine residues such as F2796.51, F3097.39, and F3127.42. In the S1P1 and CB1 receptor, which are known as typical lipid GPCRs, the position of the ligand-binding pockets is equivalent to the position of cavity I in the ghrelin receptor (Fig. 5a, b)26,36. Thus, in the ghrelin receptor, the hydrophobic environment of the ligand-binding pocket seems to accommodate the acyl acid moiety of ghrelin, just as lipid GPCRs accommodate hydrophobic moieties of their lipo-ligands.
Furthermore, the gap structure of the ghrelin receptor is composed of TM6 and TM7, whose movements are thought to change the GPCR conformation into its active form37. Thus, the interaction between the gap structure and the acyl acid moiety of ghrelin may participate in transforming the ghrelin receptor into an active conformation. Further details, obtained by structural analysis of the active form of ghrelin receptor bound to acyl-modified ghrelin, will be required to fully elucidate the mechanism of ghrelin receptor activation.
Methods
Expression and purification of the ghrelin receptor
Human GHSR cDNA, which is based on the nucleotide sequence of the human GHSR gene (encoded by GHSR; UniProt accession Q92847), was cloned into a modified pFastBac1 vector (Invitrogen) containing an expression cassette with a hemagglutinin signal sequence and a FLAG epitope tag followed by a human rhinovirus (HRV) 3C protease recognition site at the N terminus, and an HRV 3C protease recognition site followed by an enhanced green fluorescent protein (eGFP) and an 8xHis tag at the C terminus. Twenty-eight amino acids of the N terminus (residues 1–28) were replaced with the thermostabilized apocytochrome b562RIL (bRIL) from Escherichia coli (M7W, H102I, and R106L) protein38,39, and twenty amino acids were deleted from the C terminus (residues 347–366). Two mutations were introduced: T130K to improve thermostability and expression, and N188Q to avoid glycosylation. Recombinant baculovirus was prepared using the Bac-to-Bac baculovirus expression system (Invitrogen). Spodoptera frugiperda (Sf9) insect cells (Thermo Fisher Scientific) were infected with baculovirus at a cell density of 2–3.5 × 106 cells ml−1 at multiplicity of infection of 1 in PSFM-J1 medium (Wako) supplemented with 2%(v/v) fetal calf serum, 50 units ml−1 penicillin, 50 µg ml−1 streptomycin, and 0.5 µg ml−1 amphotericin B. Infected cells were harvested by centrifugation 48 h post-infection, and the cell pellets were stored at −80 °C for future use.
The cell pellets were homogenized using a Dounce homogenizer in hypotonic buffer containing 10 mM HEPES-NaOH (pH7.5), 20 mM KCl, 10 mM MgCl2, and EDTA-free protease inhibitor cocktail (Nacalai Tesque). The cell membranes were isolated by ultracentrifugation at 100,000 × g for 30 min at 4 °C. Washing of the membranes was performed by two rounds of Dounce homogenization and centrifugation in high-osmolarity buffer containing 10 mM HEPES-NaOH (pH7.5), 1.0 M NaCl, 20 mM KCl, and 10 mM MgCl2. Purified membranes were resuspended in a buffer containing 50 mM HEPES-NaOH (pH 7.5), 500 mM NaCl, 2 mg ml−1 iodoacetamide (Wako), and 5 µM YIL781 (Tocris), and incubated overnight at 4 °C in the dark. The membranes were then solubilized in a solubilization buffer containing 50 mM HEPES-NaOH (pH7.5), 500 mM NaCl, 1%(w/v) n-dodecyl-ß-D-maltopyranoside (DDM; Anatrace), 0.2% cholesteryl hemisuccinate (CHS; sigma), and 0.2% (w/v) sodium cholate (Dojindo) for 2 h at 4 °C, followed by ultracentrifugation at 100,000×g for 30 min at 4 °C. Solubilized supernatant supplemented with 5 mM imidazole was mixed with 10 ml of Ni-NTA agarose (Qiagen) in batch-binding mode for 2 h at 4 °C. After binding, the Ni resin was washed with 10 column volumes of Ni wash buffer containing 30 mM HEPES-NaOH (pH7.5), 750 mM NaCl, 0.1% DDM, 0.02% CHS, 0.02% sodium cholate, and 5 µM YIL781. The protein was eluted with three column volumes of Ni wash buffer containing 500 mM imidazole. Elution from the Ni resin was supplemented with 2 mM calcium and loaded onto M1 anti-FLAG affinity resin (Sigma). Detergent was gradually exchanged on the M1 resin from DDM to 0.01%(v/w) lauryl maltose neopentyl glycol (LMNG; Anatrace), and YIL781 was replaced with 5 µM Compound 21, a ghrelin receptor antagonist. The receptor was eluted with 3 column volumes of FLAG elute buffer containing 20 mM HEPES-NaOH (pH7.5), 100 mM NaCl, 0.01% LMNG, 0.001% CHS, 200 µg ml−1 Flag peptide (Peptide Institute Inc.), 5 mM EDTA, and 5 µM Compound 21. The N-terminal FLAG tag and C-terminal eGFP-8xHis tag were cleaved by 3 C protease overnight at 4 °C. The purified receptor was concentrated with a 50-kDa molecular weight cut-off Amicon Ultra concentrator (Millipore) and loaded onto Superdex 200 Increase 10/300 GL (GE Healthcare) in buffer containing 20 mM HEPES-NaOH (pH7.5), 100 mM NaCl, 0.01% LMNG, and 0.001% CHS. The peak fractions were collected and flash-frozen with liquid nitrogen and stored at −80 °C until use.
Protein thermostability assay
The thermal stability analysis was performed with the thiol-specific fluorochrome N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM)40. CPM at 4 mg ml−1 in DMSO was diluted 40-fold into buffer containing 20 mM HEPES-NaOH (pH 7.5), 100 mM NaCl, 0.01% LMNG, and 0.001% CHS. Purified protein (20 µg) was mixed with diluted CPM and incubated on ice for 15 min in darkness. The reaction mixture (20 µl) was heated in a controlled manner with a ramp rate of 3 °C min−1 in a MyiQ2 thermal cycler (Bio-Rad). The excitation wavelength was set at 387 nm, and the emission wavelength at 463 nm. Assays were performed over a temperature range starting from 4 °C and ending at 80 °C. The peak in the derivative of the fluorescence signal was used to determine the melting temperature.
Antibody generation
All animal experiments described in this study conformed to the guidelines outlined in the Guide for the Care and Use of Laboratory Animals of Japan and were approved by Kyoto University Animal Care Committee (approval No. Med Kyo 16043). For use as antigen, purified human ghrelin receptor [29–346] (T130K/N188Q) was reconstituted into liposomes containing chicken egg yolk phosphatidylcholine (eggPC, Avanti) and monophosphoryl lipid A (Sigma-Aldrich). Briefly, MRL/lpr mice were immunized three times at 10–14-day intervals with 100 µg purified ghrelin receptor mutant. The immunized mice were sacrificed, and single-cell suspensions were prepared from their spleens. These cells were fused with NS-1 myeloma cells using polyethylene glycol (PEG) according to conventional methods. To screen for antibodies that specifically recognize native receptors, we developed an ELISA method using proteoliposomes41. For liposome ELISA, we used purified ghrelin receptor mutant reconstituted into liposomes containing biotinyl phosphatidylethanolamine (Avanti) to maintain the protein in its native conformation and effectively immobilize liposomes onto Streptavidin-coated plates (Nunc). To eliminate antibodies recognizing the flexible loops, the N and C termini, or unstructured regions of the ghrelin receptor, we performed ELISA using ghrelin receptor denatured with 1% (w/v) SDS. The selected cells were isolated by limiting dilution to establish monoclonal hybridoma cell lines producing antibodies against the ghrelin receptor. The supernatant of the large-scale cell culture was loaded onto HiTrap Protein G HP (GE Health care) to purify mouse IgG. Fab fragments were obtained by proteolytic cleavage of IgG with papain (Worthington) and purified by Superdex200 10/60 (GE Healthcare). Purified Fab monomer (Fab 7881) was concentrated to 10 mg ml−1 using a 10-kDa molecular weight cut-off Amicon Ultra concentrator. Sequences of Fab fragments were determined according to the standard 5’-RACE method using total RNA isolated from hybridoma cells. The Fab 7881 obtained in this manner was used for crystallization of the ghrelin receptor–Fab complex.
Crystallization of the ghrelin receptor–Fab complex
Purified ghrelin receptor and the Fab 7881 were mixed in a 1:1.5 molar ratio and incubated for 1 h at 4 °C. The complex was purified by gel filtration using Superdex 200 Increase 10/300 GL. Compound 21 was added to the peak fractions at a 1:5 molar ratio, and the mixture was concentrated to >30 mg ml−1 using a 50-kDa molecular weight cut-off Amicon Ultra concentrator. The purified ghrelin receptor–Fab 7881 complex was reconstituted into the lipidic cubic phase (LCP) by mixing with a monoolein:cholesterol mixture at a mass ratio of 10:1. In the mixture, the protein solution:lipid mass ratio was fixed at 2:3 using the twin-syringe mixing method. Crystallization experiments were carried out in 96-well glass sandwich plates (Molecular Dimensions) on an NT8-LCP crystallization robot (Formulatrix) using a 30 nl protein solution cubic phase overlaid with 600 nl precipitant solution, and incubated at 20 °C. The best crystals were obtained from precipitant solution containing 100 mM MES (pH 6.6–7.0), 400 mM potassium acetate, and 36–40% polyethylene glycol (PEG) 300. The crystals were collected within 2 weeks using mesh grid loops (WAKENBTECH) and flash-cooled in liquid nitrogen. Initial crystals were obtained in hanging drops by vapor diffusion at 20 °C from the precipitant solution containing 100 mM bicine (pH 9.0), 100 mM sodium chloride, and 30% PEG550MME.
Data collection and structure determination
All diffraction datasets were collected at SPring-8 BL32XU42 using a micro-focused X-ray beam. Loop-harvested microcrystals were identified using raster scan and analysis by SHIKA43. Small wedge data, each consisting of 4–6°, were collected from single crystals, and the collected datasets were processed automatically using KAMO44 using XDS45, followed by hierarchical clustering analysis using the correlation coefficients of the normalized structure amplitudes between datasets. Finally, a group of outlier-rejected datasets were scaled and merged using XSCALE46. The structure was determined by molecular replacement with the program PHASER47, using the receptor portions of the antagonist-bound orexin receptor (PDB ID: 4S0V), bRIL (PDB ID: 1M6T), and the Fab 7881 antibody (this study, ID: 6KS2) as the search models. The SMILES string of the ligand was generated by Ligand Builder in COOT48, and the geometry restraints were generated by eLBOW in PHENIX49. The electron density map and the structural model were iteratively refined and rebuilt using PHENIX and COOT50. Data collection and refinement statistics are summarized in Supplementary Table 1. All molecular graphics were prepared using Cuemol2 (http://www.cuemol.org/).
Radioligand binding studies of the ghrelin receptor
Ligand binding assays were performed using membrane preparations from HEK293 cells (ATCC) transiently expressing wild-type or mutant ghrelin receptors. (The crystallization construct was expressed in Sf9 cells.) The ghrelin receptor–expressing cells were homogenized on ice in homogenization buffer containing 50 mM Tris-HCl (pH7.5), 5 mM EDTA, 5 mM MgCl2, and EDTA-free protease inhibitor cocktail. The homogenate was ultracentrifuged at 100,000×g for 30 min at 4 °C, and the pellet was washed twice with cold homogenization buffer. The protein concentration was determined using BCA Protein Assay (Thermo Fisher). All binding assays were conducted in ghrelin binding buffer containing 50 mM Tris-HCl (pH7.5), 5 mM EDTA, 5 mM MgCl2, and 1% (w/v) bovine serum albumin. For the saturation binding assay, 20 μg (5 µg for crystallization construct) of membrane homogenates were incubated on ice for 1 h with 0.039–5 nM [125I]-ghrelin (PerkinElmer) or 3.125–100 nM (3.125–50 nM for crystallization construct) [3H]-Compound 21 (Sekisui Medical) in the absence or presence of 5 μM unlabeled ghrelin or 100 μM unlabeled Compound 21. Bound [125I]-ghrelin was quantified on an AccuFLEX-γ 8010 (Hitachi). Bound [3H]-Compound 21 was quantified with an AccuFLEX LCS-8000 liquid scintillation counter (Hitachi) after addition of 2.5 ml of Clear-Sol liquid scintillation cocktail (Nacalai Tesque). Data were analyzed by nonlinear curve-fitting using the program GraphPad Prism 5. Binding data are reported as means ± s.d.
Mutagenesis analyses of residues in the ligand recognition
Receptor activities of mutated ghrelin receptors were examined. Mutagenesis of the ghrelin receptor was introduced using the QuikChange Site-Directed Mutagenesis Kit (Agilent). Full-length human ghrelin receptor was used for the mutagenesis. The positions of mutated amino acids and primers are listed in Supplementary Table 2. Each mutant ghrelin receptor was expressed in CHO cells, and its receptor-specific responses to ghrelin were examined using an intracellular Ca2+ concentration assay.
Intracellular Ca2+ concentration assay
For the fluorescence-based assay to detect changes in intracellular Ca2+ concentration, CHO cells (ATCC) were seeded in 6-cm dishes at a density of 3 × 105 cells/dish and transfected with the ghrelin receptor N188Q (control) or the indicated ghrelin receptor mutant. Transfections were performed using the FuGENE HD transfection reagent (Promega). The next day, the transfected cells were collected and plated into 96-well black-wall assay microplates (Corning) at a density of 3 × 104 cells/well. After a 1-day incubation, the cells were incubated for 1 h with 100 μl of calcium indicator Fluo-4 from the FLIPR Calcium 5 Assay Kit (Molecular Devices) in Hank’s BSS containing 20 mM HEPES-NaOH (pH7.5) and 2.5 mM probenecid. Fifty-microliter aliquots of samples were tested to induce changes in fluorescence due to increased calcium concentration on a FlexStation3 (Molecular Devices). Maximum [Ca2+]i changes were determined in quadruplicate (n = 4).
Cell-surface expression levels of ghrelin receptors
HEK293 cells (ATCC) were seeded in 6-well plates at a cell density of 8 × 105 cells/well and cultured for 24 h. The cells were transfected with complex of vector DNA (the FLAG-tagged N188Q ghrelin receptor (control) or the indicated FLAG-tagged ghrelin receptor mutants) and FuGENE HD transfection reagent (Promega). The next day, transfected HEK293 cells were harvested in PBS containing 1 mM EDTA, and then incubated for 30 min on ice with 2.5 μg ml−1 anti-FLAG antibody (Wako) and 5 μg ml−1 Alexa Fluor 488–conjugated anti-mouse IgG goat polyclonal antibody (Thermo Fisher Scientific) in FACS buffer (PBS, 2% FBS, 0.05% NaN3). Cell-surface expression levels were evaluated by flow cytometry on a Guava EasyCyte Plus Flow Cytometer. FLAG-positive cells were defined as cell populations with signals greater than the top 5% of MOCK cells.
Luciferase assay
To detect constitutive activity of ghrelin receptor, CHO cells were seeded in 6-cm dishes at a density of 2.5 × 105 cells/dish and cotransfected with pGL4.30[luc2P/NFAT-RE/Hygro] (2.5 μg, Promega), pRL-TK (0.1 μg, Promega), and the indicated ghrelin receptor plasmid (5 μg). Transfections were performed using the FuGENE HD transfection reagent (Promega). The next day, the transfected cells were collected and plated into 96-well white microplates (Corning) at a density of 2.5 × 104 cells/well. After a 1-day incubation, the luciferase activities were determined using Dual-Luciferase Reporter Assay System (Promega). When assessing Compound 21 activity, the transfected cells were incubated with various concentrations of Compound 21 for 1 h just before luciferase activity measurement. All firefly luciferase activities were normalized against Renilla luciferase activity. All experiments were performed in sextuplicate.
Statistical analysis
Data are presented as means ± s.e.m. The outliers on the luciferase activity were detected and removed based on the Smirnov–Grubbs test. The GraphPad Prism 8 software (GraphPad Software Inc.) was used for statistical analysis. Differences between groups were analyzed using the two-tailed Student t test. P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request. A reporting summary for this Article is available as a Supplementary Information file. The structure and corresponding structure factors have been deposited into the Protein Data Bank with the PDB 6KO5 for the human ghrelin receptor–Fab 7881 complex and PDB 6KS2 for Fab 7881. Source data are provided with this paper.
References
Howard, A. D. et al. A receptor in pituitary and hypothalamus that functions in growth hormone release. Science 273, 974–977 (1996).
Kojima, M. et al. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402, 656–660 (1999).
Cui, H., López, M. & Rahmouni, K. The cellular and molecular bases of leptin and ghrelin resistance in obesity. Nat. Rev. Endocrinol. 13, 338–351 (2017).
Yanagi, S., Sato, T., Kangawa, K. & Nakazato, M. The homeostatic force of ghrelin. Cell Metab. 27, 786–804 (2018).
Kojima, M. & Kangawa, K. Ghrelin: structure and function. Physiol. Rev. 85, 495–522 (2005).
Yang, J., Brown, M. S., Liang, G., Grishin, N. V. & Goldstein, J. L. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell 132, 387–396 (2008).
Gutierrez, J. A. et al. Ghrelin octanoylation mediated by an orphan lipid transferase. Proc. Natl Acad. Sci. USA 105, 6320–6325 (2008).
Hougland, J. L. Ghrelin octanoylation by ghrelin O-acyltransferase: Unique protein biochemistry underlying metabolic signaling. Biochem. Soc. Trans. 47, 169–178 (2019).
Ohgusu, H. et al. Ghrelin O-acyltransferase (GOAT) has a preference for n-hexanoyl-CoA over n-octanoyl-CoA as an acyl donor. Biochem. Biophys. Res. Commun. 386, 153–158 (2009).
Kaiya, H. et al. Bullfrog ghrelin is modified by n-octanoic acid at its third threonine residue. J. Biol. Chem. 276, 40441–40448 (2001).
Kaiya, H. et al. Chicken ghrelin: purification, cDNA cloning, and biological activity. Endocrinology 143, 3454–3463 (2002).
Matsumoto, M. et al. Structure-activity relationship of ghrelin: pharmacological study of ghrelin peptides. Biochem. Biophys. Res. Commun. 287, 142–146 (2001).
Holst, B., Cygankiewicz, A., Jensen, T. H., Ankersen, M. & Schwartz, T. W. High constitutive signaling of the ghrelin receptor—identification of a potent inverse agonist. Mol. Endocrinol. 17, 2201–2210 (2003).
Damian, M. et al. High constitutive activity is an intrinsic feature of ghrelin receptor protein: a study with a functional monomeric GHS-R1a receptor reconstituted in lipid discs. J. Biol. Chem. 287, 3630–3641 (2012).
Holst, B. et al. Common structural basis for constitutive activity of the ghrelin receptor family. J. Biol. Chem. 279, 53806–53817 (2004).
Yasuda, S. et al. Hot-spot residues to be mutated common in G protein-coupled receptors of class a: identification of thermostabilizing mutations followed by determination of three-dimensional structures for two example receptors. J. Phys. Chem. B 121, 6341–6350 (2017).
Hanrahan, P. et al. Substituted azaquinazolinones as modulators of GHSr-1a for the treatment of type II diabetes and obesity. Bioorg. Med. Chem. Lett. 22, 2271–2278 (2012).
Venkatakrishnan, A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013).
Yin, J., Mobarec, J. C., Kolb, P. & Rosenbaum, D. M. Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature 519, 247–250 (2015).
Pantel, J. et al. Loss of constitutive activity of the growth hormone secretagogue receptor in familial short stature. J. Clin. Invest. 116, 760–768 (2006).
White, J. F. et al. Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 (2012).
Suno, R. et al. Crystal structures of human orexin 2 receptor bound to the subtype-selective antagonist EMPA. Structure 26, 7–19 (2018).
Yin, J. et al. Structure and ligand-binding mechanism of the human OX 1 and OX 2 orexin receptors. Nat. Struct. Mol. Biol. 23, 293–299 (2016).
Shihoya, W. et al. Activation mechanism of endothelin ET B receptor by endothelin-1. Nature 537, 363–368 (2016).
Yang, Z. et al. Structural basis of ligand binding modes at the neuropeptide y Y 1 receptor. Nature 556, 520–524 (2018).
Sanna, M. G. et al. Crystal structure of a lipid G protein-coupled receptor. Science 335, 851–855 (2012).
Toyoda, Y. et al. Ligand binding to human prostaglandin E receptor EP 4 at the lipid-bilayer interface. Nat. Chem. Biol. 15, 18–26 (2019).
Fan, H. et al. Structural basis for ligand recognition of the human thromboxane A 2 receptor. Nat. Chem. Biol. 15, 27–33 (2019).
Srivastava, A. et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature 513, 124–127 (2014).
Cao, C. et al. Structural basis for signal recognition and transduction by platelet-activating-factor receptor. Nat. Struct. Mol. Biol. 25, 488–495 (2018).
Taniguchi, R. et al. Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA 6. Nature 548, 356–360 (2017).
Bender, B. J. et al. Structural model of ghrelin bound to its g protein-coupled receptor. Structure 27, 537–544 (2019).
Asada, H. et al. Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog. Nat. Struct. Mol. Biol. 25, 1–9 (2018).
Ferré, G. et al. Structure and dynamics of G protein-coupled receptor-bound ghrelin reveal the critical role of the octanoyl chain. Proc. Natl Acad. Sci. USA 116, 17525–17530 (2019).
Daina, A. et al. Rational design, synthesis, and pharmacological characterization of novel ghrelin receptor inverse agonists as potential treatment against obesity-related metabolic diseases. J. Med. Chem. 61, 11039–11060 (2018).
Hua, T. et al. Crystal structures of agonist-bound human cannabinoid receptor CB 1. Nature 547, 468–471 (2017).
Zhou, Q. et al. Common activation mechanism of class A GPCRs. eLife https://doi.org/10.7554/eLife.50279 (2019).
Chun, E. et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 20, 967–976 (2012).
Chu, R. et al. Redesign of a four-helix bundle protein by phage display coupled with proteolysis and structural characterization by NMR and X-ray crystallography. J. Mol. Biol. 323, 253–262 (2002).
Alexandrov, A. I., Mileni, M., Chien, E. Y. T., Hanson, M. A. & Stevens, R. C. Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359 (2008).
Suharni et al. Proteoliposome-based selection of a recombinant antibody fragment against the human M2 muscarinic acetylcholine receptor. Monoclon. Antib. Immunodiagn. Immunother. 33, 378–385 (2014).
Hirata, K. et al. Achievement of protein micro-crystallography at SPring-8 beamline BL32XU. J. Phys. Conf. Ser. 425, 8–12 (2013).
Hirata, K. et al. Zoo: An automatic data-collection system for high-throughput structure analysis in protein microcrystallography. Acta Crystallogr. Sect. D. Struct. Biol. 75, 138–150 (2019).
Yamashita, K., Hirata, K. & Yamamoto, M. KAMO: towards automated data processing for microcrystals. Acta Crystallogr. Sect. D. Struct. Biol. 74, 441–449 (2018).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. Sect. D. Biol. Crystallogr. 66, 133–144 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007).
Emsley, P. & Cowtan, K. & IUCr. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D. Biol. Crystallogr. 66, 213–221 (2010).
Liebschner, D. et al. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr. Sect. D. Struct. Biol. 73, 148–157 (2017).
Acknowledgements
We thank Y. Yamanaka for supporting the crystallization of Fab 7881 and evaluating the thermostability of ghrelin receptor mutants; H. Hosoda for supplying ghrelin peptides; M. Nagashima and R. Saeki for technical assistance; D. Im, T. Shimamura, R. Suno, and T. Nakagita for expert help with the purification procedure; and the beamline staff at BL32XU of SPring-8 (Hyogo, Japan) for assistance with data collection. We also thank Kurume University RMCP (Research Mind Cultivation Program) students, M. Isogai, Y. Kawai, N. Hiyoshi, and T. Yamamura for their help with making mutant receptor constructs. Diffraction experiments were performed at SPring-8 BL32XU (proposal 2017A2524) with the approval of RIKEN. This work was supported by JSPS KAKENHI Grant Numbers JP26650025, JP16K07281, and JP20K06531 (M.K.), JP14J06159 and JP16K18515 (Y.S.), JP19K20143 (A.H.), JP18K08497 (K.M.); the MEXT-Supported Program for the Strategic Research Foundation at Private Universities; and grants from the Takeda Science Foundation and the Naito Foundation (M.K.), the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics and Structural Life Science); and the Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS) from the Japan Agency for Medical Research and Development (AMED) (Grant No. 19am0101079; Support No. 1021) and JST/PRESTO. DNA sequencing analysis was performed at the Medical Research Support Center, Graduate School of Medicine, Kyoto University. Radioisotope experiments were performed at the Radioisotope Research Center, Kyoto University.
Author information
Authors and Affiliations
Contributions
Y.S. optimized the ghrelin receptor construct, developed the purification procedure, purified the receptor for crystallization, and determined the structure. S.H. and K.H. collected the diffraction data and determined the structure. Y.S. and T.U. generated the anti-ghrelin receptor–expressing hybridoma cells and performed cloning and expression of the Fab fragment. Y.S. and H.A. crystallized the ghrelin receptor–Fab 7881 complex and performed the ligand-binding assay on the ghrelin receptor mutants. Y.S., A.H., and M.T. planned and performed the mutational analyses. K.M. performed the luciferase assay and data analyses. Y.S., S.I., and M.K. wrote the manuscript, with feedback from all of the authors. T.K., S.I., and M.K. supervised the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks the anonymous reviewers for their contributions to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shiimura, Y., Horita, S., Hamamoto, A. et al. Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode. Nat Commun 11, 4160 (2020). https://doi.org/10.1038/s41467-020-17554-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-020-17554-1
This article is cited by
-
Can machine learning ‘transform’ peptides/peptidomimetics into small molecules? A case study with ghrelin receptor ligands
Molecular Diversity (2023)
-
Molecular mechanism of agonism and inverse agonism in ghrelin receptor
Nature Communications (2022)
-
Structure of SARS-CoV-2 membrane protein essential for virus assembly
Nature Communications (2022)
-
Ghrelin infusion into the basolateral amygdala suppresses CTA memory formation in rats via the PI3K/Akt/mTOR and PLC/PKC signaling pathways
Acta Pharmacologica Sinica (2022)
-
Selectively increasing GHS-R1a expression in dCA1 excitatory/inhibitory neurons have opposite effects on memory encoding
Molecular Brain (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.