Abstract
The respiratory syncytial virus (RSV) RNA polymerase, constituted of a 250 kDa large (L) protein and tetrameric phosphoprotein (P), catalyzes three distinct enzymatic activities — nucleotide polymerization, cap addition, and cap methylation. How RSV L and P coordinate these activities is poorly understood. Here, we present a 3.67 Å cryo-EM structure of the RSV polymerase (L:P) complex. The structure reveals that the RNA dependent RNA polymerase (RdRp) and capping (Cap) domains of L interact with the oligomerization domain (POD) and C-terminal domain (PCTD) of a tetramer of P. The density of the methyltransferase (MT) domain of L and the N-terminal domain of P (PNTD) is missing. Further analysis and comparison with other RNA polymerases at different stages suggest the structure we obtained is likely to be at an elongation-compatible stage. Together, these data provide enriched insights into the interrelationship, the inhibitors, and the evolutionary implications of the RSV polymerase.
Similar content being viewed by others
Introduction
Nonsegmented negative-sense (NNS) RNA viruses are a class of pathogenic and sometimes deadly viruses that include rabies, Ebola, and respiratory syncytial virus (RSV)1. RSV infection is the leading cause of severe lower respiratory tract diseases in young children, older adults, and immunocompromised patients worldwide2,3. RSV initiates viral infection by delivering into the host cell a virus-specific RNA synthesis machine required for both genome replication and gene transcription4,5. This machine comprises the nucleoprotein (N) coated genomic RNA (N:RNA) and the RNA polymerase6. The catalytic core is a 250 kDa large (L) protein that catalyzes the RNA polymerization in both replication and transcription, the cap addition, and cap methylation of nascent viral mRNAs. A tetrameric phosphoprotein (P) is essential to modulate and constitute an active RNA polymerase with L4,5.
RSV RNA synthesis is believed to follow the “start-stop model” of sequential and polar transcription7,8,9. Like all NNS RNA viruses, the RSV RNA template is N:RNA, not RNA alone. The leader (Le) or trailer complementary (TrC) sequences from the terminus of the RNA genome or antigenome serve as the promoters for the RSV RNA synthesis10,11,12,13. To copy the N:RNA template, L requires the tetrameric P to displace N14. Interestingly, the RSV polymerase not only synthesizes mRNAs but also co-transcriptionally adds a cap and a poly-A tail to each transcript. The mRNA caps are synthesized using unconventional chemical reactions: (a) the cap is formed by a polyribonucleotide transferase but not a guanylyltransferase through generating a covalent L:RNA intermediate, distinct from eukaryotes and all other virus families9,15,16,17,18; and (b) the cap is methylated at the 2′-O position first, followed by the N-7 position, the opposite order of mammalian mRNAs19,20,21. To date, the in vitro RNA polymerization assays of RSV and several other NNS RNA viruses have been established13,22,23. The RSV cap addition and cap methylation assays have not been described yet and are speculated to share similar mechanisms with vesicular stomatitis virus (VSV), of which the assays were reported16,17,18,24,25.
There are six conserved regions and three functional domains shared within L of NNS RNA viruses26,27. The domain boundaries and the active sites of the three functional domains, RNA dependent RNA polymerization (RdRp), capping (Cap), and cap methyltransferase (MT), are highlighted as GDN, HR, and GXGXG (X denotes any residues) with numbers, respectively28,29,30,31 (Fig. 1a). P is predicted to contain several disordered regions and multiple phosphorylation sites32,33,34,35,36,37,38, and P is a tetrameric protein that regulates the RNA synthesis through interacting with L, RNA-free N (N0), N, and M2-1 (Fig. 1a). The precise interactions between L and P and the RNA synthesis mechanism by the RSV polymerase remain poorly understood39. Here, we characterize the structure of the RSV polymerase (L:P) using cryo-electron microscopy (cryo-EM). Our studies reveal that the RdRp and Cap domains of the RSV L shares similar architectures of that of the VSV L40, uncover a previously unknown basis of how P interacts with L, and provide molecular insights into RNA synthesis by the RSV polymerase.
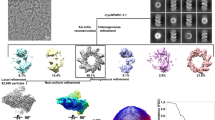
a Schematic domain representation of the human respiratory syncytial virus (RSV) RNA polymerase (L:P complex), with labeled domain boundary (Alignment details in Supplementary Fig. 4). The domains with missing density are colored in gray. b The SDS-PAGE gel shows the quality of the wild-type (wt) and mutant RSV RNA polymerase, wt L:P, and L(D811A):P, respectively (repeated ≥ 5 times). c Elution profile of the purified L:P complex on the Superose 6 Increase 10/300 GL size-exclusion column (repeated ≥ 5 times). d The RNA dependent RNA polymerization assays show that the purified wt polymerase is active and can synthesis RNA with specific RNA templates (lane 6) compared with that of catalytically inactive polymerase L(D811A):P (lane 3). e Representative 2D class averages from the 200 kV cryo-EM dataset, selected amongst 125 classes. Scale bar: 100 Å. f The final cryo-EM density map with the model (colored as a) in two orientations.
Results
Cryo-electron microscopy structure determination
We co-expressed and co-purified the recombinant RSV polymerase (L:P) from Sf21 insect cells. Gel filtration and SDS-PAGE indicated pure and full-length wild-type (wt) L:P and mutant L(D811A):P complexes (Fig. 1b, c). Using an established RdRp assay13,41, we demonstrated the characteristic RNA products of both de novo initiation (≤25 nt) and back primer (>25 nt) activities by the wt polymerase L:P (lane 6), but not the catalytically inactive polymerase L(D811A):P (lane 3) using a short trailer complementary 25 (TrC25) RNA template. The Le or TrC sequences of the genome serve as the promoters for the RSV polymerase. The trailer 25 (Tr25) sequence, which is the transcription product of TrC25, is not a natural template and was included as a negative control. As expected, we did not observe RNA products when using Tr25 as a template (lane 5) (Fig. 1d). Therefore, the prepared wt RSV polymerase is stable and catalytically active.
Cryo-EM analysis was conducted using a 200 kV Talos Arctica microscope with a BioQuantum/K2 direct electron detector. An initial dataset of 1349 movies resulted in 4.3 Å reconstruction, and one additional dataset of 1251 movies was collected. A total of 2600 movies gave rise to the final 3.67 Å map, refined with 253,372 particles selected from multiple rounds of 2D and 3D classifications (Fig. 1e, f, Supplementary Figs. 1–2, and Supplementary Table 1).
We performed an atomic model building on the final 3.67 Å map with COOT, assisted by the structure of the VSV L protein40 (PDB: 5A22). We refined the model coordinates with PHENIX. The cryo-EM map revealed the characteristic ring-like core RdRp domain and an unconventional Cap domain of the L protein that only exists in NNS RNA viruses. The map also showed the typical helix bundles of the oligomerization domain of P (Fig. 1f). Further data analysis suggested intrinsic flexibility and structural rearrangements could be attributed to the missing densities of L and P domains and will be discussed in later sections.
The overall structure of the RSV polymerase complex
The final atomic model contains the RdRp domain (blue) and the Cap domain (green) of the RSV L protein, and the oligomerization domain (POD, red) and C-terminal domain (PCTD, orange) of tetrameric P proteins (Fig. 2a, representative model fitting with the map, Supplementary Fig. 3). We have assigned residues to domains as follows: RdRp, 1-945; Cap, 946-1461; POD, 128-157; PCTD, 158-241. The RdRp domain adopts a conventional “fingers-palm-thumb” right-hand fold as many other RNA and DNA polymerases. The Cap domain that is distinct from host cells or other types of viruses shares a similar architecture as that of VSV L40. POD is fully assembled into a four-helix bundle as expected. Most PCTD is flexible, and only the regions that interact with L can be visualized and modeled (Fig. 2a).
a The cartoon diagrams of RSV polymerase complex is shown in three orientations. Domains are color-coded as in Fig. 1a, and the RdRp active site (D811) is shown in the magenta sphere. The boxes indicate the magnified views of the interactions between L and P. Magnified views of interaction interfaces between the L and POD (b), L and PCTD (c, d), and representative residues are shown. The same color scheme for the cartoons and side chains. The interacting residues of L on P are underlined.
Sequence alignments and secondary structure predictions suggest that the RSV L shares five well-organized domains (two domains visible and modeled in this study) among NNS RNA viruses, while the P protein is flexible and contains many disordered regions21,32,33,34,42,43,44,45 (Supplementary Fig. 4). The SDS-PAGE gel (Fig. 1b) and mass spectrometry (Supplementary Fig. 5) showed that both L and P proteins are intact. However, the 3.67 Å cryo-EM map revealed no extra density for the connector domain (CD), the MT domain and the C-terminal domain (CTD) of L as well as the N-terminal domain (NTD) of P. We speculate that the missing density of L is due to significant structural rearrangements of L, and the missing density of P is primarily due to the intrinsic flexibility of P.
Structural insights into RSV L and P interactions
The RSV L primarily uses the “fingers” motif to interact with POD and PCTD. The interactions between L and a tetrameric P can be divided into three parts: (1) two of four-helix bundles of POD and the RdRp; (2) two flexible PCTD chains wrap around the surface of RdRp and stabilize the base of the tetrameric POD; (3) one flexible PCTD chain extends to the positively charged “palm” motif of RdRp and the edge of the putative NTP entrance channel (Fig. 2b–d). These interactions bury surface area of about 1101.1 Å2, 1542.9 Å2, and 884.6 Å2, respectively. P is negatively charged overall, and the calculated isoelectric points of POD and PCTD are 4.82 and 4.34, respectively. Indeed, the interaction between L and P is dictated partly by electrostatic complementarity (Supplementary Fig. 6).
Our structure of the RSV L:P agrees with previous biochemical studies that PCTD is critical to interact with L and also identifies the previously unknown role of POD in interacting with L32,43 (Fig. 2). On the POD, residues that interact with L include K141, E144, H150, and V154 (Fig. 2b). Interestingly, four chains of PCTD adopt four different conformations and interact broadly with the RdRp domain of L (Fig. 2b–d and Supplementary Fig. 6). There are extensive interactions between L and two of the PCTD chains near the POD, including residues G158, R167, D168, R174, E176, and N189 (Fig. 2c). There is a composite surface on the L protein that accommodates one chain of PCTD, and these residues serve as a strong base to stabilize the POD four-helix bundles. One PCTD chain extends and uses residues D209, T210, D212, L216, T219, and L223 to interact with L (Fig. 2d). The observations of such extensive interactions are consistent with the conserved regions by the sequence alignments of L and P, respectively (Supplementary Fig. 4).
Structural comparison of the L and P proteins
When the structure of the RSV L:P overlays with that of the VSV L40 (PDB: 5A22), the CD, MT, and CTD domains (gray) extends from the top part of the complex (Fig. 3a, b). The RSV RdRp domain is comparable to many other RNA polymerases40,46,47,48,49,50,51 (Fig. 3c and Supplementary Fig. 7). The RSV Cap domain exhibits a similar overall fold to that of VSV L40 (Fig. 3e and Supplementary Fig. 8).
a Superimposition of the RNA dependent RNA polymerase (RdRp) and capping (Cap) domains of the L protein from RSV (this work) and VSV (PDB: 5A22). RSV L is colored the same as Fig. 2a, and VSV L is colored in gray. b The cartoon representation of the RSV L and P. The missing domains are colored in gray. c Overview of the RdRp domain. d Magnified view showing the core residues (GDN, magenta for RSV, orange for VSV) and the missing supporting helix (orange, VSV L) in the RdRp domain. e Overview of the Cap domain. f Magnified view showing the core residues (HR, magenta for RSV, orange for VSV) in the Cap domain. The priming-loop-like element of the RSV L and the VSV L is highlighted in magenta and orange, respectively. A major shift (~37 Å) from the tip of the priming-loop-like element is shown as the black arrow. g Superimposition of the oligomerization domain of P (POD) from RSV (this work) and HMPV (PDB: 4BXT). The HMPV P is in gold.
However, there are notable differences: (1) There are additional motifs at the N-terminal regions of the RSV RdRp; (2) The RSV PdRp has a missing connecting helix (residues 660–691) (equivalent to VSV residues 571–597) adjacent to the active site (Fig. 3d and Supplementary Fig. 8); (3) The RSV priming-loop-like element (residues 1265–1282, magenta) of the Cap domain shows a significant shift of 146° and 37.2 Å compared to its equivalent motif of VSV L (residues 1155–1174, orange), and the pivot points for the shift are the residues 1264 and 1283 (refs. 40,50,52) (Fig. 3f and Supplementary Fig. 8).
We also compared the structures of the RSV P with P from other NNS RNA viruses. P is the most variable protein and shares the lowest sequence identities among NNS RNA viruses. Strikingly, despite the low sequence conservation, all P (or VP35 in Filoviridae) share a common feature and exist as an oligomer (dimer, trimer, or tetramer) in solution53,54,55,56,57,58,59,60 (Fig. 3g and Supplementary Fig. 9). Although the precise role of the oligomerization of P remains unclear, the comparison suggests the oligomerization of P plays critical roles in enhancing the interactions with L and bridging multiple sets of co-factors (i.e., N and M2-1) during RNA synthesis.
Conformational transitions of L:P
The significant flexibility of P leads to a weaker density for many regions of P (visualized at a lower σ). Further 3D classification revealed an almost equal number (63,912 vs. 82,400) of the particles that show less P density and yielded a cryo-EM map at 4.86 Å resolution. Compared with a 4.54 Å map, the RdRp and Cap domains of L are virtually identical, but the density for P is almost depleted (Supplementary Fig. 10). Focused refinement of P regions does not yield a higher resolution map, confirming the intrinsic flexibility and mobility of P.
The substantial conformational variability of CD, MT, and CTD of L leads to a missing density throughout the initial image processing. A larger size 3D class produced a 7.2 Å resolution map of reconstruction. Additional density blobs appear on the top part of the polymerase, agree with the potential location of the missing domains. However, no well-defined density for these domains was observed in the cryo-EM map (Supplementary Fig. 10).
Discussion
This work provides structural insights into the polymerase (L:P) of RSV, a significant NNS RNA virus pathogen, and offers a framework for understanding the coordination of the enzymatic activities of L within structurally distinct but functionally coordinated domains. Further investigations are required to discover whether the RSV L alone exhibits the same way as VSV L. Interestingly, all RNA polymerases of NNS RNA viruses require P (or VP35 in Filoviridae), but P exists as diverse oligomeric protein (Supplementary Fig. 9) and shares low sequence identity53,54,55,56,57,58,59,60. In most cases, the protomers, the basic building blocks of oligomeric proteins, assemble into a higher-order oligomer following a defined repeating or symmetry rule. However, the nonsymmetric structures of PCTD suggest that P likely plays a nonsymmetric structural role more than previously appreciated. Except for the POD, every chain of the P tetramer appears to either interact with different sites of L, such as the “palm” motif of the RdRp domain, which may be important for regulating the polymerase activities or remain flexible waiting for interacting with other binding partners. These pronounced structural differences attest to a high degree of versatility in L upon binding of P.
Indeed, it was suggested that L bears much movement during RNA synthesis. We compared the priming-loop-like element of the RSV L to that of the VSV L (Supplementary Fig. 8). The priming-loop-like element of the VSV Cap domain has been demonstrated to play critical roles in transcription initiation and capping61, and it was thought to be at the initiation stage because the overlay of the structure of the initiation complex of other RdRP (such as reovirus polymerase) revealed that this priming-loop-like element of the VSV L occupied the same location as the priming loop in the reovirus polymerase40,50. In addition, the comparisons of the VSV L and the initiation/elongation complex of the reovirus polymerase suggest that if the RNA product extends, there will be not enough space to accommodate additional newly synthesized RNA products, and this priming-loop-like element is likely to move away from this initiation stage40,50 (Supplementary Fig. 11c–d). We also superposed the elongation complexes of other RdRPs (such as FluB, polio, or rotavirus polymerase) with the RSV L:P, and the position of the priming-loop-like element does not “conflict” with the locations of either RNA template or RNA product52,62,63 (Supplementary Fig. 11). Together, the missing connecting helix (residues 660–691) of the RSV RdRp domain and the significant shift of the priming-loop-like element of the RSV Cap domain suggest that the structure we obtained is likely to be at an elongation-compatible stage. Based on the structures of the VSV L (preinitiation stage, PDB: 5A22) and influenza polymerase (elongation stage, PDB: 6QCV), we modeled the RNA template and the transcript into our RSV polymerase structure40,50,52,62,63 (Fig. 4a, b and Supplementary Fig. 11).
a, b The RSV polymerase with modeled RNA template and transcript (PDB: 6QCV). The same color scheme for RSV proteins as Fig. 2a. The modeled RNA template and RNA transcript are shown in yellow and pink, respectively. (Supplementary Fig. 11 for additional comparisons). The 3′ RNA template enters the RdRp from the bottom, and 5′ RNA transcript exits from the top. The upstream 3′ RNA template is drawn in black line.
Besides, our study has implications for understanding current RSV polymerase inhibitors. Previous studies showed that resistance to the nucleoside analog inhibitor ALS-8112 is conferred by QUAD resistance mutations (M628L, A789V, L795I, and I796V) in the RdRp domain of L, while a nonnucleoside inhibitor AZ-27 that inhibited the RdRp activity could be escaped by a Y1631H resistance mutation64. In our structure, the QUAD mutation sites (yellow) of ALS-8112 are in approximate close location to the active site (D811, magenta), and the QUAD mutations can potentially alter the microenvironment of RNA synthesis and the conformation of the RdRp active sites (Supplementary Fig. 12). However, Y1631 is not visible in the structure, and the inhibition mechanism of AZ-27 remains unclear.
Further, our study has evolutionary implications: How did three distinct enzymatic activities (RNA polymerization, RNA capping, and RNA cap methylation) for RNA synthesis integrate within a single polypeptide (L)? How are those functional domains evolved/related to multiple much complex counterparts in the eukaryotic cell and many other viruses? Further studies will reveal whether this “compact” mechanism illustrates the evolutionary pressure applied for the RNA synthesis machinery in general.
During the review process of our manuscript, another structure of the RSV polymerase (L:P) complex in an apo state was published65. We superposed the published structure (PDB: 6PZK) with our structure (PDB: 6UEN) described here, and they share similar overall fold with high similarities (RMSD = 1.450 Å). Interestingly, the superpositions of the individual RdRp domain, Cap domain, and the P tetramers yield lower RMSD values of 1.240, 1.021, and 0.991 Å, respectively. The structural comparisons suggest that the individual domains are mostly the same, but the inter-domain arrangements of the two structures have slight differences. In the closer dissection of both structures, we identified minor shifts between the interface of the L:P complex, in particular, the P tetramers and the two helixes of L that interact with P (dock A and dock B) if fixed the position of the RdRp domain. It appears that the P tetramers slide closer to L, and the docks A and B shift towards the RdRp domain and adopt more compact packing to accommodate closer interactions with P (6PZK) than that of this work (6UEN) (Supplementary Fig. 13 and arrows indicate the shift direction). This observation suggests plasticity in the L:P interaction, and that this interface may adopt a larger degree of conformational rearrangements during RNA synthesis.
Outstanding questions remain: (1) It is known that the phosphorylation of P regulates the activities of L. Previous studies suggested many potential phosphorylation sites reside in the PNTD (1–127) but are not visible in this study36,37,38. (2) How other co-factors such as M2-1, N0, or N:RNA influence (or stabilize) the polymerase conformations? To determine the structures of L:P:RNA or L:P:M2-1 complexes may be a future research focus.
Methods
Expression and purification of the RSV polymerase (L:P)
Codon-optimized RSV (strain A2) L and P genes (the DNA sequences and primers are listed in Supplementary Table 2) were subcloned into the pFastBac Dual vector (MacroLab), and the virus was prepared using the Bac-to-Bac system (Invitrogen). PCR-based site-directed mutagenesis was used for the construction of mutant L(D811A) with the plasmid encoding the wt L:P complex as the template (the primers are listed in Supplementary Table 2c). N-terminal 6x His-tagged L containing a TEV cleavage site and no-tagged P was co-expressed in baculovirus-mediated transduction of Sf21 suspension cell cultures. Cells were lysed by Dounce homogenization in lysis buffer (50 mM sodium phosphate pH 7.4, 300 mM NaCl, 6 mM MgSO4, 10% glycerol, 0.2% NP-40, and EDTA-free protease inhibitor), followed by Co2+-NTA agarose beads (GoldBio), washed with wash buffer (50 mM sodium phosphate pH 7.4, 300 mM NaCl, 6 mM MgSO4, 10% glycerol, and 10 mM imidazole), and eluted with elution buffer (50 mM sodium phosphate pH 7.4, 300 mM NaCl, 6 mM MgSO4, 10% glycerol, and 250 mM imidazole). The eluted sample was treated with TEV enzyme in TEV cleavage buffer (50 mM sodium phosphate pH 7.4, 300 mM NaCl, 6 mM MgSO4, 10% glycerol, and 0.2% NP-40, 1.4 mM 2-Mercaptoethanol) overnight at 16 °C and further applied to Co-NTA. The flow-through sample was applied to the Heparin column (Buffer A: 50 mM Tris-HCl pH 8.0, 50 mM NaCl, 10% Glycerol, and Buffer B: 50 mM Tris-HCl pH 8.0, 1.5 M NaCl, 10% Glycerol) and followed by size-exclusion chromatography using the gel filtration buffer (25 mM HEPES pH 7.4, 300 mM NaCl, 6 mM MgSO4, and 0.5 mM TCEP) with a Superose 6 Increase 10/300 GL (GE Healthcare). The quality of purified proteins was analyzed by SDS-PAGE gel. The bands migrating ~250 and ~35 kDa were confirmed to be RSV L and P polypeptides by liquid chromatography-mass spectrometry (LC/MS, Supplementary Fig. 5). The proteins were tested active for RNA synthesis activity. The pure proteins were flash-frozen in liquid nitrogen and stored in 30 μl aliquots at −80 °C for further use. The mutant L(D811A):P complex was expressed, purified, and stored in the same manner as the wt L:P complex.
In vitro RNA synthesis assay
The terminal trailer complementary (TrC25: 5′-UAGUUUUUGACACUUUUUUUCUCGU-3′) template and trailer (Tr25: 5′-ACGAGAAAAAAAGUGUCAAAAACUA-3′) product sequences of the RSV genome were used in the in vitro RdRp assay (detailed procedures described in Noton et al.13). RNA oligonucleotides were chemically synthesized from IDT. Radioactive isotope-labeled nucleotides [α-32P] GTP was purchased from Perkin Elmer. The reaction mixtures contained 2 µM RNA template, RSV L:P complexes (~300 ng RSV L), NTPs (ATP, CTP, and UTP each at 1.25 mM and GTP at 50 µM with 5 µCi of [α-32P]GTP), and reaction buffer (50 mM Tris-HCl at pH 7.4, 8 mM MgCl2, 5 mM DTT, 10% glycerol) in a final volume of 20 µl. The reactions were incubated at 30 °C for 5 h, heated to 90 °C for 5 min, and then stopped by adding 5 µl stop buffer (90% formamide, 20 mM EDTA, 0.02% bromophenol blue). The RNA products were analyzed using a 20% polyacrylamide gel containing 7 M urea in a TBE buffer, followed by autoradiography. The RNA ladder used was generated by incubating [ɣ-32P] ATP and 7-nt, 14-nt, 21-nt, and 25-nt RNA trailer sequences (Tr7, Tr14, Tr21, and Tr25) with T4 PNK (NEB). The sequences of the RNA oligos used to generate the ladders are as follows: Tr7 (5′-ACGAGAA-3′), Tr14 (5′-ACGAGAAAAAAAGU-3′), Tr21 (5′-ACGAGAAAAAAAGUGUCAAAA-3′), and Tr25 (5′-ACGAGAAAAAAAGUGUCAAAAACUA-3′).
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Protein samples (~3 μg) were treated with 1 mM dithiothreitol (DTT) at room temperature (RT) for 30 min, followed by 5 mM iodoacetamide at RT for 30 min in the dark. Proteins were digested overnight with 0.5 μg lysyl endopeptidase (Wako) at RT and further digested overnight with 1 μg trypsin (Promega) at RT. Resulting peptides were desalted with an HLB column (Waters). Peptides were analyzed with a Q Exactive™ Plus Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Fisher Scientific). Mass spectrometry data were analyzed using Proteome Discoverer 2.1 against the UniProt database of human respiratory syncytial virus A (strain A2) and Spodoptera frugiperda (Fall armyworm).
Cryo-EM sample preparation and data acquisition
A total of 3.0 μl of the purified RSV polymerase (L:P) complex at a concentration of 0.33 mg/ml were applied to a glow-discharged Quantifoil holey carbon grid (R1.2/1.3, Cu, 400 mesh) (SPI). Grids were blotted for 3 s at ~90% humidity at RT and plunge-frozen in liquid ethane using a Cryoplunge 3 System (Gatan). Cryo-EM data were recorded on a Talos Arctica 200 kV (TEM) with BioQuantum/K2 direct electron detector (Thermo Fisher) at Emory University. All cryo-EM movies were recorded in counting mode using EPU (Thermo Fisher). The nominal magnification of ×130,000 corresponds to a calibrated pixel size of 1.045 Å on the specimen. The dose rate was set to 1.365 e-/Å2/frame. The total exposure time of each movie was 10 s, leading to a total accumulated dose of 55 e−/Å2, fractionated into 40 frames (250 ms per frame). All movies were recorded in a defocus range between −1.25 and −2.5 μm. A total of 2600 micrographs were collected from two separate data collection sessions.
Image processing and 3D reconstruction
Drift correction, beam-induced motion, and dose-weighting were performed for dose-fractionated movies by the program MotionCor266 with a 5 × 5 patch, resulting in corrected movies and summed images. All 40 frames in each movie were summed with the dose-weighted scheme. The summed images were used in all image processing steps. The contrast transfer function (CTF) was estimated using the program CTFFIND467. To generate RSV polymerase complex templates for automatic picking and the initial model, around 50,000 particles were manually picked and classified. The data were initially processed in two datasets, one for each data collection session (1349 and 1251 micrographs) and then merged into a total of 2600 micrographs. A total of 792,070 particles were picked, and the box size of 200 pixels was used to extract the particles. Particle picking and screening, 2D classification, as well as the initial 3D model building, 3D classification, 3D refinement, CTF refinement, and polishing were performed using RELION 3.0.768. The final refinement was validated using cisTEM69, using the best class as the initial model. The global search was performed once without the mask followed by another global search using a soft mask (six-pixel soft edge) that was generated in RELION. All reported resolutions were based on gold-standard refinement procedures and the Fourier shell correlation (FSC) = 0.143 criterion. Local resolution was estimated using ResMap70.
Model building and refinement
The 3.67 Å resolution map was used for model building and refinement. To obtain better side-chain densities for model building, B-factor was used for map sharpening. The coordinates of the VSV L protein (PDB: 5A22) and the homology models of the RSV L protein by Phyre271 and I-TASSER72 were positioned into the map as the initial guides using UCSF Chimera73 and COOT74. The structure model was manually built by COOT. Structure factors were calculated by PHENIX75,76, and the full structure was subjected to multiple cycles of global real-space refinement with rotamer, Ramachandran plot restraints enabled in PHENIX. FSCs were calculated between the two half maps, the model against the working map and the other (free) half map and full (sum) map. The confidence maps and locally sharpened maps were calculated to facilitate the model building77,78. MolProbity79 was used to validate the geometries of the model.
Figure preparation
All the figures representing model and electron density maps were generated using COOT74, UCSF Chimera73, and PyMOL80. Multiple sequence alignments were performed using Multalin81 and ESPript82.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The 3D cryo-EM density maps and atomic coordinates generated and analyzed during the current study are available on the Electron Microscopy Data Bank (Accession code: EMD-20754) and Protein Data Bank (Accession code: 6UEN), respectively.
References
Knipe, D. M. & Howley, P. M. Fields Virology 6th edn (Lippincott Williams & Wilkins, 2013).
Collins, P. L., Fearns, R. & Graham, B. S. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr. Top. Microbiol. Immunol. 372, 3–38 (2013).
Shi, T. et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390, 946–958 (2017).
Whelan, S. P., Barr, J. N. & Wertz, G. W. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283, 61–119 (2004).
Conzelmann, K. K. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu Rev. Genet. 32, 123–162 (1998).
Grosfeld, H., Hill, M. G. & Collins, P. L. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69, 5677–5686 (1995).
Ball, L. A. & White, C. N. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl Acad. Sci. USA 73, 442–446 (1976).
Abraham, G. & Banerjee, A. K. Sequential transcription of the genes of vesicular stomatitis virus. Proc. Natl Acad. Sci. USA 73, 1504–1508 (1976).
Testa, D., Chanda, P. K. & Banerjee, A. K. Unique mode of transcription in vitro by Vesicular stomatitis virus. Cell 21, 267–275 (1980).
Fearns, R., Peeples, M. E. & Collins, P. L. Mapping the transcription and replication promoters of respiratory syncytial virus. J. Virol. 76, 1663–1672 (2002).
Noton, S. L. & Fearns, R. Initiation and regulation of paramyxovirus transcription and replication. Virology 479–480, 545–554 (2015).
Tremaglio, C. Z., Noton, S. L., Deflube, L. R. & Fearns, R. Respiratory syncytial virus polymerase can initiate transcription from position 3 of the leader promoter. J. Virol. 87, 3196–3207 (2013).
Noton, S. L., Deflube, L. R., Tremaglio, C. Z. & Fearns, R. The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog. 8, e1002980 (2012).
Albertini, A. A., Schoehn, G., Weissenhorn, W. & Ruigrok, R. W. Structural aspects of rabies virus replication. Cell Mol. Life Sci. 65, 282–294 (2008).
Ogino, T., Yadav, S. P. & Banerjee, A. K. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl Acad. Sci. USA 107, 3463–3468 (2010).
Ogino, T. & Banerjee, A. K. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 25, 85–97 (2007).
Li, J., Rahmeh, A., Brusic, V. & Whelan, S. P. Opposing effects of inhibiting cap addition and cap methylation on polyadenylation during vesicular stomatitis virus mRNA synthesis. J. Virol. 83, 1930–1940 (2009).
Furuichi, Y. & Shatkin, A. J. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55, 135–184 (2000).
Gupta, K. C. & Roy, P. Alternate capping mechanisms for transcription of spring viremia of carp virus: evidence for independent mRNA initiation. J. Virol. 33, 292–303 (1980).
Testa, D. & Banerjee, A. K. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J. Virol. 24, 786–793 (1977).
Paesen, G. C. et al. X-ray structure and activities of an essential Mononegavirales L-protein domain. Nat. Commun. 6, 8749 (2015).
Morin, B., Rahmeh, A. A. & Whelan, S. P. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 31, 1320–1329 (2012).
Morin, B., Liang, B., Gardner, E., Ross, R. A. & Whelan, S. P. An in vitro RNA synthesis assay for rabies virus defines ribonucleoprotein interactions critical for polymerase activity. J. Virol. 91, https://doi.org/10.1128/JVI.01508-16 (2017).
Li, J., Wang, J. T. & Whelan, S. P. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl Acad. Sci. USA 103, 8493–8498 (2006).
Rahmeh, A. A., Li, J., Kranzusch, P. J. & Whelan, S. P. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 83, 11043–11050 (2009).
Poch, O., Sauvaget, I., Delarue, M. & Tordo, N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8, 3867–3874 (1989).
Poch, O., Blumberg, B. M., Bougueleret, L. & Tordo, N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71, 1153–1162 (1990).
Sleat, D. E. & Banerjee, A. K. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J. Virol. 67, 1334–1339 (1993).
Fix, J., Galloux, M., Blondot, M. L. & Eleouet, J. F. The insertion of fluorescent proteins in a variable region of respiratory syncytial virus L polymerase results in fluorescent and functional enzymes but with reduced activities. Open Virol. J. 5, 103–108 (2011).
Ferron, F., Longhi, S., Henrissat, B. & Canard, B. Viral RNA-polymerases–a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 27, 222–224 (2002).
Li, J., Rahmeh, A., Morelli, M. & Whelan, S. P. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 82, 775–784 (2008).
Sourimant, J. et al. Fine mapping and characterization of the L-polymerase-binding domain of the respiratory syncytial virus phosphoprotein. J. Virol. 89, 4421–4433 (2015).
Galloux, M. et al. Identification and characterization of the binding site of the respiratory syncytial virus phosphoprotein to RNA-free nucleoprotein. J. Virol. 89, 3484–3496 (2015).
Castagne, N. et al. Biochemical characterization of the respiratory syncytial virus P-P and P-N protein complexes and localization of the P protein oligomerization domain. J. Gen. Virol. 85, 1643–1653 (2004).
Pereira, N. et al. New insights into structural disorder in human respiratory syncytial virus phosphoprotein and implications for binding of protein partners. J. Biol. Chem. 292, 2120–2131 (2017).
Villanueva, N., Hardy, R., Asenjo, A., Yu, Q. & Wertz, G. The bulk of the phosphorylation of human respiratory syncytial virus phosphoprotein is not essential but modulates viral RNA transcription and replication. J. Gen. Virol. 81, 129–133 (2000).
Asenjo, A., Calvo, E. & Villanueva, N. Phosphorylation of human respiratory syncytial virus P protein at threonine 108 controls its interaction with the M2-1 protein in the viral RNA polymerase complex. J. Gen. Virol. 87, 3637–3642 (2006).
Asenjo, A. & Villanueva, N. Phosphorylation of the human respiratory syncytial virus P protein mediates M2-2 regulation of viral RNA synthesis, a process that involves two P proteins. Virus Res. 211, 117–125 (2016).
Cowton, V. M., McGivern, D. R. & Fearns, R. Unravelling the complexities of respiratory syncytial virus RNA synthesis. J. Gen. Virol. 87, 1805–1821 (2006).
Liang, B. et al. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327 (2015).
Cressey, T. N., Noton, S. L., Nagendra, K., Braun, M. R. & Fearns, R. Mechanism for de novo initiation at two sites in the respiratory syncytial virus promoter. Nucleic Acids Res. 46, 6785–6796 (2018).
Lu, B. et al. Identification of temperature-sensitive mutations in the phosphoprotein of respiratory syncytial virus that are likely involved in its interaction with the nucleoprotein. J. Virol. 76, 2871–2880 (2002).
Khattar, S. K., Yunus, A. S. & Samal, S. K. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P and P-L interactions using a minigenome system. J. Gen. Virol. 82, 775–779 (2001).
Mason, S. W. et al. Interaction between human respiratory syncytial virus (RSV) M2-1 and P proteins is required for reconstitution of M2-1-dependent RSV minigenome activity. J. Virol. 77, 10670–10676 (2003).
Mallipeddi, S. K., Lupiani, B. & Samal, S. K. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P interaction using a two-hybrid system. J. Gen. Virol. 77, 1019–1023 (1996).
Choi, K. H. et al. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl Acad. Sci. USA 101, 4425–4430 (2004).
Lesburg, C. A. et al. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6, 937–943 (1999).
Reich, S. et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516, 361–366 (2014).
Gerlach, P., Malet, H., Cusack, S. & Reguera, J. Structural insights into Bunyavirus replication and its regulation by the vRNA promoter. Cell 161, 1267–1279 (2015).
Tao, Y., Farsetta, D. L., Nibert, M. L. & Harrison, S. C. RNA synthesis in a cage–structural studies of reovirus polymerase lambda3. Cell 111, 733–745 (2002).
Lu, X. et al. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 16, 1678–1688 (2008).
Kouba, T., Drncova, P. & Cusack, S. Structural snapshots of actively transcribing influenza polymerase. Nat. Struct. Mol. Biol. 26, 460–470 (2019).
Bruhn, J. F. et al. Crystal structure of the marburg virus VP35 oligomerization domain. J. Virol. 91, e01085–16 (2017).
Bruhn, J. F. et al. Crystal structure of the nipah virus phosphoprotein tetramerization domain. J. Virol. 88, 758–762 (2014).
Leyrat, C., Renner, M., Harlos, K. & Grimes, J. M. Solution and crystallographic structures of the central region of the phosphoprotein from human metapneumovirus. PLoS ONE 8, e80371 (2013).
Cox, R. et al. Structural and functional characterization of the mumps virus phosphoprotein. J. Virol. 87, 7558–7568 (2013).
Communie, G. et al. Structure of the tetramerization domain of measles virus phosphoprotein. J. Virol. 87, 7166–7169 (2013).
Ivanov, I., Crepin, T., Jamin, M. & Ruigrok, R. W. Structure of the dimerization domain of the rabies virus phosphoprotein. J. Virol. 84, 3707–3710 (2010).
Ding, H., Green, T. J., Lu, S. & Luo, M. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J. Virol. 80, 2808–2814 (2006).
Tarbouriech, N., Curran, J., Ruigrok, R. W. & Burmeister, W. P. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7, 777–781 (2000).
Ogino, M., Gupta, N., Green, T. J. & Ogino, T. A dual-functional priming-capping loop of rhabdoviral RNA polymerases directs terminal de novo initiation and capping intermediate formation. Nucleic Acids Res. 47, 299–309 (2019).
Gong, P. & Peersen, O. B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl Acad. Sci. USA 107, 22505–22510 (2010).
Ding, K. et al. In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release. Nat. Commun. 10, 2216 (2019).
Deval, J. et al. Biochemical effect of resistance mutations against synergistic inhibitors of RSV RNA polymerase. PLoS ONE 11, e0154097 (2016).
Gilman, M. S. A. et al. Structure of the respiratory syncytial virus polymerase complex. Cell 179, 193–204 (2019).e114.
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Avramov, T. K. et al. Deep learning for validating and estimating resolution of cryo-electron microscopy density maps. Molecules 24, 1181 (2019).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Yang, J. et al. The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Beckers, M., Jakobi, A. J. & Sachse, C. Thresholding of cryo-EM density maps by false discovery rate control. IUCrJ 6, 18–33 (2019).
Jakobi, A. J., Wilmanns, M. & Sachse, C. Model-based local density sharpening of cryo-EM maps. eLife 6, e27131 (2017).
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Rigsby, R. E. & Parker, A. B. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem. Mol. Biol. Educ. 44, 433–437 (2016).
Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Acknowledgements
This study was supported by the US National Institute of General Medical Sciences (NIGMS), National Institutes of Health (NIH) under award number R01GM130950, and the Research Start-Up Fund at Emory University School of Medicine. We thank Dr Martin Moore for providing the helper plasmids of RSV L and P. We thank the assistance and services provided at the Robert P. Apkarian Integrated Electron Microscopy Core (IEMC) at Emory University. We also thank Dr Pritha Bagchi at Emory Integrated Proteomics Core (EIPC) at Emory University. The mass spectrometry work was supported in part by the Emory Integrated Proteomics Core and Parker H. Petit Institute for Bioengineering and Biosciences at Georgia Tech. We thank Ryan Youngs, Amaal Abdi, Erica Lee, Masthan Shaik, Ziheng Ma, Sarah Franklin, and Jasmine Berry for the help of manual particle picking. We also thank the members of the Liang lab for critical discussions and helpful support.
Author information
Authors and Affiliations
Contributions
D.C., Y.G. and B.L. conceived the project. D.C., C.R., S.R., P.D., L.Z., J.S., M.D., A.A., S.R., S.K. and G.F. contributed to the cell culture. D.C., C.R., C.R., S.R., P.D., L.Z., J.S. and B.L. carried out the protein purification. D.C., Y.G., P.J. and B.L. prepared cryo-EM grids. D.C., P.J. and B.L. collected cryo-EM data. B.L. carried out cryo-EM image processing. D.C. and B.L. built and refined atomic models. D.C., Y.G. and B.L. analyzed data. B.L. wrote the manuscript. D.C. and B.L. revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks Aartjan te Velthuis, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cao, D., Gao, Y., Roesler, C. et al. Cryo-EM structure of the respiratory syncytial virus RNA polymerase. Nat Commun 11, 368 (2020). https://doi.org/10.1038/s41467-019-14246-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-019-14246-3
This article is cited by
-
Structures of the promoter-bound respiratory syncytial virus polymerase
Nature (2024)
-
An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles
Nature Structural & Molecular Biology (2023)
-
Structure of the Newcastle Disease Virus L protein in complex with tetrameric phosphoprotein
Nature Communications (2023)
-
Conserved allosteric inhibitory site on the respiratory syncytial virus and human metapneumovirus RNA-dependent RNA polymerases
Communications Biology (2023)
-
Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.