Abstract
Inflammatory bowel disease (IBD) includes Crohn’s disease and ulcerative colitis. Each disease is characterized by a diverse set of potential manifestations, which determine patients’ disease phenotype. Current understanding of phenotype determinants is limited, despite increasing prevalence and healthcare costs. Diagnosis and monitoring of disease requires invasive procedures, such as endoscopy and tissue biopsy. Here we report signatures of heterogeneity between disease diagnoses and phenotypes. Using mass cytometry, we analyze leukocyte subsets, characterize their function(s), and examine gut-homing molecule expression in blood and intestinal tissue from healthy and/or IBD subjects. Some signatures persist in IBD despite remission, and many signatures are highly represented by leukocytes that express gut trafficking molecules. Moreover, distinct systemic and local immune signatures suggest patterns of cell localization in disease. Our findings highlight the importance of gut tropic leukocytes in circulation and reveal that blood-based immune signatures differentiate clinically relevant subsets of IBD.
Similar content being viewed by others
Introduction
Inflammatory bowel disease (IBD) is increasingly prevalent1,2. Despite available therapies, response remains challenging largely due to heterogeneity of clinical phenotypes3. IBD is divided into Crohn’s disease (CD) and ulcerative colitis (UC), both chronic relapsing/remitting immune-mediated conditions thought to be triggered by environmental factors in genetically predisposed individuals4. Within IBD, there is significant heterogeneity, especially amongst CD patients. CD can affect any part of the gastrointestinal tract, and disease lesions can present as ulcers, strictures, or penetrate from the lumen to the fat wrapping the intestines5. CD can be classified by disease behavior—inflammatory (B1; non-stricturing nonpenetrating), stricturing (B2), or penetrating (B3)6. CD can also be categorized by intestinal region(s) affected, grouped commonly by ileal, ileocolonic, or colonic disease. The majority of CD patients with colonic and/or rectal involvement develop perianal disease7. However, there are no objective measures of these disease phenotypes for use as markers of disease, tools to follow disease course, and/or enhance understanding of pathogenesis.
Compared to CD, UC is a relatively homogenous disease restricted to the colon, generally characterized by ulcerations and pseudopolyps5. In the absence of treatment effects, UC always involves the rectum, and disease is typically classified as proctitis (E1; restricted to the rectum), left-sided (E2; involvement from the rectum to the splenic flexure), or pan colitis (E3; extensive disease beyond the splenic flexure)8,9. Conversely, CD is typically patchy with skip lesions10. Thus, UC includes a more uniform set of disease phenotypes.
CD and UC can be distinguished clinically by disease location (UC is restricted to the colon, while CD can involve any part of the gastrointestinal tract) and disease phenotype (stricturing or penetrating can indicate CD), using endoscopy and imaging5. However, gathering this information can be invasive and costly, and still up to 20% of IBD cases with colonic disease are indeterminate10. About 10% of patients who undergo colectomy for perceived UC are subsequently diagnosed with CD previously confined to the colon10,11. Because CD and UC can each present with a variety of disease phenotypes, identification of better markers using less invasive methods would enable earlier diagnosis as well as improved monitoring and treatment of disease.
Despite roles of host genetics, the immune system, the microbiota, and the environment in the pathogenesis of IBD4, factors responsible for the breadth of disease manifestations between patients (heterogeneity) are not well defined. Identifying and monitoring the symptoms, severity, behavior, and therapeutic response that characterize each disease manifestation (phenotype) is critical for improving health, yet frequently requires the use of invasive procedures, such as endoscopy.
In the tissue, IBD is characterized by intestinal barrier breakdown, allowing microbiota to prime the immune system. The ensuing inflammatory response involves recruitment of leukocytes from the periphery to the gut and is associated with dissemination of activated cells in circulation2,12,13. Since leukocytes that traffic to the gut from the periphery mediate this inflammatory response, we hypothesized that, by enriching for gut tropic cells in the blood, we could detect CD- and UC-specific as well as phenotype-specific signatures and investigate their relationship to the tissue. Since blood is easily accessible, identification of immune signatures in circulation that represent intestinal immunity would be highly informative and provide less invasive tools to assess disease.
We use mass cytometry (CyTOF) to resolve single cells and characterize their lineage, gut tropism, and function14. Consistent with our hypothesis, we identify blood signatures of disease diagnosis, phenotype, and state (flare/remission), heavily represented by gut tropic cell populations and reflective of the greater clinical heterogeneity observed for CD than UC. We report a multi-parameter generalized linear model to classify patients by disease diagnosis (CD/UC), suggesting that blood-based assays could reduce the delay, risk, discomfort, and cost associated with more invasive procedures.
Results
Study design
We collected blood and biopsies from IBD patients (Fig. 1a) and selected 68 subjects with gastroenterologist-confirmed IBD diagnoses or matched healthy controls (HC) (Table 1). Cohort 1 included blood samples from 56 subjects (Supplementary Table 1), and cohort 2 had paired blood and biopsy samples from 12 subjects (Supplementary Table 2). By cohort, disease-group demographics were matched as best as possible for sex and reports of extraintestinal manifestations (Table 1; Supplementary Tables 1, 2). There was no significant difference in disease-state proportions (flare/remission for cohort 1), sex, age, age at onset, disease duration, extraintestinal manifestations, or tissue-state proportions (inflamed/uninflamed for cohort 2) between CD and UC patients by cohort (Table 1). Thus, it was unnecessary to adjust data based on these parameters. Disease phenotypes were specific to each diagnosis and could not be matched. IBD samples included subjects in clinical remission or flare (determined by IBD specialists; see the Methods section), and a spectrum of disease activity scores were represented in each group.
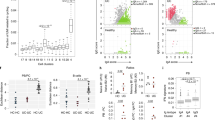
Circulating leukocytes distinguish CD and UC. a Schematic of the study conducted on cohort 1. Created with BioRender. b viSNE using CD11c, CD56, CD16, CD8, CD3, CD123, CD27, CD24, CD14, CD19, CD4, CD20, and CD45RO for clustering with samples from cohort 1. Coefficient of variation was calculated for 2208 parameters per sample by disease group. Statistics: unpaired two-tailed Student’s T test (CD remission vs. HC, t = 12.43, df = 4412; CD remission vs. UC remission, t = 14.12, df = 4406; UC flare vs. HC, t = 6.994, df = 4403; UC flare vs. UC remission, t = 8.621, df = 4397). Sample sizes: CD flare = 13; CD remission = 11; UC flare = 10; UC remission = 10; HC = 12. c Features distinguished all CD and UC. Statistics: BH FDR-corrected unpaired two-tailed Student’s T test using Morpheus (see the Methods section; CCR9+GPR15+CD56+ B cells, t = 2.58; α4β7+CCR1+CD56+ plasmablasts, t = 2.74). Sample sizes: CD = 23, UC = 18. d Features differentiating CD and UC identified by hypothesis-driven tests. Statistics: unpaired two-tailed Student’s T test (Basophils [% of live singlets]: all CD vs. UC, t = 2.57, df = 42; CD vs. UC flare, t = 3.34, df = 21; CD flare vs. HC, t = 2.79, df = 23; CD flare vs. remission, t = 2.87, df = 22; all UC vs. HC, t = 3.88, df = 30; UC flare vs. HC, t = 4.02, df = 20; UC flare vs. remission, t = 6.91; df = 18. Basophils [median pCREB]: all CD vs. UC, t = 2.53, df = 42; CD vs. UC flare, t = 3.17; df = 21. pDCs [% of DCs]: all CD vs. UC, t = 2.61, df = 42; CD vs. UC flare, t = 2.97, df = 21; UC flare vs. remission, t = 4.03; df = 18. α4β7+ mDCs [% of mDCs]: all CD vs. UC, t = 2.07, df = 39; CD vs. UC flare, t = 3.30, df = 19; CD flare vs. remission, t = 2.33, df = 21. Effector memory CD4 T cells [median pCREB]: all CD vs. UC, t = 2.27, df = 42; CD vs. UC flare, t = 3.13, df = 21; CD flare vs. remission, t = 2.92; df = 22. IgD−CD27− B cells [% of CD19+CD20+]: all CD vs. UC, t = 2.15, df = 42; CD vs. UC flare, t = 2.77, df = 21; UC flare vs. remission, t = 3.47, df = 18; UC flare vs. HC, t = 5.05, df = 20). Sample sizes: all CD = 24; CD flare = 13; CD remission = 11; all UC = 20; UC flare = 10; UC remission = 10; HC = 12 (23, 13, 10, 18, 8, 10, and 12, respectively, for α4β7+ mDCs). Center lines = mean; whiskers = standard deviation. Source data are provided as a Source Data file
We analyzed viably cryopreserved leukocytes from blood and tissue by CyTOF using panels with surface and intracellular antigens (Supplementary Table 3; Supplementary Figs 1, 2). We used four trafficking molecules to identify gut tropic cells: α4β7, a pan-gut-trafficking molecule and target of the therapeutic antibody vedolizumab13; CCR1, a trafficking molecule identified in GWAS studies and a marker of activity in CD15,16; CCR9, a lymphocyte trafficking molecule associated with small intestine tropism13; and GPR15, a T cell trafficking molecule that we and others showed to be important for trafficking to the colon13,17,18. While our CyTOF panels included phosphoproteins and functional markers, we found in pilot studies that cell stimulation was unnecessary to resolve differences in phospho-signaling between sample groups. Trafficking receptor expression patterns in tissue and blood shed light on local and peripheral immune responses since little is known about leukocyte trafficking to the gut in human, especially in the context of disease.
Blood leukocytes demonstrate increased heterogeneity in CD
We conducted targeted analysis of CyTOF data by manually gating and calculating median protein expression levels to compile 2208 parameters per sample, as well as unbiased analysis using viSNE, CITRUS, and Spade algorithms. Coefficients of variation (CVs) for each parameter were used as a proxy for disease group population diversity19, supporting clinical observations that CD includes more heterogenous disease manifestations than UC (Fig. 1b). Samples from CD remission had significantly higher CVs than samples from UC remission or HCs, suggesting greater heterogeneity in CD remission compared to UC remission and HCs. Samples from CD flare had higher CVs but exhibited a much higher standard deviation and were not significantly different than from CD remission, UC flare, or HCs, suggesting an overall increase in heterogeneity amongst CD flare. Conversely, samples from UC flare had significantly higher CVs than samples from UC remission or HCs, suggesting distinct states of flare and remission in UC where flare is inherently more heterogeneous than remission regardless of diagnosis.
Gut-homing molecule expression distinguishes CD from UC
Manually calculated parameters were compared between all CD and UC blood samples in cohort 1, which revealed statistically significant differences in abundance of CCR9+GPR15+ CD56+ B cells and α4β7+CCR1+ CD56+ plasmablasts after correction for multiple testing (Fig. 1c). These data show that gut destined, or tropic, B cell subsets, including CCR9 and GPR15 co-expressing cells (Supplementary Fig. 6), appear to be relevant for disease distinction. Although expression of CD56 (neural cell adhesion molecule) is typically associated with NK cells, the protein is also expressed on other leukocytes, including activated B cells in lymphoma patients20 and T helper 1 (Th1) responses21. CD56 expression is consistent with elevated CD56+ B cells and plasmablasts here in CD, where Th1 responses are known to contribute to disease22.
Based on gene expression deconvolution analysis23 (Supplementary Fig. 8) and knowledge of IBD, we conducted a refined set of additional hypothesis-driven tests to compare CD and UC. This approach revealed six parameters in the blood significantly different between all CD and UC samples (Fig. 1d), as well as four significantly different for flare (Supplementary Fig. 3). The increase of basophils in CD compared with UC is consistent with gene expression deconvolution data (Supplementary Fig. 8). Moreover, median phospho-cyclic AMP-responsive element-binding protein (pCREB) expression by basophils was significantly higher in CD compared with UC, which is consistent with the association between CD activity and blood expression of the total CREB24 (Fig. 1d). Circulating pDCs, α4β7+ mDCs, and memory effector CD4+ T cells were also significantly increased in CD compared with UC (Fig. 1d). IgD−CD27− B cells were significantly increased among the total CD19+CD20+ B cells in UC as compared with CD, suggesting an antigen-driven B cell response (Fig. 1d). These IgD−CD27− memory B cells are larger and more granular than IgD+CD27− naive B cells25, class switched, and somatically hypermutated26. Expansion of IgD−CD27− B cells was reported in systemic lupus erythematosus27, HIV28,29, and rotavirus30 infections. In summary, we identified eight cellular features from a combination of unbiased and hypothesis-driven analyses that were significantly different between all CD and UC patients.
Blood heterogeneity is conserved in CD flare and remission
Next, we compared IBD patients in remission to HCs to identify signatures of subclinical disease persistent in remission, as well as patients in flare to those in remission to identify features specific to disease activity. We found few significant trends across CD patients (Fig. 2a, b). CD remission compared with HC revealed an increase in two gut tropic B cell populations identified by GPR15 and CCR9 expression (Fig. 2a). We compared CD flare to remission and found an increased abundance of highly activated CD38+HLA-DR+ CD8 T cells (Fig. 2b). CD flare was associated with increased median total IκBα expression in IgD+ memory B cells and multiple NK cell populations, indicating enhanced NF-κB inhibition. Comparison of CD flare with remission produced a speckled Pearson correlation heatmap based on vectors of all significant feature values for individual patients (shown on the left), indicating high relative heterogeneity among CD in both flare and remission (Fig. 2b). Since each patient had a different baseline, correlation coefficients allowed us to detect relative heterogeneity when it was not possible to discern absolute heterogeneity.
Some circulating leukocyte signatures differentiate disease flare and remission while others persist in remission. Significant differences between CD remission (N = 11) and HC (N = 12) samples (a), CD flare (N = 13) and CD remission (N = 11) samples (b), UC remission (N = 10) and HC (N = 12) samples (c), and UC flare (N = 10) and UC remission (N = 10) samples (d) with p ≤ 0.05 after correction for multiple testing. On the right of (b, d) are Pearson correlations based on vectors that represent all significant feature values for individual patients shown in the heatmaps immediately to the left. Statistics: BH FDR-corrected unpaired two-tailed Student’s T test using Morpheus (see the Methods section; t-statistics shown in heatmap legends). Source data are provided as a Source Data file
Unlike CD, we found more significant trends across UC patients in flare compared with remission (Fig. 2c, d). Many cellular differences identified here also displayed gut tropism (Fig. 2d), with significantly lower circulating α4β7+ cells in UC flare than remission, possibly because of higher tissue localization during flare. The distinction from CD is further illustrated by the clearly delineated Pearson correlation map (Fig. 2d), revealing homogeneous phenotypes among UC flare and remission.
These findings reinforce observations of greater heterogeneity in CD, which is conserved among patients in remission. Signatures identified relied heavily on trafficking molecule expression, which supports our hypothesis that trafficking molecules allow for non-invasive monitoring of gut-specific immune responses.
Blood leukocytes reflect clinical stratifications of disease
To identify blood signatures responsible for disease heterogeneity, we separated subjects by disease behavior and location. These stratifications are important for monitoring and treatment. When we compared CD patients with inflammatory versus fistulizing, inflammatory versus stricturing, and fistulizing versus stricturing disease (excluding two patients with history of both fistulizing and stricturing disease), we found one B cell signature (Fig. 3a), three signatures of monocyte and B cell subsets (Fig. 3b), and one dendritic cell (DC) signature (Fig. 3c), respectively, that distinguished these disease phenotypes. Interestingly, when we compared CD patients with and without perianal disease, we found a single difference in gut tropic Tregs (Fig. 3d). When we compared CD patients with only ileal versus only colonic disease, we noted a significant decrease in circulating α4β7+CCR9+ subpopulations of mature NK and CD45RO+ NKT cells in ileal CD (Fig. 3e). Since there were no significant differences in the abundance of the total mature NK cells or total CD45RO+ NKT cells, differences in gut tropic subsets suggest enhanced tissue localization in ileal CD, although we did not have sufficient samples from patients with only ileal or only colonic CD in cohort 2 to investigate this further. In UC, only one highly activated GPR15 and CCR9 co-expressing CD4+ T cell population was increased in pan-colonic compared with left-sided disease (Fig. 3f). In summary, gut-tropism signatures in blood differentiated clinical disease subsets.
Circulating leukocytes reflect disease heterogeneity. Features significantly different between inflammatory (N = 7) and fistulizing (N = 7) CD (a), inflammatory (N = 8) and stricturing (N = 7) CD (b), fistulizing (N = 7) and stricturing (N = 7) CD (c), and the presence (N = 7) and absence (N = 16) of perianal disease in CD patients (d). Statistics: BH FDR-corrected unpaired two-tailed Student’s T test using Morpheus (see the Methods section; IgD−IgA− CD19+CD20+ B cells [median α4β7], t = 3.69; CD14+ cells, t = −4.36; IgD−CD27− B cells, t = 3.91; IgD−IgA− CD19+CD20+ B cells [% of CD19+CD20+], t = 4.38; α4β7+CCR9+ HLA-DRlo DCs, t = 3.87; α4β7+GPR15+ Tregs, t = 2.29). Features that distinguish ileal (N = 5) and colonic (N = 4) CD (e), left-sided (N = 6) and pan-colonic (N = 11) UC (f), and colonic CD (N = 4) and UC (N = 18, or 20 for naïve IgD+ B cells) (g) identified by hypothesis-driven testing. Statistics: unpaired two-tailed Student’s T test (α4β7+CCR9+ mature NK cells, t = 3.18, df = 7; α4β7+CCR9+ CD45RO+ NKT cells, t = 7.15, df = 7; CCR9+GPR15+ CD38+HLA-DR+ CD4 T cells, t = 2.20, df = 15; GPR15+ Tregs, t = 2.20; df = 20; naive IgD+ B cells, t = 2.18, df = 22; GPR15+ naive IgD+ B cells, t = 2.23; df = 20; CCR9+GPR15+ plasmablasts, t = 2.28; df = 20). Center lines = mean; whiskers = standard deviation. Source data are provided as a Source Data file
GPR15 expression distinguishes colonic CD from UC
CD can involve the colon in up to 60% of patients31. In a subset where disease is confined to the colon without involving other areas, CD can be difficult to distinguish from UC11. This has important implications, particularly when surgery is considered; surgery can be curative for UC (total colectomy), but not CD32,33. We compared samples from patients with colonic CD (Crohn’s colitis) to UC. GPR15+ Tregs, naive IgD+ B cells, and GPR15+ naive IgD+ B cells were increased while CCR9+GPR15+ plasmablasts were decreased in colonic CD compared with UC (Fig. 3g). Three of these four significantly different cell subsets were enriched for GPR1517,18, demonstrating that trafficking molecule expression by blood leukocytes facilitates complex disease differentiation. These four blood-based features could help address the unmet need to differentiate colonic CD from UC.
Blood and tissue cells are largely but not entirely distinct
We analyzed paired blood and biopsy samples from cohort 2 (Fig. 4a, b) and found two signatures previously identified in cohort 1 blood samples consistent with tissue for all CD versus UC (Fig. 4c). The reduction of basophil frequency we observed in UC biopsies compared with CD suggests an overall reduction of basophils in UC, since this trend is consistent with the blood (Figs 1d, 4c). Moreover, tissue inflammation (identified by endoscopist and confirmed by blinded pathologist) appeared to decrease basophil frequency further in UC, but not in CD (Fig. 4c). Plasmablast frequency was significantly increased in UC tissue compared with CD, which was also observed in the blood when comparing the frequency of α4β7+ plasmablasts between CD and UC flare, suggesting an overall increase of plasmablast abundance in UC flare (Fig. 4c; Supplementary Fig. 3). Basophils and plasmablasts in blood and tissue were consistent with predicted cell frequencies for basophils and plasma cells from gene expression deconvolution of publicly available microarray data sets (Supplementary Fig. 8; plasmablasts were not identifiable via deconvolution, but our plasmablast gate included plasma cells).
Tissue contains immune responses distinct from blood. a Schematic of the study conducted on cohort 2. Blood was drawn and biopsies were collected from study subjects, peripheral blood mononuclear cells (PBMCs) and tissue leukocytes were isolated and cryopreserved, and samples were analyzed in batches by CyTOF. Created with BioRender. b viSNE based on 15 core lineage antigens (CD11c, CD11b, CD56, CD16, CD8, CD3, CD123, CD27, CD24, CD14, CD19, CD4, CD20, TCRγδ, and CD45RO) for samples from cohort 2. Sample sizes: CD blood = 6; UC blood = 6; CD inflamed tissue = 11; CD uninflamed tissue = 12; UC inflamed tissue = 5; UC uninflamed tissue = 13. c Significant differences between disease tissues consistent with trends observed in the blood from cohort 1. Statistics: unpaired two-tailed Student’s T test (Basophils: all CD vs. all UC tissue, t = 1.83, df = 39; UC inflamed vs. uninflamed tissue, t = 2.43, df = 16. Plasmablasts: all CD vs. all UC tissue, t = 2.51, df = 39). Sample sizes: CD blood = 6 (5 for α4β7+); UC blood = 6; all CD tissue = 23; CD inflamed tissue = 11; CD uninflamed tissue = 12; all UC tissue = 18; UC inflamed tissue = 5; UC uninflamed tissue = 13. Center lines = mean; whiskers = standard deviation. d T peripheral helper (Tph) cells, defined as CD3+CD4+CD45RO+CXCR5−PD-1+, in paired blood and tissue samples. Statistics: unpaired two-tailed Student’s T test (all blood vs. tissue, t = 2.65, df = 51; all CD vs. UC tissue, t = 2.25, df = 39; CD inflamed vs. uninflamed tissue, t = 0.75, df = 21). Sample sizes: all blood = 12; all tissue = 41; all CD tissue = 23; all UC tissue = 18; CD inflamed tissue = 11; CD uninflamed tissue = 12. Center lines = mean; whiskers = standard deviation. Source data are provided as a Source Data file
Because B cells and plasmablasts were common among signatures identified, we analyzed CD3+CD4+CD45RO+CXCR5−PD-1+ peripheral helper T (Tph) cells, which drive B cells and plasmablasts in autoimmunity34. We found no Tph abundance differences in blood between CD and UC patients in cohort 2, but there were significantly higher frequencies in tissue compared with blood in both diseases (Fig. 4d). Although there was no difference in Tph cell frequency between CD and UC tissues, Tph cells had significantly higher median pSTAT3 expression in CD than UC tissues (Fig. 4d). Median pSTAT3 expression by Tph cells was not different between inflamed and uninflamed tissues, suggesting that higher pSTAT3 in this population may be a hallmark of CD regardless of inflammation state (Fig. 4d). In summary, Tph cell activation as indicated by pSTAT3 expression but not abundance was significantly higher in CD than UC tissue.
Blood and tissue leukocyte correlations
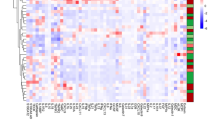
Due to expected differences in cell frequencies and median protein expression levels between blood and tissue, we investigated immune phenotype correlates. Since it was unlikely that marker expression or cell abundance would be equal between blood and tissue because of their distinct cellular environments, we studied paired blood and tissue samples from the ileum, colon, or rectum to identify blood signatures reflective of tissue. We found that 795 of 2145 targeted parameters were significantly different between blood and tissue for all patients in cohort 2. Of these 795 significant features, 55.35% were higher in blood and 44.65% were higher in tissue (Fig. 5a). Pearson correlation based on significant features demonstrated relative homogeneity amongst blood samples compared with tissue diversity (Fig. 5a), suggesting distinct immune responses across regions of the gut and inflammation states.
Mapping paired samples reveals blood correlates of tissue immunity. a Of 2145 manually gated cell frequencies and median expression levels, 795 were significantly different (adjusted p ≤ 0.05; data not shown) between blood (N = 12) and tissue (N = 41). Of significantly different parameters, 55.35% were higher in blood (red) and 44.65% were higher in the tissue (purple). Significantly different parameters were used to construct the Pearson correlation map. Statistics: BH FDR-corrected unpaired two-tailed Student’s T test using Morpheus (see the Methods section). b Of 2145 parameters, three were correlated (green box and green wedge in pie chart) between blood and ileum tissue samples. Statistics: Pearson correlation coefficient (r) and p-values from Pearson correlation tests are shown. P-values for these three correlations were not corrected for multiple testing because hypothesis-driven tests were conducted based on biological insight after independent preliminary analysis. Sample sizes: ten blood/tissue pairs for CD3+ cells and central memory CD4 T cell median pCREB expression; nine blood/tissue pairs for CCR9+ switched memory B cells. Solid line = linear regression; dotted lines = 95% confidence interval. Source data are provided as a Source Data file
Since we did not have sufficient samples to separate uninflamed and inflamed tissues by region (ileum, colon, and rectum), we focused on correlations between blood and biopsies from each region regardless of inflammation due to expected variation by location. Since the heterogeneity of tissue and the breadth of parameters tested would preclude identification of significant correlates by directly comparing all pairs and correcting for all comparisons, we first compared subsets based on disease diagnosis, tissue location, and/or tissue inflammation state with adjustment for multiple comparisons. We combined this screen with biological insight from disease mechanisms to inform a refined set of hypothesis-driven tests without adjustment using all samples divided only by region. With this approach, paired blood and tissue samples revealed that CD3+ frequency of CD14−CD11c+HLA-DR+ cells, median pCREB expression by central memory CD4 T cells, and CCR9+ switched memory B cell frequency were correlated between blood and ileum (Fig. 5b). There were no significant correlations between blood and colon or rectum in cohort 2 amongst 2145 parameters tested, suggesting greater variance in samples from these regions. Surprisingly, all significant correlations were positive; inverse correlations between blood and tissue indicative of cellular trafficking were not found, possibly due to downregulation of trafficking molecules after homing to the tissue and/or the single collection timepoint and location of the samples analyzed here (biopsies represent a small area of the tissue). The limited number of blood and tissue pairs available for correlation analyses after stratifying by disease diagnosis, tissue location, and/or tissue inflammatory state as well as the breadth of parameters tested with stringent adjustment for multiple comparisons could account for the low number of parameters significantly correlated (Fig. 5b); in the future additional samples may reveal more subset-specific correlations.
Upon closer inspection of blood/tissue correlates without correction for multiple testing, median pCREB expression by α4β7+ central memory CD4 T cells was significantly lower in all UC compared with CD or HC blood samples from cohort 1 and higher in inflamed UC tissues compared with inflamed CD tissues in cohort 2, suggesting greater tissue localization and/or activation in UC (Supplementary Fig. 5a). CREB promotes Th17 cell differentiation and inhibits iTreg survival, which is consistent with IBD pathophysiology35,36,37. CCR9+ switched memory B cell frequency was significantly higher in the circulation of UC flare compared with remission in cohort 1 (Supplementary Fig. 5b). While CCR9+ switched memory B cell frequency was not different in blood of CD and UC patients, the frequency of these cells was significantly higher in UC than CD tissue (Supplementary Fig. 5b). The increase of CCR9+ B cells we observed in the blood and tissue of UC flare was surprising, given the association of CCR9 with trafficking to the small intestine under homeostasis. This could reflect CCR9+ B cells shown in mice to co-express CCR10 and traffic to the colon, where the CCR10 ligand CCL28 is highly expressed38,39. The finding might also be related to inflammation since elevated intestinal expression of the CCR9 ligand CCL25 (normally restricted to the small intestine) has been reported and correlated with disease activity in UC patients40, and CCR9 antagonists are being pursued for the treatment of UC41. In summary, we identified specific cellular signatures in the blood as a proxy for intestinal immune phenotypes relevant for disease group stratification.
Tissue-based immune signatures distinguish CD and UC
We took an analogous approach to that used on cohort 1 blood samples for cohort 2 tissue samples to identify trends that distinguished all CD from UC regardless of tissue inflammation (Fig. 6a). After correction for multiple testing of 2145 manually computed parameters, we found 16 leukocyte signatures significantly different between all CD and UC tissue (Table 2a; Supplementary Fig. 5c). Interestingly, activated subsets of NK and NKT cells with higher median pCREB and pSTAT3 were significantly elevated in CD compared with UC tissues, which is consistent with blood trends. When comparing only inflamed CD to inflamed UC tissues, we identified 15 signatures significantly different, including three gut tropic CD45RO+ B cell populations (antigen experienced, previously reported in CD42,43) increased in inflamed CD tissue (Table 2b; Supplementary Fig. 5d). Interestingly, elevated pSTAT3 across cell subsets in CD compared with UC was the most conserved trend when comparing all tissue samples or only inflamed tissue samples, supporting both pro- and anti-inflammatory roles of STAT3 in IBD44,45, although further studies are needed to understand its disease-specific role(s).
Tissue leukocytes distinguish diseases and inflammation states. a Schematic of tissue comparisons. Created with BioRender. b Paired CD tissue from inflamed and uninflamed areas of the same region (N = 4 pairs consisting of samples 239_1 and 239_4, 249_3 and 249_5, 252_1 and 252_3, and 255_1 and 255_3; see Supplementary Table 2). Statistics: ratio paired T test (df = 3; CD45RO+ CD4 T cells, t = 5.48; CD27+CD45RO− CD4 T cells, t = 5.74; CD43+ T cells, t = 3.76; CD25+CD19+CD20+ B cells, t = 3.51). P-values in b were not corrected for multiple testing because hypothesis-driven tests were conducted based on biological insight after independent preliminary analysis. Lines connect paired samples from the same subject. Source data are provided as a Source Data file
Activated leukocytes distinguish tissue inflammation in CD
We compared inflamed to uninflamed tissues from CD patients to explore what factors might contribute to the discontinuous nature of inflamed regions in CD unlike continuous patterns in UC. Increased frequencies of CD45RO+ B cell subsets and decreased frequencies of highly activated T cells, NK cells, and antigen-presenting B cells were associated with inflammation in CD tissues (Table 2c; Supplementary Fig. 5e). Since CD is characterized by heterogeneous, patchy disease, we evaluated four pairs of uninflamed and inflamed tissues from similar locations in CD by hypothesis-driven comparisons. We identified four significant differences between paired inflamed and uninflamed areas of the same anatomical region (i.e., ileum or colon) (Fig. 6b). There were higher frequencies of CD45RO+ memory CD4 T cells in paired inflamed tissues, which suggests chronic antigen-driven responses. Interestingly, there was greater median expression of pSTAT3 in CD27−CD45RO− CD4+ T cells and higher frequencies of activated CD43+ T cells in uninflamed tissues, which could reflect subclinical disease in uninflamed tissues, immune exhaustion and senescence in inflamed regions, and/or an anti-inflammatory effect of these cells. Consistent with the above potential anti- and pro-inflammatory roles of STAT3, we found greater median pSTAT3 expression by regulatory phenotype CD25+ B cells in uninflamed tissues, which may indicate a protective role for these cells in CD. Overall, activated and antigen-driven T and B cell responses as well as pSTAT3 expression and NK cell expansion were associated with tissue inflammation in CD, which is consistent with trends identified in blood. However, we found no differences between inflamed and uninflamed UC tissues, highlighting distinctions between CD and UC tissues. Alternatively, these findings might be due to limited tissue sample sizes and the relatively mild disease of UC patients in cohort 2.
Blood-based signatures classify IBD patients
We utilized differences identified between total CD and UC blood for classification of samples by disease as a potential non-invasive blood-based diagnostic to reduce the required frequency of endoscopy (Fig. 7a). Unbiased approaches for biomarker discovery, such as CITRUS analysis using SAM, PAMR, or LASSO/GLMNET algorithms, did not produce robust results because dividing cohorts into cross-validation folds reduced disease group sample sizes. We constructed generalized linear models (GLMs) using blood signatures we identified throughout the study to classify patients into one of two disease groups (CD versus UC). Using eight blood features significantly different between all CD and UC samples in cohort 1 (Fig. 1c, d), we created a GLM to classify patients as CD or UC (Fig. 7b). Discriminatory performance of the GLM was assessed using receiver-operating characteristic (ROC) analysis, in which the true-positive rate (sensitivity) is plotted versus the false-positive rate (1-specificity). The area under the curve (AUC) reflects the probability that the model will rank a randomly chosen positive (CD) sample higher than a randomly chosen negative (UC) sample. This approach revealed an AUC of 0.845 (95% CI, 0.742–0.948) when using the model with all eight signatures combined to discriminate CD from UC amongst all blood samples in the study, which was higher than any single-signature model (Fig. 7b; Supplementary Table 4). A cutoff of 0.4874743 for the model identified with the Youden index method to maximize sensitivity and specificity (see Methods) yielded a sensitivity of 0.80 and a specificity of 0.85 (PPV = 0.86, NPV = 0.79).
Signatures in the blood classify IBD patients by clinical subsets. a Schematic of the approach for non-invasive classification of CD vs. UC based on blood. Blood was drawn from study subjects, peripheral blood mononuclear cells (PBMCs) were isolated and cryopreserved, and samples were analyzed in batches by CyTOF. Created with BioRender. b Generalized linear models (GLMs) were created for eight parameters significantly different between all CD and UC samples. Corresponding receiver-operating characteristic (ROC) curves are shown for single feature and eight feature models. All CD and UC samples were used to plot ROC curves. UC was used as baseline for the purposes of the GLMs, such that a true-positive indicates correct classification of a CD sample. Statistics: generalized linear models were constructed using glm in R (see the Methods section). Intercepts and parameter coefficients for each model are provided in Supplementary Table 4. Cutoffs and associated performance characteristics are discussed in the text. Source data are provided as a Source Data file
We performed multivariate analysis to determine if subject characteristics confounded the eight parameters utilized for classification of CD and UC. We used analysis of covariance (ANCOVA) to assess the interaction between age, sex, or age and sex with each of the eight parameters (Supplementary Table 5). There were no significant interactions for cohort 1 or 2. Thus, it was unnecessary to adjust the data based on age or sex, and we did not include these in the classification model.
CD and UC patients are treated with different medications, often as combination therapies. Most subjects in this study were on multiple medications with distinct mechanisms of action. We sought to determine whether or not medications confounded the eight parameters utilized for disease classification. There was no standard method to correct for medications as confounding factors in the context of our sample sizes or the numerous combination and single-therapy regimens. We stratified all subjects on versus not on each therapy (Supplementary Fig. 7a) and reevaluated the performance of the same classification model built using all samples. The most common medications were TNF antagonists, glucocorticoids, 5-aminosalicylates (5-ASA), and 6-mercaptopurines (6-MP). Methotrexate was used for a small number of subjects as a therapy for preventing immunogenicity of a biologic therapy. IL-12/23 and α4β7 antagonists were used for three and four subjects, respectively. We used the same GLM presented in Fig. 7b and found that stratification by TNF antagonists, glucocorticoids, 5-ASA, or 6-MP had a negligible effect on model performance (Supplementary Fig. 7b–e). Since discrimination between CD and UC for subjects on each medication was close to that for subjects not on corresponding medications, it was unnecessary to adjust for therapy in the classification model. This is ideal for evaluating patients on diverse treatment regimens encountered in the clinic.
Discussion
Animal models of IBD provide insight into disease pathogenesis and opportunities to study therapeutic leads, yet very few successfully treat the human disease and there remains a dramatic unmet clinical need for long-lasting treatments. Many cytokine based-therapies extrapolated from experimental models have failed in clinical trials46,47,48. Translation of basic research into clinical practice is also hampered by the heterogeneity of human disease. Although the gut is accessible, endoscopy is invasive and has risks. We sought to address these challenges and improve future studies by identifying blood signatures that objectively differentiate disease type, state, and behavior; represent local immune responses; and classify patient subsets. We performed CyTOF with lineage, activation, and trafficking markers. This allowed us to identify cellular signatures and phenotypes associated with IBD subsets, highlighting gut tropic leukocytes. It will also be interesting for other investigators to apply this approach to other tissues with corresponding trafficking molecules.
We utilized a diverse cohort of IBD and HCs and reported findings that relate peripheral and tissue immune responses across distinct disease states. Prior knowledge of leukocyte trafficking in the field is largely from animals; our study demonstrates significance and opportunities for trafficking molecules and gut tropic cells. Decreases in gut tropic cells we observed in UC flare compared with remission and HC might indicate an increase in trafficking or tissue localization, which could explain why UC patients often respond better to the anti-α4β7 vedolizumab49,50. Furthermore, our flow-cytometric validation of CyTOF data demonstrates the potential for others to apply these results.
Our findings are consistent with distinct responses in circulation and tissue as well as CD4+ T cells in CD51. We identified additional cells, many accessible in the blood, that reflected disease group distinctions. Although disease-specific leukocytes reside mostly in affected tissues51, using gut-trafficking molecules we enriched for disease-specific cells in blood and demonstrated their utility in defining disease groups. We uncovered signatures of disease diagnosis (CD vs. UC), state (flare vs. remission), and phenotypes (behavior and location). The prevalence of elevated pSTAT3 in CD suggests pSTAT3 as a marker of CD in subjects studied here. Elevated pSTAT3 in CD tissue Tph cells is consistent with increased B cell subsets34. STAT3 also signals in the IL-23/Th17 pathway, which was identified in IBD GWAS studies and is a therapeutic target3,52. Expression of pSTAT3 in CD25+ B cells was higher in uninflamed than paired inflamed CD tissue of the same region, consistent with the regulatory effects of these cells53. Regulatory B cells are associated with IL-10 and TGF-β, both upstream of pSTAT354,55. Thus, observed pSTAT3 levels might indicate a protective role for CD25+ B cells in CD.
Gut tropic CD45RO+ B cell subsets were elevated in inflamed CD tissue compared with UC. These cells were previously described as a biomarker correlated with CD activity index43 and intestinal permeability42. B cell expression of CD45RO is associated with late-stage differentiation and antigen stimulation43, activation, and IgVH mutation56, suggesting antigen-driven B cell responses in CD tissue. Further elucidating B cell subsets in the pathophysiology of IBD represents a significant advance in the field.
Our analysis has several limitations. First, some subclinical active disease processes may be ongoing in patients in clinical remission. We used IBD specialist-defined flare and remission for patients based on clinical criteria utilized for standard of care with intent to treat at the time of sample collection, although there was a spectrum of quantitative disease activities and phenotypes for each group. Some cohort 2 subjects in clinical remission exhibited histologic features of mild disease activity and might not fit deep remission states. That we identified distinct signatures between CD and UC patients even in remission suggests that while flares of either disease might respond to similar therapies as evidenced by their clinical utility, different maintenance therapies might be necessary for subsets of patients. Second, comparison with tissue samples from healthy subjects is lacking. Third, our findings were guided by our CyTOF panels, and can only suggest cell localization based on knowledge of trafficking largely from mouse studies. Short-term homing assays where labeled cells are transferred are currently not feasible in human.
Consistent with clinical observations, we found greater cellular heterogeneity among CD compared with UC. Blood signatures could be developed as non-invasive, cost-effective, and safe markers for disease diagnosis and monitoring. Although our classification model could be over specified and there exist cellular features not tested here, these limitations do not interfere with its potential utility. Future validation using larger independent cohort(s) will be necessary.
Since the vast majority of patients in academic and/or tertiary centers are not treatment naive, the breadth of drug regimens represented here reflects the reality encountered. Our future studies will address limitations of this work and focus on evaluating disease classification models in larger and more diverse independent cohorts, include HC samples and additional disease comparators, and track individuals longitudinally to examine phenotype stability over time.
Methods
Patient samples
All blood and tissue samples were collected using a study protocol approved by the Stanford Institutional Review Board. Informed consent was obtained from subjects aged 18–75 with an IBD-specializing gastroenterologist-confirmed diagnosis of inflammatory bowel disease (except healthy controls), excluding those who were pregnant, had other autoimmune or inflammatory diseases (except for extraintestinal manifestations of IBD), had a malignancy, had an active infection at the time of enrollment, had undergone surgery within 1 month of enrollment, had a blood transfusion within 1 month of enrollment, had received an organ or bone marrow transplant, or were unable to provide informed consent. All clinical data for subjects were current at the time of sample collection. Flare was determined by IBD-specializing physicians for standard of care and intent to treat using all available information, including bloodwork, clinical assessments, patient history, and/or endoscopy. When available (for most patients), recent endoscopy reports were reviewed to confirm disease state. Disease activity was defined using HBI (CD) and Partial Mayo (UC) scores.
Blood samples were collected by standard of care venipuncture; three vacutainers with sodium heparin (BD, cat. #366480) were filled with blood and kept at room temperature until processing, which occurred within 2 h. Tissue samples were collected during standard of care endoscopic procedures (within 2 h of paired blood sample collection) using biopsy forceps rinsed in sterile saline to remove any residual formalin. Two to four tissue bites per site were collected and deposited in 3 mL of sterile PBS without calcium or magnesium in a 5-mL Eppendorf tube and kept at room temperature if processed within 30 min or kept on ice if processed within 2 h. Biopsies were excluded if they were sampled from sites exposed to methylene blue during endoscopic procedures. Samples from inflamed and uninflamed tissues were initially identified by the endoscopist and subsequently confirmed by a blinded pathologist.
Blood leukocyte isolation and cryopreservation
Blood was centrifuged in vacutainers used for collection at 2000 rpm for 10 min. Plasma was aspirated from the top and frozen at −80 °C in 1-mL aliquots in cryovials (Thermo Fisher Scientific, cat. #375418) using a freeze controller (Bel-Art Products, cat. #F18844-0000) pre-chilled to −4 °C according to the manufacturer’s instructions. The remaining blood was diluted 1:1 in PBS without calcium or magnesium, layered over 15 mL of Ficoll-Paque (GE Healthcare, cat. #17-1440-03) in an Accuspin tube (Sigma-Aldrich, cat. #A2055), and centrifuged at 2000 rpm for 20 min at 21 °C with acceleration at five and break at zero. The buffy coat leukocyte layer was collected and washed twice in 50 mL of PBS without calcium or magnesium by centrifuging at 2000 rpm for 10 min. Cells were counted, washed again, and resuspended in the Recovery Cell Culture Freezing Medium (Thermo Fisher Scientific, cat. #12648010) at 3.5–10 × 106 cells/mL in 1-mL aliquots, transferred to a freeze controller (Bel-Art Products, cat. #F18844-0000) pre-chilled to −4 °C according to the manufacturer’s instructions, stored at −80 °C for 1–7 days, and then transferred to liquid nitrogen for storage.
Tissue leukocyte isolation and cryopreservation
Combined biopsy bite tissue samples from the same site were washed in HBSS without calcium or magnesium supplemented with 2% BSA and then transferred to 5 mL of RPMI with HEPES, 5% BSA, collagenase IV at 0.7 mg/mL (Sigma-Aldrich, cat. #C5138), and DNase I at 50 µg/mL (Worhington Biochemical, cat. #LS002060) at 37 °C on a magnetic stirrer at 400 rpm for 40 min in a small glass jar with a magnetic stirrer. The cell suspension was strained through a 100-µm filter (Falcon, cat. #352360), quenched with 5 mL of RPMI with HEPES and 5% BSA, centrifuged at 560 g for 10 min, and kept on ice. The remaining undigested tissue was again resuspended in 5 mL of RPMI with HEPES, 5% BSA, collagenase IV at 0.7 mg/mL (Sigma-Aldrich, cat. #C5138), and DNase I at 50 µg/mL (Worhington Biochemical, cat. #LS002060) at 37 °C on a magnetic stirrer at 400 rpm for 40 min in a small glass jar with a magnetic stirrer. The material was again strained through a 100-µm filter (Falcon, cat. #352360), quenched with 5 mL of RPMI with HEPES and 5% BSA, and centrifuged at 560 g for 10 min. The combined cell suspensions were then resuspended in 8 mL of 40% Percoll, which was made by preparing a mixture of 10% 10X PBS and 90% Percoll (GE Healthcare, cat. #17-0891-01) and then diluting this in RPMI with HEPES and 5% BSA. The 40% Percoll cell suspension was overlaid on 2 mL of 80% Percoll (prepared in a manner analogous to that previously described for 40%), centrifuged at 560 g for 20 min with acceleration of four and break of one at room temperature. The buffy coat leukocyte layer was collected and washed in 15 mL of RPMI with HEPES and 5% BSA. Cells were counted, washed again, resuspended in 500 µL of the Recovery Cell Culture Freezing Medium (Thermo Fisher Scientific, cat. #12648010) per tissue sample site, transferred to a freeze controller (Bel-Art Products cat. #F18844-0000) pre-chilled to −4 °C according to the manufacturer’s instructions, stored at −80 °C for 1–7 days, and then transferred to liquid nitrogen for storage.
Mass cytometry
Phospho CyTOF was conducted at the Stanford Human Immune Monitoring Center using viably cryopreserved leukocyte samples according to published methods57, unless otherwise noted, in batches of 10–20 samples per day on the same Helios instrument using the same operator. All antibody conjugates were validated for accurate detection of their respective antigens and to ensure minimal isotope spillover by us, the Stanford Human Immune Monitoring Center, and/or in the literature using flow cytometry with antibody clones and mass cytometry with antibody–metal conjugates (Supplementary Table 6). Beads (Fluidigm, cat. #201078) were spiked into each sample for subsequent normalization using the Helios instrument software, and no cell stimulation or barcoding were used.
In brief, cells were thawed, washed twice in 10 mL of complete RPMI with 1:10,000 benzonase (Pierce Antibodies, cat. #88701), and washed again in complete RPMI. Cells were counted and 1 × 106 live cells were used for staining; for tissue samples with less than 1 × 106 live cells recovered, thawed mouse splenocytes (processed as described above for human blood) were spiked in to reach 1 × 106 live cells per sample. Cells were transferred to deep-well plates, washed in RPMI and then incubated for 3 min at room temperature in 100 µL of 1:5000 cisplatin live/dead stain (Fluidigm, cat. #201064) in RPMI. Cells were washed twice with complete RPMI, resuspended in 200 µL of complete RPMI, and rested for 1 h at 37 °C in a CO2 incubator. For surface staining, cells were washed with CyFACS buffer, stained with anti-α4β7 antibody in 25-µL total volume per sample for 20 min at room temperature, washed twice with CyFACS, and fixed with 200 µL of 2% PFA in PBS for 10 min at room temperature. Cells were washed twice with 1 mL of CyFACS per well and centrifuged at 2000 rpm for 8 min at 4 °C. Samples were stained with surface antibody cocktails in a total staining volume of 20 µL per sample for 30 min at room temperature, washed twice with 1 mL of CyFACS buffer per well, and centrifuged at 974 g for 8 min at 4 °C. Cells were fixed again in 100 µL of 4% PFA in PBS for 10 min at room temperature, washed with PBS, permeabilized with 600 µL of −20 °C methanol per sample, and stored overnight at −80 °C. The next day, samples were resuspended in 1 mL of CyFACS buffer and centrifuged at 974 g for 10 min at 4 °C, and then washed again in PBS. Samples were stained with intracellular antibody cocktails in a total staining volume of 20 µL per sample for 30 min at room temperature, washed in 1 mL of CyPBS, and 300 µL of Ir-intercalator (Fluidigm, cat. #201192B) diluted according to the manufacturer’s instructions was added to each sample for 20 min at room temperature. Samples were washed once with PBS, twice with water, spiked with beads according to the manufacturer’s instructions, and then analyzed on a Helios instrument. Approximately 100,000 or all possible events (whichever lower) were acquired for each sample.
Flow cytometry
Flow cytometry was conducted at the Stanford Shared FACS Facility (SSFF) on a BD LSRII instrument in accordance with standard methods. In brief, cells were thawed, washed once in complete RPMI, incubated in 1 mL of complete RPMI with 2.5 mM MgCl2 (Thermo Fisher, cat. #AM9530G) and 0.5 mg/mL DNase I (Worthington Biochemical, cat. #LS002060) for 10 min at room temperature, and washed in 10 mL of complete RPMI. Cells were counted and 1 × 106 or 2 × 106 live cells were aliquoted for staining. Cells were washed with FACS buffer (HBSS without calcium or magnesium and supplemented with 2% BSA). Cells were resuspended in 100 µL of 1:500 Zombie Green fixable viability stain (BioLegend, cat. #423111) in PBS without calcium or magnesium and incubated for 15 min at room temperature in the dark. Cells were washed with FACS buffer, resuspended in a master mix of fluorochrome-conjugated antibodies (Supplementary Fig. 4b) using the supplier-recommended 5 µL of each antibody per 1 × 106 cells, and incubated for 30 min at 4 °C in the dark. Cells were washed with FACS buffer, resuspended in 100 µL of FACS buffer, and kept on ice in the dark until sample analysis. Approximately 1 × 106 or all possible events (whichever lower) were acquired for each sample. For single-color compensation controls, one drop of negative control and one drop of anti-mouse compensation beads (BD, cat. #552843) were incubated in 100 µL of FACS buffer and 5 µL of antibody for 15 min at room temperature in the dark and then kept on ice in the dark until analysis.
Data analysis
Bead normalized sample files were obtained from the Helios instrument using on-board software. FlowJo was used for cleaning up files, concatenating files, and calculating manual gates and statistics. Doublets were carefully gated out in all samples (Supplementary Figs 1, 2). Although several reported cell subpopulations are not canonical populations, they have been reported in the literature or were validated here using flow cytometry (Supplementary Table 6; Supplementary Fig. 4c–f). VorteX58 was used to find the optimal cluster number for use in subsequent analyses. Cytobank was used to perform viSNE, CITRUS, and Spade analyses. viSNE analyses were run on live human single cells concatenated from individual samples by group. In total, 5000 events were randomly subsampled from each concatenated file, and clustering was run on all concatenated files in parallel using a random seed, 1000 iterations, perplexity of 30, and theta of 0.5. GraphPad PRISM 7 was used to plot some figures and conduct some targeted statistical tests, and Microsoft Excel was used for some basic data maneuvering. Morpheus was utilized for constructing heatmaps and Pearson correlation maps based on significant parameters selected using T-tests with 10,000 permutations and Benjamini and Hochberg (BH) FDR correction for multiple testing (documentation available at: https://software.broadinstitute.org/morpheus/documentation.html). R was used for additional statistical analyses (see below). There were three patients in cohort 1 and one patient in cohort 2 excluded from analyses involving trafficking receptors because they were treated with an α4β7 antagonist.
Statistics
All p-values were derived from two-tailed unpaired T tests without adjustment for multiple testing unless otherwise noted. Most p-values were corrected for multiple testing as indicated using the BH FDR correction method in Morpheus (see above). When correcting for multiple testing manually, the p.adjust function with the BH correction method (also known as FDR) was used in R according to its documentation to account for the expected proportion of false discoveries among the rejected hypotheses. Correlation tests were based on Pearson correlations using cor.test in R according to its documentation. For paired blood and tissue correlations, each biopsy location was paired with blood from the same individual, and Pearson correlations were calculated for each pair. The mean value for tissue was used if there was more than one tissue sample from a given location. P-values varied with the number of samples in each subgroup being compared, as p.adjust incorporates the number of comparisons being made. Correlated parameters were eliminated if driven by an outlier or less than three data points. For receiver-operating characteristic (ROC) analysis, generalized linear models (GLM) were constructed in R using the glm package according to its documentation using features significantly different between disease groups. GLMs were calculated for each individual parameter and then compared with a model including all significant parameters. For each ROC curve, the area under the curve and 95% confidence intervals were calculated using the ROCR package in R according to its documentation. Optimal cutoff values and associated sensitivity and specificity values for GLMs were calculated using the OptimalCutpoints package in R with the Youden index method according to its documentation. Analysis of covariance (ANCOVA) was performed using the aov package in R according to its documentation in order to test for significant correlation between each cellular parameter and age, sex (female = 1; male = 0), or age and sex.
Gene expression deconvolution
Gene expression deconvolution analyses were conducted according to published methods23 using the immunoStates basis matrix for major cell lineages. Only major cell lineages were identifiable using this approach due to the availability of sorted cell gene expression data used in the basis matrix. In brief, we measured the mean expression of each gene in the 312-gene matrix for major cell lineages and performed deconvolution with support vector regression using the CIBERSORT algorithm59. We then compared the resulting estimated cell-type frequencies between classes and subsequently calculated the effect sizes. We downloaded all data sets from Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo/) using the MetaIntegrator package from CRAN60.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data sets generated during the current study are available from the corresponding author on reasonable request that does not include confidential patient information. The source data underlying Figs 1b–d, 2a–d, 3a–g, 4c, d, 5b, 6b, 7b, and Table 2a–c, as well as Supplementary Figs 3a, b, 4e, f, 5a–e, 6a–c, 7b–e and 8a, b are provided as a Source Data file.
Code availability
The computer code generated and used during this study is available from the corresponding author on reasonable request that does not include confidential patient information. Web-based tools utilized in this study are freely accessible for the foreseeable future (https://software.broadinstitute.org/morpheus/).
References
Molodecky, N. A. et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142, 46–54 (2012).
Abraham, C. & Cho, J. H. Inflammatory bowel disease. N. Engl. J. Med 361, 2066–2078 (2009).
Bilsborough, J., Targan, S. R. & Snapper, S. B. Therapeutic Targets in Inflammatory Bowel Disease: Current and Future. Am. J. Gastroenterol. Suppl. 3, 27–37 (2016).
Ananthakrishnan, A. N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 12, 205–217 (2015).
Marsal, J. & Agace, W. W. Targeting T-cell migration in inflammatory bowel disease. J. Intern Med 272, 411–429 (2012).
Gasche, C. et al. A simple classification of Crohn’s disease: report of the Working Party for the World Congresses of Gastroenterology, Vienna 1998. Inflamm. Bowel Dis. 6, 8–15 (2000).
Safar, B. & Sands, D. Perianal Crohn’s disease. Clin. Colon Rectal Surg. 20, 282–293 (2007).
Silverberg, M. S. et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can. J. Gastroenterol. 19(Suppl A), 5A–36A (2005).
Satsangi, J., Silverberg, M. S., Vermeire, S. & Colombel, J. F. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 55, 749–753 (2006).
Yantiss, R. K. & Odze, R. D. Diagnostic difficulties in inflammatory bowel disease pathology. Histopathology 48, 116–132 (2006).
McGovern, V. J. & Goulston, S. J. Crohn’s disease of the colon. Gut 9, 164–176 (1968).
Islam, S. A. & Luster, A. D. T cell homing to epithelial barriers in allergic disease. Nat. Med 18, 705–715 (2012).
Habtezion, A., Nguyen, L. P., Hadeiba, H. & Butcher, E. C. Leukocyte Trafficking to the Small Intestine and Colon. Gastroenterology 150, 340–354 (2016).
Bendall, S. C., Nolan, G. P., Roederer, M. & Chattopadhyay, P. K. A deep profiler’s guide to cytometry. Trends Immunol. 33, 323–332 (2012).
Dobre, M. et al. Mucosal CCR1 gene expression as a marker of molecular activity in Crohn’s disease: preliminary data. Rom. J. Morphol. Embryol. 58, 1263–1268 (2017).
McGovern, D. P., Kugathasan, S. & Cho, J. H. Genetics of inflammatory bowel diseases. Gastroenterology 149, 1163–1176 e1162 (2015).
Kim, S. V. et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 340, 1456–1459 (2013).
Nguyen, L. P. et al. Role and species-specific expression of colon T cell homing receptor GPR15 in colitis. Nat. Immunol. 16, 207–213 (2015).
Morrison, R. W. & De Jong, K. A. Measurement of population diversity. Artif. Evol. 2310, 31–41 (2002).
Muroi, K. et al. CD56 expression in B-cell lymphoma. Leuk. Res 22, 201–202 (1998).
Van Acker, H. H., Capsomidis, A., Smits, E. L. & Van Tendeloo, V. F. CD56 in the immune system: more than a marker for cytotoxicity? Front Immunol. 8, 892 (2017).
Brand, S. Crohn’s disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn’s disease. Gut 58, 1152–1167 (2009).
Vallania, F. et al. Leveraging heterogeneity across multiple data sets increases accuracy of cell-mixture deconvolution and reduces biological and technical biases. Nat. Commun. 9, 4735 (2018).
Qiao, Y. Q. et al. LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn’s disease. J. Biomed. Sci. 20, 87 (2013).
Wu, Y. C., Kipling, D. & Dunn-Walters, D. K. The relationship between CD27 negative and positive B cell populations in human peripheral blood. Front Immunol. 2, 81 (2011).
Fecteau, J. F., Cote, G. & Neron, S. A new memory CD27-IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. J. Immunol. 177, 3728–3736 (2006).
Fleischer, S. J. et al. Increased frequency of a unique spleen tyrosine kinase bright memory B cell population in systemic lupus erythematosus. Arthritis Rheuma. 66, 3424–3435 (2014).
Wei, C. et al. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J. Immunol. 178, 6624–6633 (2007).
Moir, S. et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med 205, 1797–1805 (2008).
Rojas, O. L., Narvaez, C. F., Greenberg, H. B., Angel, J. & Franco, M. A. Characterization of rotavirus specific B cells and their relation with serological memory. Virology 380, 234–242 (2008).
Mills, S. & Stamos, M. J. Colonic Crohn’s disease. Clin. Colon Rectal Surg. 20, 309–313 (2007).
Frizelle, F. A. & Burt, M. J. Review: the surgical management of ulcerative colitis. J. Gastroenterol. Hepatol. 12, 670–677 (1997).
Hedrick, T. L. & Friel, C. M. Colonic crohn disease. Clin. Colon Rectal Surg. 26, 84–89 (2013).
Rao, D. A. et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017).
Wang, X. et al. Cyclic AMP-responsive element-binding protein (CREB) is critical in autoimmunity by promoting Th17 but inhibiting Treg cell differentiation. EBioMedicine 25, 165–174 (2017).
Egwuagu, C. E. STAT3 in CD4+ T helper cell differentiation and inflammatory diseases. Cytokine 47, 149–156 (2009).
Galvez, J. Role of Th17 cells in the pathogenesis of human IBD. ISRN Inflamm. 2014, 928461 (2014).
Jaimes, M. C. et al. Maturation and trafficking markers on rotavirus-specific B cells during acute infection and convalescence in children. J. Virol. 78, 10967–10976 (2004).
Wang, W. et al. Identification of a novel chemokine (CCL28), which binds CCR10 (GPR2). J. Biol. Chem. 275, 22313–22323 (2000).
Trivedi, P. J. et al. Intestinal CCL25 expression is increased in colitis and correlates with inflammatory activity. J. Autoimmun. 68, 98–104 (2016).
Bekker, P. et al. CCR9 antagonists in the treatment of ulcerative colitis. Mediat. Inflamm. 2015, 628340 (2015).
Yacyshyn, B. R. & Meddings, J. B. CD45RO expression on circulating CD19+ B cells in Crohn’s disease correlates with intestinal permeability. Gastroenterology 108, 132–137 (1995).
Yacyshyn, B. R. & Pilarski, L. M. Expression of CD45RO on circulating CD19+ B-cells in Crohn’s disease. Gut 34, 1698–1704 (1993).
Nguyen, P. M., Putoczki, T. L. & Ernst, M. STAT3-activating cytokines: a therapeutic opportunity for inflammatory bowel disease? J. Interferon Cytokine Res 35, 340–350 (2015).
Pickert, G. et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med 206, 1465–1472 (2009).
Reinisch, W. et al. Anrukinzumab, an anti-interleukin 13 monoclonal antibody, in active UC: efficacy and safety from a phase IIa randomised multicentre study. Gut 64, 894–900 (2015).
Danese, S. et al. Tralokinumab for moderate-to-severe UC: a randomised, double-blind, placebo-controlled, phase IIa study. Gut 64, 243–249 (2015).
Hueber, W. et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700 (2012).
Feagan, B. G. et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med 369, 699–710 (2013).
Sandborn, W. J. et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N. Engl. J. Med 369, 711–721 (2013).
van Unen, V. et al. Mass cytometry of the human mucosal immune system identifies tissue- and disease-associated immune subsets. Immunity 44, 1227–1239 (2016).
Lees, C. W., Barrett, J. C., Parkes, M. & Satsangi, J. New IBD genetics: common pathways with other diseases. Gut 60, 1739–1753 (2011).
Kessel, A. et al. Human CD19(+)CD25(high) B regulatory cells suppress proliferation of CD4(+) T cells and enhance Foxp3 and CTLA-4 expression in T-regulatory cells. Autoimmun. Rev. 11, 670–677 (2012).
Vadasz, Z., Haj, T., Kessel, A. & Toubi, E. B-regulatory cells in autoimmunity and immune mediated inflammation. FEBS Lett. 587, 2074–2078 (2013).
Bing, X., Linlang, L. & Keyan, C. Decreased Breg/Th17 Ratio Improved the Prognosis of Patients with Ulcerative Colitis. Can. J. Gastroenterol. Hepatol. 2018, 5760849 (2018).
Jackson, S. M. et al. CD45RO enriches for activated, highly mutated human germinal center B cells. Blood 110, 3917–3925 (2007).
Fernandez, R. & Maecker, H. Cytokine-stimulated phosphoflow of PBMC Using CyTOF mass cytometry. Bio Protoc 5, e1496 (2015).
Samusik, N., Good, Z., Spitzer, M. H., Davis, K. L. & Nolan, G. P. Automated mapping of phenotype space with single-cell data. Nat. Methods 13, 493–496 (2016).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Haynes, W. A. et al. Empowering multi-cohort gene expression analysis to increase reproducibility. Pac. Symp. Biocomput 22, 144–153 (2017).
Acknowledgements
We thank Y. Yang, Y. Wei, A. Fan, and A.R. Ji for technical assistance, S. Toppo and M. Adelus for clinical assistance with patient recruitment, N. Sigal and H. Maecker for their mass cytometry expertise, A. McMillan for expert biostatistics consultation and guidance, L. Stell for general statistical advice, and the patients and healthcare providers without whom this study would not be possible. We also thank the Stanford Human Immune Monitoring Center and the National Institutes of Health S10 Shared Instrumentation Grant (S10RR027431-01) for access to mass cytometers and expert technical services. This work was supported by the National Institutes of Health (R01 DK101119) and the Ann and Bill Swindells Charitable Trust as well as Leslie and Douglas Ballinger. S.J.S. Rubin thanks W.D. Rubin for invaluable discussions and inspiration, W.H. Robinson for mentorship and guidance, as well as the National Science Foundation Graduate Research Fellowship and the Stanford Graduate Fellowship for their generous support. This material is based in part upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE—1656518. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Anti-human α4β7 integrin (Act-1) (cat#11718) from Dr. A.A. Ansari.
Author information
Authors and Affiliations
Contributions
S.J.S.R. and A.H. conceived and designed the study, performed data analysis, and wrote the paper; S.J.S.R. designed and performed all experiments (some in conjunction with the Stanford Human Immune Monitoring Center) and analyzed all data, unless otherwise indicated; L. Bai performed some data analysis for correlation and classification; Y.E.H. designed and performed some experiments with paired blood and tissue samples; G.G. coordinated procurement of clinical samples and data retrieval; C.Y. designed some pilot studies; L. Becker, S.E.S., S.R.S. and A.H. provided clinical data and patient samples.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature communications would like to thank anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rubin, S.J.S., Bai, L., Haileselassie, Y. et al. Mass cytometry reveals systemic and local immune signatures that distinguish inflammatory bowel diseases. Nat Commun 10, 2686 (2019). https://doi.org/10.1038/s41467-019-10387-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-019-10387-7
This article is cited by
-
Insights gained from single-cell analysis of chimeric antigen receptor T-cell immunotherapy in cancer
Military Medical Research (2023)
-
Selective oxidative protection leads to tissue topological changes orchestrated by macrophage during ulcerative colitis
Nature Communications (2023)
-
ADT-OH improves intestinal barrier function and remodels the gut microbiota in DSS-induced colitis
Frontiers of Medicine (2023)
-
Mass cytometry reveals immune atlas of urothelial carcinoma
BMC Cancer (2022)
-
Protective Effect of Saffron in Mouse Colitis Models Through Immune Modulation
Digestive Diseases and Sciences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.