Abstract
Maize was domesticated from lowland teosinte (Zea mays ssp. parviglumis), but the contribution of highland teosinte (Zea mays ssp. mexicana, hereafter mexicana) to modern maize is not clear. Here, two genomes for Mo17 (a modern maize inbred) and mexicana are assembled using a meta-assembly strategy after sequencing of 10 lines derived from a maize-teosinte cross. Comparative analyses reveal a high level of diversity between Mo17, B73, and mexicana, including three Mb-size structural rearrangements. The maize spontaneous mutation rate is estimated to be 2.17 × 10−8 ~3.87 × 10−8 per site per generation with a nonrandom distribution across the genome. A higher deleterious mutation rate is observed in the pericentromeric regions, and might be caused by differences in recombination frequency. Over 10% of the maize genome shows evidence of introgression from the mexicana genome, suggesting that mexicana contributed to maize adaptation and improvement. Our data offer a rich resource for constructing the pan-genome of Zea mays and genetic improvement of modern maize varieties.
Similar content being viewed by others
Introduction
Maize (Zea mays ssp. mays) is one of the most important crops globally, with an annual production of >1 billion tons1, and has been a model for biological studies for over a century. It is also an incredibly diverse species as up to half of the genome is not shared between any two maize varieties2, 3. Structural variations (SVs) including copy number variations (CNVs), presence/absence variations (PAVs), inversions, and translocations contribute to the diversity of the maize genome4, and also play an important role in phenotypic variation5, 6. Multiple reference genomes are, therefore, required in order to represent all of the genomic content for maize and to understand its domestication, adaptation, and improvement. On the basis of available archaeological and molecular evidence, it is believed that maize domestication started approximately 9000 years ago in Southwest Mexico, from the lowland teosinte Zea mays ssp. parviglumis (hereafter, parviglumis)7, 8. Zea mays ssp. mexicana (hereafter, mexicana) is distributed across the cooler higher elevations of the Mexican Central Plateau, and can easily cross with maize9, such that gene flow from mexicana has contributed to maize local adaptation and improvement8, 10. However, this process has not been investigated on a genome-wide basis, because no mexicana genome was available up to now10, 11.
In this study, a meta-assembly strategy combining a new genetic design (Fig. 1a, b), based on selections from an interspecific cross between Mo17 and mexicana was used to assemble the contigs and scaffolds of the parental maize mexicana genomes, and the scaffolds were anchored with genetic map and publicly available genotyping-by-sequencing data6. This design allowed us to estimate the maize and teosinte spontaneous point mutation rate in each generation based on the identity by descent (IBD) regions of ten sequenced individuals. We compared large structural variations among Zea genomes, and hundreds of genes under positive selection were identified. In addition, the introgression of mexicana to maize was investigated. All the above analysis provides a resource for understanding maize adaptation history and the value of its wild relatives.
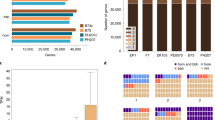
Mo17-mexicana population development and characterization of the assembled genomes. a The construction of Mo17-mexicana BC2F7 population. Only a single F1 seed was used. b The genetic design for meta-assembly of the Mo17 and mexicana genomes. Ten well-chosen TM lines that genetically divided the Mo17 and mexicana genomes into different fragments. The fragment indicated by the dash lines are used to estimate the mutation rate in mexicana. c Schematic illustration of the meta-assembly pipeline. The Illumina reads were filtered and adapters were removed for each library and the assembly can be divided into three major steps: Step 1, three strategies were used to assemble contigs, including de novo assembly of 10 individuals, reference-based assembly based on B73 genome and de novo assembly of unmapped reads. Step 2, the contigs obtained from the three strategies were merged, and further connected or extended with Pacbio long reads. Long mate-pair libraries were then added to assemble scaffolds. Step 3, the NRGene scaffolds were integrated, and the merged scaffolds were the final scaffolds. The number represents contig length before diagonal and scaffold length after diagonal
Results
Genome assembly via a new genetic design
A selfed backcross (BC2F7) population (hereafter, TM population) was derived from a single F1 cross between Mo17 and mexicana, and consisted of 191 individuals, which were genotyped with the Illumina 56 K MaizeSNP50 array12 (Fig. 1a). Ten selected individuals covering the whole Mo17 genome and ~ 96% of the mexicana genome were sequenced, and the Mo17 and mexicana genomes were represented by 211 and 176 distinct chromosomal segments, respectively (Fig. 1b and Supplementary Data 1, Supplementary Data 2). A meta-assembly strategy (Fig. 1c) integrating contigs from Illumina reads (1.17 TB, Supplementary Table 1), PacBio long reads (9.29 GB, Supplementary Table 2), and DenovoMAGIC 2TM (445.83 GB, Supplementary Table 3, NRGene, Israel) was adopted to separately assemble Mo17 and mexicana genomes (Supplementary Note 1 and Supplementary Fig. 1). The two final assembled genomes were 2.04 Gb for Mo17 and 1.20 Gb for mexicana, with scaffold N50s of 3 Mb and 107 kb, respectively (Table 1). We anchored and ordered 1973 Mb (96.6%) and 1072 Mb (88.8%) of the Mo17 and mexicana scaffolds with the genetic map of the TM population, and an additional 36.4 Mb (1.8%) of the Mo17 genome and 85.5 Mb (7.1%) of the mexicana genome were anchored with the publicly available genotyping-by-sequencing anchors from 14,129 maize inbred lines6 (Supplementary Note 2).
Curated repeat libraries and de novo prediction were used to annotate the transposable elements (TEs) that comprised 79.7% of Mo17 and 72.8% of the mexicana genome (Table 1 and Supplementary Table 4). Long terminal repeat (LTR) elements were the major class of TEs in Mo17 and mexicana, similar to the B73 reference genome (Supplementary Fig. 2a, b). A total of 40,003 and 31,387 high-confidence protein-coding gene models were predicted for Mo17 and mexicana, by combining ab initio prediction and evidence-based approaches (protein, EST, and RNA-seq of three tissues) (Supplementary Table 5 and Supplementary Note 3). Comparative analysis with B73, rice, and sorghum revealed that a core set of 10,878 gene families were shared among all five genomes (Supplementary Fig. 3a). Almost 2,000 conserved protein families were identified among B73, Mo17, and mexicana genomes, similar to the comparison of B73 and other grass species (i.e., rice and sorghum)4, 13, 14.
The quality of the assembled genomes was confirmed using the following approaches: (1) 93% of the Plantae BUSCO15 data set could be aligned to the assembled Mo17 genome, which was similar to the B73 reference genome4. For mexicana, 86% of the BUSCO gene set was successfully aligned; (2) 96.0% (88.7%) and 90.7% (83.1%) of the partial (complete) core eukaryotic genes mapping approach (CEGMA) gene set from six model organisms16 could be mapped to the assembled Mo17 and mexicana genomes, with coverage > 70%; (3) the highly conserved core gene families (coreGFs) weighted score17 for Mo17 and mexicana was 94.0% and 87.6%, respectively (Supplementary Table 6); (4) 98.2% and 81.5% of the genes from B73 reference genome4 (RefGen_v2 release 5b) could be mapped to Mo17 and mexicana genomes, respectively, with coverage > 80%; (5) two Mo17 bacterial artificial chromosome (BAC) clones that had previously been sequenced for Vgt1 (Accession: EF659467) and ZmWAK cloning5, 18 showed alignments of 99.9% and 99.8% with our Mo17 assembled genome (Supplementary Fig. 3b, c), and one Mo17 scaffold sequence19 also aligned to our Mo17 genome with an identity of 99.9% (Supplementary Fig. 3d); and (6) four Mo17 BACs were randomly selected from a newly constructed BAC library and sequenced on PacBio RSII platform. All the four BACs showed high consistence with our assembly (Supplementary Fig. 3e–h). In summary, these results support the conclusion that the assembled quality of Mo17 is acceptable. Because the sequencing coverage of mexicana is much lower than Mo17, the assembled genome length of mexicana is not as complete as Mo17, only half of mexicana genome was assembled, and most of the un-assembled regions were repetitive sequences. However, the protein-coding regions in Mo17 and mexicana genomes are both well assembled (Supplementary Table 6).
Zea genomes contain large structural variations
Structural variations are widespread in maize genomes and are important for local adaptation and genetic improvement20, 21. Considering that the mexicana genome is incomplete, we only compared the presence/absence variations (PAVs, ≥ 100 bp) between B73 and Mo17 by using comparatively genomic and read-mapping methods (Methods section). Consequently, 220,860 (~88.8 Mb) PAVs between B73 and Mo17 were identified (Supplementary Fig. 4a, b and Supplementary Table 7). These PAVs were unevenly distributed across the genome (Fig. 2). The rate of TE-related PAVs (PAVs containing ≥ 80% TE) is ~ 34.6%, more than in other species22, 23, indicating that they are closely related to transposition events. Full-length LTR retrotransposons in PAV regions were also identified (Supplementary Table 7), indicating recent transposition events. A total of 1,293 PAV genes between B73 and Mo17, were identified, and these PAV genes tended to be shorter than other annotated genes and the underlying mechanism is still unclear (Supplementary Fig. 4c).
Comparative analysis of Mo17 and mexicana genomes. Tracks from outer to inner circles indicate: SNPs, PAV density (window size of 1 Mb, the outer and inner layers indicate Mo17 (Blue) and mexicana (Orange)respectively), and a heatmap of introgression, indicating number of lines containing the corresponding introgression region (low = blue; high = red), hotspots are labeled with red arrows
Three PAVs larger than 1 Mb were identified between Mo17 and B73. PAVs in gene regions are increasingly considered to be one of the origins of phenotypic variations across/within species4, 24,25,26,27. For example, PAVs in maize are causal variants for disease resistance21, 28, including a large one on chromosome 6 between Mo17 and B73, which is putatively responsible for variation in sugarcane mosaic virus resistance28 (Supplementary Fig. 5a). In the present study, we validated this PAV and found that the deletion in Mo17 was ~ 2.9 Mb, ~ 300 kb larger than previous estimates21 (Supplementary Table 8 and Supplementary Fig. 5b). In addition, two new large insertions ( > 1 Mb) were identified in Mo17 genome compared with B73. One ~2.2 Mb insertion, containing 10 annotated genes, was located at 69.7–71.9 Mb of Mo17 chromosome 6 (Supplementary Table 9). Consequently, this region could be PCR amplified from Mo17, but not from B73 (Supplementary Table 8 and Supplementary Fig. 5a–c). Another ~1.4 Mb insertion was located at 143.6–145.1 Mb of Mo17 chromosome 4, and contained 8 annotated genes (Supplementary Table 9) that were also validated by PCR amplification (Supplementary Table 8 and Supplementary Fig. 5d, e). Six of the 18 annotated genes within these two large insertions were expressed in one or more tissues (Supplementary Table 9); however, the phenotypic effects of these two large PAVs remain to be established.
An inversion (Inv9d) showing no crossovers within a ~30 Mb region on the short arm of chromosome 9 was identified between modern maize (B73/Mo17) and mexicana, and was confirmed by the TM genetic map and linkage disequilibrium (LD) analysis based on 95 mexicana accessions genotyped with the Illumina MaizeSNP50 array12 (Fig. 3a, b). This inversion was found only in some mexicana accessions and not in parviglumis in a previous study20. The Inv9d homologous region of B73 (~27 Mb) was shorter than that of mexicana (~38 Mb); therefore, a distinct arc in the synteny dot-plot was observed within the inversion region, and the gradient of gene density increase towards chromosome ends was disrupted in the B73 homologous region (Fig. 3c). This phenomenon may be caused by changes in rates of DNA loss and gain in regions that switch from chromosome ends to the near pericentromeric contexts29. We further compared the gene co-linearity between rice and mexicana/B73 using MCScan30, 31 and found that the Inv9d region showed a well co-linearity between mexicana and rice in the synteny block, whereas there was an inversion between B73 and rice (Fig. 3d, e). Rice has largely preserved the ancestral karyotype of the grass common ancestor, with no major changes in genome structure after it separated from other clades, and has been least affected by transposon activities and accumulation30. It implied that the mexicana genome was closer to the ancestor state and the inversion in modern maize genome was derived from selection. To explore the biological impact of Inv9d, we found a large ear leaf width QTL within the Inv9d region (Fig. 3b, f), which was consistently identified in six environments in our TM population. The inversion causes a wider ear leaf width that can increase the leaf area, thus increasing the photosynthesis. Furthermore, Inv9d showed altitudinal clines in environmental association analysis in a previous study20, emphasizing its potential role in local adaptation to ecological factors. However, further efforts are needed to identify the underlying genes and possible application value in maize breeding.
The relationship between Inv9d and phenotype. a LD reveals the large inversion in Zea mays. mexicana based on 56 K SNPs in 95 mexicana accessions. LD (r2 ≥ 0.1, red; r2 <0.1, pink; P < 0.1, black; P ≥ 0.1, gray) for pairs of SNPs are shown across chromosome 9. Dashed black lines delineate the likely boundaries of structural variants discussed in the text. b QTLs for ear-leaf width located in Inv9d have been detected in 6 environments. The dashed line indicates the cutoff of LOD value (LOD ≥ 2.5). c The dot-plot of gene co-linearity between B73 and mexicana on chromosome 9. The histogram indicated the gene density in a 1 Mb window. The green block covered the gene density in the Inv9d region. d The gene co-linearity between chromosome 9 of B73 and chromosome 6 of rice. e The gene co-linearity between chromosome 9 of mexicana and chromosome 6 of rice. f The phenotype of ear-leaf width of the 10 sequenced lines. The phenotype of mexicana haplotype with Inv9d (orange) and Mo17 haplotypes without Inv9d (green) are shown. The length of scale bar is 10 cm
Mexicana genome contributes to maize adaptation
It is interesting to ask how maize domesticated from lowland parviglumis and adapted to highland environments. This may have been facilitated by gene flow (introgression) from highland mexicana 8. Combining our new genomic resource and the hapmap332 of maize, the introgression region in each individual varied from 0.005% to 0.724% as determined by investigation of 895 inbreds, and this is likely to be an underestimate (Supplementary Fig. 6b), since marker density was low (Fig. 2) (Methods section). By summing the unique introgression regions from all investigated individuals, we found that as much as 10.7% of the maize genome showed evidence of introgression from mexicana. However, based on a comparison of the current genome data (B73, Mo17 and mexicana), the B73 and Mo17 genome only contained ~1.2% (~24.5 Mb) and ~0.34% (~7.1 Mb) of putative introgression regions from mexicana, respectively (Fig. 2) (Methods section). These results could provide a better resolution as compared with a previous estimation10 (Supplementary Data 3). Several introgression hot spots were identified, and one of them located on chromosome 8 and is shared among 312 lines (Fig. 2). However, nearly half of the introgressed regions are specific to a given inbred (Supplementary Fig. 6c). Leaf macrohairs and pigment intensity are regarded as adaptive traits under highland conditions, and QTLs for those traits have been identified using a segregating parviglumis-mexicana population33. At least three of these adaptation traits QTLs overlap with introgression regions were identified in the present study (Supplementary Data 4). The highland Mexican maize have more sympatric populations with mexicana, and their expected introgression proportion should be higher than that in lowland maize. A new genotype by sequencing (GBS) data set of 4,022 landraces34 in which 2900 landraces had their elevation information were further used to assess the introgression proportion in highland Mexican maize. We found that maize from highland indeed have significantly higher introgression proportion than that in lowland Mexico (Supplementary Fig. 7a, P = 0.002). Furthermore, we found no matter where the maize came from (Mexico, Honduras, Guatemala, Panama Costa Rica, etc), the introgression proportion was significantly correlated (P = 9.703e-06) with the elevation (n = 482) (Supplementary Fig. 7b). This result indicated that some introgression regions should associate with highland adaptation, and had been kept when maize expand to other places.
Patterns of spontaneous mutations in the maize genome
Mutation is the foundation of evolution and the origin of phenotypic variation in populations35. The design of this study allowed a direct estimate of the spontaneous mutation rate on a genomic scale, using the Mo17 segments shared among the 10 selected TM individuals with 10 continuous generation selection inherited from a single macrospore (Fig. 1a, b). In total, 7960 spontaneous point mutations were identified and the mutation rate was estimated to be 3.87 × 10−8 per site per generation, using strict criteria of excluding all possible regions of residual heterozygosity (Supplementary Fig. 8a, b and Methods section). For further excluding the influences of remaining heterozygosity and/or different Mo17 individual being used for developing TM population, a large mexicana fragment on chromosome 7 (38,378,754–105,766,898 bp) covered by 5 individuals were selected to estimate the mutation rate in mexicana (Fig. 1b). Using the same filtering criterions, we estimated the mutation rate of mexicana was 2.17 × 10−8 (73 point mutations in ~ 67 Mb region). Therefore, we estimated the spontaneous mutation rate in the maize genome is 2.17~ 3.87 × 10−8, which is slightly higher than the previous indirect estimation using IBD regions in modern maize breeding (1.63 × 10−8)36, and is also higher than the mutation rate in Arabidopsis thaliana (5.9~ 7.1 × 10−9)37.
The global landscape of mutations demonstrated a remarkably nonrandom distribution in the genome (P = 1.2 × 10−20, KS test, Fig. 4a), a phenomenon also observed in viruses, bacteria, and many eukaryotes, including yeast, humans, and Arabidopsis 38, 39. At the chromosome level, the mutation rate had weak correlation with chromosome length (P = 0.02, r = 0.86), and chromosome 2 seemed to have a higher mutation rate than others (Supplementary Table 10). The mutation rate was about 3.5 fold higher in genic regions (8.84 × 10−8, including upstream regions (−5 kb), downstream regions ( + 5 kb), UTRs, exons, and introns) than in the intergenic regions (2.50 × 10−8) (Fig. 4c and Supplementary Table 11). A nonrandom distribution was also observed within the genic regions, with the highest mutation rate in the gene bodies, and the upstream regions had a higher mutation rate than the downstream regions40 (Fig. 4b and Supplementary Fig. 8c). A lower mutation rate was observed in the hypermethylated centromere regions41, 42, opposite to the findings in Arabidopsis 37. Hypermethylation may affect the mutation rate, but may be masked by other effects, such as recombination, especially in the chromosome arms where recombination is promoted. In coding regions, amino acid substitutions are expected to have a large impact on the biological function of the protein. In total, 279 new mutations were found in protein-coding regions (nonsynonymous/synonymous = 151/128), including 37 predicted deleterious mutations43. The mutation rate of 3.18 × 10−8 (19 mutations) in the protein-coding regions of pericentromeric genes was lower than in the chromosome arms (4.55 × 10−8, 260 mutations). However, the ratio of nonsynonymous to synonymous changes was higher in the pericentromeric regions (11/8≈1.38) than that in the chromosome arms (140/120≈1.17), and the deleterious mutation rate in the pericentromeric regions was about two-fold higher (5/19≈0.26 vs. 32/260≈0.12) (Fig. 4c). Deleterious mutations may be more common in the pericentromeres due to the lower recombination rate, which reduces their possibility of elimination44, 45. A context-dependent effect44, 46, namely the dependence of the substitution rate at a site on two flanking nucleotides, was also observed (Fig. 4d). The triplets “ASA” and “CSG” (S = “C” or “G”) were more common (1.7-fold more than the mean of other triplets), and may be explained by context-dependent DNA replication errors, cytosine deamination, or biased gene conversion46. The fact that CG is the most common type of methylation may also explain part of this phenomenon41. Transitions were 2.34 times more frequent than transversions, and GC > AT transition was the most frequent (Fig. 4e). However, the rate of transitions/transversions in genic regions was 2.25, lower than the rate at the whole genome level (Supplementary Table 11). We also observed a higher than expected occurrence of very-closely clustered mutations (P = 2.3 × 10−16), which is a common phenomenon in many organisms44, 46. Multiple mechanisms, including compound mutation and adaptive natural selection, have been proposed to explain this phenomenon, but none have been directly confirmed38.
The characteristics of spontaneous mutations. a The distribution of mutations was not random. The distances between adjacent mutations (observed, orange) against a random distribution of mutation across the genome (expected, gray). b The proportion of deleterious mutations in centromeres (the inner circle) and chromosome arms (the outer circle) are shown in orange. c The distribution of mutations by functional classification. The relative positions of mutations in gene regions were normalized referring to an average length (4 kb) based on B73 gene models4. d Variation in mutation rate of different triplets. e The mutation number of transitions and transversions
Discussion
The sequencing and assembly of large, highly repetitive, and heterozygous genomes rich in copy number variants (CNVs) is extremely challenging. In this study, we de novo assembled one maize and one mexicana genome based on a new genetic design (Fig. 1) that eliminates assembly problems associated with the heterozygosity of the mexicana parent. This design divided the two genomes into 211 and 176 unique chromosomal segments originating from selected individuals of a maize Mo17 × mexicana segregating population, derived from a single F1 individual. The meta-assembly approach included high-coverage Illumina, DenovoMAGIC 2TM sequencing, low-coverage PacBio sequencing, and a high-density linkage map of the TM population. The contribution of each method is shown (Fig. 1c). However, the genetic design of the BC2F7 population made the sequencing depth of the Mo17 genome about 7 times higher, which resulted in lower coverage of the mexicana genome. Double haploid or other balanced populations could be used in the future to overcome this. The present Mo17 and mexicana draft genomes are considered “version 1.0”, due to the technical limitations of short read assembly and Zea genome complexity4 and could be improved by using additional long-read assembly strategies47. Through comparative genomic analysis, 220,860 (~ 88.8 Mb PAVs between B73 and Mo17 including 1293 PAV genes were identified. However, given the sequence bias of Illumina and the low coverage of PacBio, the number of PAVs might be slightly overestimated.
Our genetic design provided an opportunity to study spontaneous mutation rates at a genome-wide level. In the present study, the mutation rate was 2.17 × 10−8 ~3.87 × 10−8 per site per generation, slightly higher than in other species37, 48, which may contribute to the abundant diversity that exists in the maize genome49. Our estimate is also higher than a previous one based on IBD analysis from maize inbreds35, possibly due to the high overall heterozygosity and divergent parents in the early TM population generations49. A higher deleterious mutation rate was observed in pericentromeric regions, where recombination was suppressed, and helps to explain the excess residual heterozygosity in pericentromeric regions50, since heterozygosity increases tolerance to harmful mutations. Transcription-related processes could explain the higher mutation rate in genic regions51. When DNA is single-stranded state, there is no protection of nucleosomes, therefore, it might be mutated more easily52. A nonrandom distribution of mutations was also observed outside of the centromere regions similar to other species. The underlying mechanisms are still unclear, though a number of hypotheses have been proposed46, 49, 53. A high-resolution recombination maps constructed on the basis of single-microspore sequencing54 will help in the study of the relationships between mutation and recombination38, 55 and of gene conversion56.
We estimated that about 10.7% of maize genomic regions were introgressed from mexicana, however, the size and number of introgressed regions for each individual maize line may have been underestimated due to limited marker density. Introgression can be associated with maize adaptation from lowland to highland environments, and may have contributed to genetic improvement by increasing yield potential and other important agronomical traits. The TM population provided a unique resource to identify a number of introgression regions that potentially affect phenotypes and important agronomic traits that were supported by previous and present QTL studies. Because limited mexicana genomic resources were available, the QTL detection power and the phenotypic effects of the introgression regions may be underestimated. The developed TM population provided a good resource to identify the underlying genes for introgression and other important agronomic traits derived from the mexicana genome that had been largely ignored in the past. Future studies will enhance our understanding of introgression, adaptation, and breeding.
Methods
Plant material and DNA sequencing
The BC2F7 population (containing 191 individuals) was derived from a single F1 seed from a cross between Mo17 and Zea. mays. ssp. mexicana (PI 566673); each individual theoretically containing 12.5% (actually, 11.22%) mexicana and 87.5% (actually, 88.78%) Mo17 genome fragments. Ten individuals (Fig. 1a) derived from the TM population were chosen for genome assembly, to cover as much as possible of mexicana genome, based on the high-resolution linkage map. These individuals contain DNA segments covering the whole Mo17 genome and approximately 96% mexicana genome (16.48% on average for each selected individual). We assigned the assemblies of the two parental genomes into separate bins (Fig. 1a and Supplementary Data 1). Young leaves in each individual were used for paired-end-tag DNA sequencing using Illumina HiSeq2000 platform. Millions of reads were generated from libraries with different fragment sizes (175 bp, 300 bp, 500 bp, 3 kb, 9 kb, and 12 kb, Supplementary Table 1). Moreover, Mo17, TM24, and TM104 were sequenced on a Pacific Biosciences (PacBio) platform with 10 Single Molecule Real-Time (SMRT) cells (Supplementary Table 2), and TM104 was further sequenced and assembled using DenovoMAGIC 2TM (http://nrgene.com/products-technology/denovomagic/) (Supplementary Table 3). See the Supplementary Note 4 for additional details.
Genome assembly with a meta-assembly pipeline
A meta-assembly pipeline was developed to assemble Mo17 and mexicana genomes (Fig. 1c). We filtered the Illumina reads and removed adapters for each library. The assembly can be divided into three major steps: Step 1, three strategies were executed to assemble contigs, including de novo assembly of 10 individuals, reference-based assembly based on B73 genome, and de novo assembly of unmapped reads. Each of the 10 individuals was de novo assembled with SOAPdenovo257 (V2.04). Reads of all libraries were aligned to the maize B73 reference genome (RefGen_v2 release 5b)4, the mapped reads were assembled with MaSuRCA58 (V.2.1.0) in each bin, and unmapped reads were de novo assembled with MaSuRCA58 (V.2.1.0); Step 2, the contigs of the above three strategies were merged, and were further connected or extended with Pacbio long reads (Supplementary Fig. 1, Supplementary Note 1). The PacBio long reads were filtered and aligned to B73 genome4 by using BLASR59. After integrating the contigs, long mate-pair libraries were added to assemble scaffolds with SSPACE60. SOAP GapCloser57 (V1.12), GapFiller61, and Pbjelly62 were used to bridge scaffold gaps with paired-end reads and PacBio long reads to obtain scaffolds; and Step 3, the NRGene scaffolds were integrated, and the merged scaffolds were the final scaffolds (Supplementary Note 1).
Anchoring of the assembled scaffolds to genetic map
A high-density genetic linkage map was developed using the TM population with 191 recombinant inbred lines and genotyped with the Illumina MaizeSNP50 array12. In addition, the 10 sequenced individuals were genotyped using a maize 600k Affymetrix SNP array63. First, scaffolds were ranked using B73 reference position by aligning the probes of the 600k SNP array to scaffolds. Second, the genetic linkage maps of TM population and integrated B73 and Mo17 (IBM) population were used to adjust structure variations and mis-assembly of Mo17 and mexicana genomes. Finally five scaffolds of Mo17 from NRGene were corrected by comparison with the genetic and physical maps (Supplementary Table 12). The remaining scaffolds were anchored using genotype by sequencing (GBS) probes6.
Identification of repetitive elements
Transposable element (TE) libraries were constructed with RepeatModeler64, and were applied to mask the Mo17 and mexicana genomes by using RepeatMasker software (http://www.repeatmasker.org/).
Gene prediction and annotation
An integrated approach combining de novo prediction with evidence-based data (ESTs, protein homology, and RNA-seq) analysis was employed by using the PASA65 and EVM66 pipeline. The predicted gene models from EVM66 were then updated by PASA65 assembly alignments. Gene functions were assigned according to the best alignment using BLASTP67 (E value < 10−5) to the UniProt database68. InterProScan69 was used to identify gene ontology terms, motifs, and domains of gene models. See the Supplementary Note 3 for additional details.
PAV detection
Two approaches were employed to identify PAVs between B73 and Mo17, (Supplementary Fig. 5a). The sequences uniquely present in Mo17 compared with the B73 genome were identified using approach 1, consisting of the following steps: (1) Mo17 scaffolds were aligned to the B73 genome with NUCmer70 (-c 90 -l 40–maxmatch); (2) Unaligned sequences in Mo17 were extracted and gap regions containing undetermined N bases were filtered out; (3) The filtered Mo17 sequences (>100 bp) were aligned to the B73 genome with BLASTN67 (-evalue 1e-5 -perc_identity 90); and (4) Unaligned sequences were kept as variations uniquely present in Mo17.
The sequences uniquely present in B73 as compared with Mo17 were identified using approach 2, consisting of the following steps: (1) Mo17 reads were mapped to the B73 genome with bwa71 (V0.7.4); (2) Regions (≤50 bp) with coverage depth ≤ 2 were merged; (3) Genomic sequences in the merged regions were extracted and aligned to the other genomes with BLASTN67; and (4) Unaligned regions (>100 bp) without N bases were kept as the unique PAVs.
SNP calling and cross validation
Sequencing reads from 10 individuals of the TM population were mapped to the maize B73 reference genome (RefGen_v2 release 5b)4 using bwa71 (V0.7.4) with default parameters. Only reads mapping to unique sites were retained for SNP calling. We used the rmdup function in SAMtools72 to filter PCR duplicates. SNPs were first called using UnifiedGenotyper of GATK73 (V 2.7–2), the results were used as the input files to the local realignment tool to minimize the number of mismatch bases. The calmd function of SAMtools72 was used to improve SNP specificity. Finally, SNPs were called using UnifiedGenotyper of GATK73 and mpileup of SAMtools72. The final SNPs were filtered as follows: (1) the low mapping quality SNPs (MQ < 30) were removed; (2) the read depths of high-quality SNPs were in the range of 5 ~ 40; (3) the distance of two adjacent SNPs was ≥ 5 bp; and (4) the depth of minor allele was ≥ 5.
In order to verify the reliability of SNPs, the commercially available maize 600k Affymetrix SNP array63 was used to identify SNPs of the 10 selected TM lines. The overall mean concordance rate was 99.5%, and the mean SNPs heterozygous rate was lower than 5% (Supplementary Table 13).
Mutation identification
The 10 sequenced individuals from BC2F7 population were derived from a single F1 individual and covered Mo17 genomes more than 8 times, which provided the opportunity to detect the spontaneous mutations during the 10 continuous generation sections by using the shared Mo17 IBD regions. A preliminary candidate mutation was called if one individual had a different allele compared with other individuals in the Mo17 IBD regions. Rigorous filtering criteria were employed to ensure the accuracy of the mutation identification: the candidate mutation must be supported by at least 5 uniquely mapped high-quality reads (MQ ≥ 50 for each of the two alleles & the paired reads were also well mapped onto the same chromosome). Excluded from analysis were those regions with unexpected and clustered SNPs identified that could be caused by residual heterozygosity of Mo17, which was used three times and from different individuals each time for producing the TM population (Fig. 1a). In total, nearly 2000 mutations were detected in two or more individuals in the 10 sequenced lines (Supplementary Fig. 9), which were also excluded from analysis. This might be due to the preexistent mutations in the parent of Mo17, as mentioned above.
Introgression analysis
We combined the data of the present two genomes and public genomic data (including 2 mexicana, 18 parviglumis and 895 maize31) to detect the introgression regions by using the population approach74. First, combining our SNPs of mexicana with the SNPs of teosintes obtained from Hapmap339, the IBD regions of maize to the mexicana and parviglumis subpopulation were identified via fastIBD75. The genome was divided into 100 kb size windows and the number of recorded IBD tracts between maize and the mexicana and parviglumis subpopulations was calculated in each window, respectively. The calculated numbers were normalized from 0 (no IBD detected) to 1 (IBD shared by all individuals within a subpopulation), considering that the total number of pairwise comparisons differed between the subpopulations. The normalized IBD between maize and mexicana subpopulation (nIBD mex ) and the normalized IBD between maize and parviglumis subpopulation (nIBD par ) were then used to calculate the relative IBD between the compared groups (rIBD = nIBD mex − nIBD par ). The detection of mexicana introgression was based on patterns of allele sharing and IBD, which may be biased toward low diversity regions. The bins whose SNPs density was lower than the average were filtered. A bin with \(\frac{{(\rm{\it rIBD} - \mu )}}{{\sigma \times\rm{\it rIBD}}} >2\) was regarded as a putative introgression region (Supplementary Fig. 6a), μindicated the mean of rIBD andσindicated the standard deviation of rIBD. However, we used 500 kb-sized window to detect the introgression regions in landraces because of the low SNP density (SNP number: 946,072).
Phenotyping and QTL mapping
Field trials of TM population (191 individuals) were conducted in 10 environments. Each line was grown in a single 3 m row with planting density of 45,000 plants/ha. Five randomly selected plants in each line were used for measuring ear-leaf width and kernel row number. The BLUP (best linear unbiased prediction) values for both traits were used for QTL mapping.
All the TM lines and their parents were genotyped using the Illumina MaizeSNP50 array12. To construct the genetic linkage map, a method was developed integrating the Carthagene software76 in a Linux system with in-house Perl scripts (https://github.com/panqingchun/linkage_map). Markers that completely co-localized were assigned into a chromosomal bin. Each bin was considered one marker. 12,390 polymorphic markers were obtained in total, which were incorporated into 1,282 unique bins (markers) in this study77. QTL mapping using the composite interval mapping method78 was performed in the package QTL Cartographer version 2.579.
Data availability
The Mo17 and mexicana genome have been submitted to GenBank under the accession codes LMUZ00000000 and LMVA00000000. A website to access the data has also been made available at http://mmgdb.hzau.edu.cn/maize/. The authors declare that all other data supporting the findings of this study are available from the corresponding authors upon request.
References
FAO. FAOSTAT, Production. Cited 5 April 2016. Available at: http://faostat3.fao.org/browse/Q/QC/E (2014).
Fu, H., Zheng, Z. & Dooner, H. K. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl Acad. Sci. USA 99, 1082–1087 (2002).
Brunner, S., Fengler, K., Morgante, M., Tingey, S. & Rafalski, A. Evolution of DNA sequence nonhomologies among maize inbreds. Plant. Cell. 17, 343–360 (2005).
Schnable, P. S. et al. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 (2009).
Zuo, W. L. et al. A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 47, 151–157 (2015).
Lu, F. et al. High-resolution genetic mapping of maize pan-genome sequence anchors. Nat. Commun. 6, 6914 (2015).
Piperno, D. R., Ranere, A. J., Holst, I., Iriarte, J. & Dickau, R. Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. USA 106, 5019–5024 (2009).
van Heerwaarden, J. et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc. Natl. Acad. Sci. USA 108, 1088–1092 (2011).
Hufford, M. B., Bilinski, P., Pyhäjärvi, T. & Ross-Ibarra, J. Teosinte as a model system for population and ecological genomics. Trends Genet. 28, 606–615 (2012).
Hufford, M. B. et al. The genomic signature of crop-wild introgression in maize. PLoS Genet. 9, e1003477 (2013).
Hufford, M. B. et al. Comparative population genomics of maize domestication and improvement. Nat. Genet. 44, 808–811 (2012).
Ganal, M. W. et al. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE. 6, e28334 (2011).
Kawahara, Y. et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4 (2013).
Paterson, A. H. et al. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Parra, G., Bradnam, K. & Korf, I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007).
Van Bel, M. et al. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158, 590–600 (2012).
Salvi, S. et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl Acad. Sci. USA 104, 11376–11381 (2007).
Liu, H. et al. Gene duplication confers enhanced expression of 27-kDa γ-zein for endosperm modification in quality protein maize. Proc. Natl Acad. Sci. USA 113, 4964–4969 (2016).
Pyhäjärvi, T., Hufford, M. B., Mezmouk, S. & Ross-Ibarra, J. Complex patterns of local adaptation in teosinte. Genome Biol. Evol. 5, 1594–1609 (2013).
Springer, N. M. et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5, e1000734 (2009).
Mills, R. E. et al. Mapping copy number variation by population-scale genome sequencing. Nature 470, 59–65 (2011).
Zhang, Z. et al. Genome-wide mapping of structural variations reveals a copy number variant that determines reproductive morphology in cucumber. Plant Cell. 27, 1595–1604 (2015).
Żmieńko, A., Samelak, A., Kozłowski, P. & Figlerowicz, M. Copy number polymorphism in plant genomes. Theor. Appl. Genet. 127, 1–18 (2014).
Sutton, T. et al. Boron toxicity tolerance in barley arising from efflux transporter amplification. Science 318, 1446–1449 (2007).
Todd, J. J. & Vodkin, L. O. Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant. Cell. 8, 687–699 (1996).
Xu, K. et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708 (2006).
Tao, Y. et al. Combined linkage and association mapping reveals candidates for Scmv1, a major locus involved in resistance to sugarcane mosaic virus (SCMV) in maize. BMC Plant. Biol. 13, 162 (2013).
Bertioli, D. J. et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446 (2016).
Wang, X. et al. Genome alignment spanning major Poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol. Plant 8, 885–898 (2015).
Tang, H. et al. Synteny and collinearity in plant genomes. Science 320, 486–488 (2008).
Bukowski, R. et al. Construction of the third generation Zea mays haplotype map. Preprint at http://biorxiv.org/content/early/2015/09/16/026963 (2015).
Lauter, N., Gustus, C., Westerbergh, A. & Doebley, J. The inheritance and evolution of leaf pigmentation and pubescence in teosinte. Genetics 167, 1949–1959 (2004).
Romero Navarro, J. A. et al. A study of allelic diversity underlying flowering-time adaptation in maize landraces. Nat. Genet. 49, 476–480 (2017).
Wade Michael. Evolutionary Genetics, The Stanford Encyclopedia of Philosophy Fall 2008 edn (ed. Edward N. Zalta) available at: http://plato.stanford.edu/archives/fall2008/entries/evolutionary-genetics/ (Metaphysics Research Lab, Stamford University, 2008)
Jiao, Y. et al. Genome-wide genetic changes during modern breeding of maize. Nat. Genet. 44, 812–815 (2012).
Ossowski, S. et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327, 92–94 (2010).
Schrider, D. R., Hourmozdi, J. N. & Hahn, M. W. Pervasive multinucleotide mutational events in eukaryotes. Curr. Biol. 21, 1051–1054 (2011).
Wang, J. et al. Evidence for mutation showers. Proc. Natl Acad. Sci. USA 104, 8403–8408 (2007).
McLaren, W. et al. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics 26, 2069–2070 (2010).
Regulski, M. et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23, 1651–1662 (2013).
Palmer, L. E. et al. Maize genome sequencing by methylation filtration. Science 302, 2115–2117 (2003).
Choi, Y., Sims, G. E., Murphy, S., Miller, J. R. & Chan, A. P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7, e46688 (2012).
Morton, B. R. et al. Variation in mutation dynamics across the maize genome as a function of regional and flanking base composition. Genetics 172, 569–577 (2006).
Kono, T. J. et al. The role of deleterious substitutions in crop genomes. Mol. Biol. Evol. 33, 2307–2317 (2016).
Hodgkinson, A. & Eyre-Walker, A. Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet. 12, 756–766 (2012).
Gordon, D. et al. Long-read sequence assembly of the gorilla genome. Science 352, aae0344 (2016).
Campbell, C. D. et al. Estimating the human mutation rate using autozygosity in a founder population. Nat. Genet. 44, 1277–1281 (2012).
Yang, S. et al. Parent–progeny sequencing indicates higher mutation rates in heterozygotes. Nature 523, 463–467 (2015).
McMullen, M. D. et al. Genetic properties of the maize nested association mapping population. Science 325, 737–740 (2009).
Morton, B. R. & Wright, S. I. Selective constraints on codon usage of nuclear genes from Arabidopsis thaliana. Mol. Biol. Evol. 24, 122–129 (2007).
Boulikas, T. Evolutionary consequences of nonrandom damage and repair of chromatin domains. J. Mol. Evol. 35, 156–180 (1992).
Francioli, L. C. et al. Genome-wide patterns and properties of de novo mutations in humans. Nat. Genet. 47, 822–826 (2015).
Li, X., Li, L. & Yan, J. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat. Commun. 6, 6648 (2015).
Lercher, M. J. & Hurst, L. D. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18, 337–340 (2002).
Williams, A. L. et al. Non-crossover gene conversions show strong GC bias and unexpected clustering in humans. Elife 4, e04637 (2015).
Luo, R. B. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18 (2012).
Zimin, A. V. et al. The MaSuRCA genome assembler. Bioinformatics 29, 2669–2677 (2013).
Chaisson, M. J. & Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinforma. 13, 238 (2012).
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D. & Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011).
Nadalin, F., Vezzi, F. & Policriti, A. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinforma. 13, S8 (2012).
English, A. C. et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE 7, e47768 (2012).
Unterseer, S. et al. A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k SNP genotyping array. BMC Genome 15, 823 (2014).
Smit, A. F. A. & Hubley, R. RepeatModeler Open 1,0 available at: www.repeatmasker.org (2008).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015).
Zdobnov, E. M. & Apweiler, R. InterProScan–An integration platform for the signature- recognition methods in InterPro. Bioinformatics 17, 847–848 (2001).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Sun, L. et al. GmHs1-1, encoding a calcineurin-like protein, controls hard-seededness in soybean. Nat. Genet. 47, 939–943 (2015).
Browning, B. L. & Browning, S. R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–471 (2013).
de Givry, S., Bouchez, M., Chabrier, P., Milan, D. & Schiex, T. CARHTA GENE: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21, 1703–1704 (2005).
Pan, Q. et al. Genome-wide recombination dynamics are associated with phenotypic variation in maize. New. Phytol. 210, 1083–1094 (2016).
Zeng, Z. B. Precision mapping of quantitative trait loci. Genetics 136, 1457–1468 (1994).
Wang, S., Basten, C. J. & Zeng, Z. B. Windows QTL Cartographer 2.5 (Department of Statistics, North Carolina State University, Raleigh, NC, 2012).
Acknowledgements
We greatly appreciate Prof. Jiansheng Li at China Agricultural University for significant contribution in developing the TM population and Drs Edward Buckler and Fei Lu at Cornell University for evaluating the assemble quality using GBS anchors. This research was supported by the National Key Research and Development Program of China (2016YFD0101003), the National Natural Science Foundation of China (31525017 and 31571351, 91435205), the National Youth Top-notch Talent Support Program, and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
J.Y. and L-L.C.: designed and supervised this study. Li.C. and X.Y.: developed the materials. N.Y., X.-W.X., R.-R.W., W.-L.P., J.-M.S., X.L., L.N., L.C., J.L. L.W., and D.J.: performed the data analysis. Y.W., M.J., and M.D.: conducted the PCR validation experiments. W.L.: managed the field work. Q.P. and F.L.: constructed the linkage map. N.Y., X.-W.X., L.-L.C., and J.Y.: prepared the manuscript. D.J. and X.Y.: helped to edit the manuscript. All the authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare thet they have no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, N., Xu, XW., Wang, RR. et al. Contributions of Zea mays subspecies mexicana haplotypes to modern maize. Nat Commun 8, 1874 (2017). https://doi.org/10.1038/s41467-017-02063-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-017-02063-5
This article is cited by
-
How is leaf herbivory related to agriculture? Insights from the Mexican center of crop origin
Arthropod-Plant Interactions (2024)
-
Population genomics unravels the Holocene history of bread wheat and its relatives
Nature Plants (2023)
-
The role of transposon inverted repeats in balancing drought tolerance and yield-related traits in maize
Nature Biotechnology (2023)
-
QTL mapping for flowering time in a maize-teosinte population under well-watered and water-stressed conditions
Molecular Breeding (2023)
-
Genome analyses reveal population structure and a purple stigma color gene candidate in finger millet
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.