Abstract
The histone deacetylase HDAC3 is a critical mediator of hepatic lipid metabolism, and liver-specific deletion of HDAC3 leads to fatty liver. To elucidate the underlying mechanism, here we report a method of cross-linking followed by mass spectrometry to define a high-confidence HDAC3 interactome in vivo that includes the canonical NCoR–HDAC3 complex as well as Prospero-related homeobox 1 protein (PROX1). HDAC3 and PROX1 co-localize extensively on the mouse liver genome, and are co-recruited by hepatocyte nuclear factor 4α (HNF4α). The HDAC3–PROX1 module controls the expression of a gene program regulating lipid homeostasis, and hepatic-specific ablation of either component increases triglyceride content in liver. These findings underscore the importance of specific combinations of transcription factors and coregulators in the fine tuning of organismal metabolism.
Similar content being viewed by others
Introduction
Hepatic lipid homeostasis is critical for the maintenance of normal liver physiology and organismal metabolism. Lipid composition and accumulation in the liver is controlled by a complex network of interconnected metabolic pathways such as lipid synthesis, lipolysis, β-oxidation, secretion, and storage, and the dysregulation of even one of these pathways can lead to lipid accumulation in liver or hepatic steatosis1, 2. Non-alcoholic fatty liver disease (NAFLD), defined by excess fat in the liver, is of growing clinical relevance in industrialized countries and is a major risk factor for the development of non-alcoholic steatohepatitis, liver fibrosis, and cirrhosis3. In addition, NAFLD has been linked to cardiovascular disease, metabolic syndrome, insulin resistance, and hepatocellular carcinoma2. Thus, the mechanisms governing liver lipid homeostasis are of broad importance to understanding the development of NAFLD and to identify targets for therapeutic intervention.
These pathways of lipid homeostasis are regulated in liver by a host of transcription factors including the nuclear receptors HNF4α4, 5, Rev-erbα6, LXRs7, PPARs8, and the E-box binding proteins SREBP and ChREBP9 among others. In addition to these sequence-specific DNA-binding factors, numerous cofactors and coregulators have been shown to influence the expression of genes controlling lipid metabolism10. Previous work has demonstrated histone deacetylase 3 (HDAC3) to be an important epigenomic coregulator in liver6, 11, and deletion of HDAC3 in adult liver results in remarkable hepatic steatosis12. However, the fundamental mechanisms of how HDAC3 controls metabolic gene transcription in liver are not completely understood.
HDAC3 is unique among the class I histone deacetylases as it requires binding to the nuclear receptor corepressor (NCOR1)13 or the silencing mediator for retinoic acid and thyroid hormone receptors (SMRT or NCOR2)14 for its enzymatic activity15,16,17. Together with transducing β-like 1X-linked and receptor 1 (TBL1X and TBL1XR1)18 and the G-protein suppressor 2 (GPS2)19, these proteins form the core of the NCoR transcriptional repressor complex20. The NCoR complex has been shown to be a major corepressor complex for the nuclear receptor family of transcription factors13, 14, 20, 21. Deletion of individual components of the NCoR complex results in an increase in liver triglycerides12, 22, 23, highlighting the importance of these proteins acting together as a functional complex to regulate liver metabolic gene transcription.
The vast number of transcriptionally relevant complexes highlights the important roles protein–protein interactions play in the control of gene expression. For HDAC3, there are important questions about which transcription factors recruit it to the genome, and which HDAC3-associated proteins act as downstream effectors to impact lipid gene regulation and hepatic steatosis.
Here, we describe NEAT ChIP-MS (Nuclear Extraction Affinity Tag), an improved chromatin cross-linking method followed by nano liquid chromatography-tandem mass spectrometry (nLC-MS/MS) analysis to identify in vivo interactions in liver and define a high-confidence interactome for HDAC3. We find a strong association between HDAC3 and the Prospero-related homeobox 1 protein (PROX1), which co-localize at the genome and at some sites are completely dependent on the nuclear receptor HNF4α for their co-recruitment. Interestingly, depletion of PROX1 in liver results in increased hepatic triglycerides similar to loss of HDAC3. Our results suggest an important role for an HDAC3–PROX1 corepression module in regulating the transcription of a gene program important for the maintenance of lipid homeostasis.
Results
In vivo screen for HDAC3 interactors
To elucidate nuclear interactors of HDAC3 in vivo, we developed NEAT ChIP-MS, a cross-linking proteomic interaction method that allowed for the confident identification of HDAC3 interactors in adult liver (Fig. 1a). Hdac3 fl/fl animals were tail vein-injected with either AAV virus expressing epitope-tagged HDAC3 (AAV8 TBG HDAC3-HA) in conjunction with AAV8 TBG Cre to deplete endogenous HDAC3 in hepatocytes as described12, or with control virus expressing epitope-tagged green fluorescent protein (AAV8 TBG HA-EGFP). Subsequent to nuclear isolation, samples were cross-linked and EGFP or HDAC3 was immunoprecipitated with anti-HA resin. Associated proteins were analyzed by nLC-MS/MS and the results of the HDAC3 and EGFP interactomes were compared to remove non-specific interactions. We applied stringent significance (P < 0.01) and fold enrichment (10-fold) cutoffs using label-free quantification values estimated by the intensity-based absolute quantification (iBAQ)24 algorithm to the resultant interactome to identify a set of high-confidence HDAC3 interacting proteins in vivo in mouse liver (Fig. 1b and Supplementary Data 1).
NEAT ChIP-MS reveals the HDAC3 liver nuclear interactome. a Schematic illustration of the NEAT ChIP-MS protocol. HA-tagged HDAC3 or EGFP was expressed with or without Cre in Hdac3 fl/fl mice specifically in hepatocytes via adeno-associated virus (AAV8 TBG). After isolation of nuclei and cross-linking with formaldehyde, HDAC3 or EGFP control were captured by anti-HA immunoprecipitation. Protein complexes and associated DNA sequences were analyzed by mass spectrometry or high-throughput sequencing, respectively. b Volcano plot of mass spectrometry analysis of HDAC3 interacting proteins (HDAC3 n = 11, EGFP n = 13). The x axis indicates log2 ratio of normalized intensity (iBAQ) of proteins discovered in HDAC3 to EGFP control. Red box indicates fold-change (10-fold) and P-value (0.01) cutoffs for interactors. Core NCoR complex components (blue), selected high scoring interactors (red), and Rev-erbα (green) are indicated. c Heatmap of normalized intensity (iBAQ) of HDAC3 interactors from b in the presence or absence of micrococcal nuclease (MNase) or Benzonase nuclease. Each lane represents an independent experiment. d Co-IP experiments confirming interaction of HDAC3 with PROX1 from liver expressing tagged HDAC3 (top) or endogenous IPs (bottom). e Protein–protein interaction network analysis. Circle color represents enrichment over control and the size of the circle represents −log10(P-value). Nodes displayed met eightfold enrichment cutoff after Benzonase treatment and lines indicate validated interactions (STRING, active interaction sources include experiments and databases, minimum interaction score 0.6). Interactors are grouped by known type and circles indicate known functional complexes
As expected, we observed an enrichment of all the components of the previously identified NCoR complex (sequence coverage indicated in parentheses) consisting of NCOR1/2 (11.14%/12.14%), TBL1X (46.62%), TBL1XR1 (42.52%), GPS2 (4.33%), and HDAC3 (42.85%) itself. In addition, our screen confirmed interaction of HDAC3 with a number of transcription factors known to interact with the NCoR complex, including the circadian nuclear receptor Rev-erbα6. One of the most significantly enriched HDAC3 interactors, but not considered a core component of the NCoR complex, was the Prospero-related homeobox 1 (PROX1). A highly conserved transcription factor in vertebrates, PROX1 was previously shown to be critical for the development of several organs including the lymphatic system25, lens26, liver27, 28, pancreas28, heart29, and skeletal muscle30. In addition, PROX1 has been implicated in regulating the functions of several nuclear receptors31,32,33,34,35,36 and has been shown to act as both a transcriptional repressor31,32,33 and activator26, 37 in different biological contexts.
To assess whether the proteins discovered in our chromatin bound complexes purified with HDAC3 directly through protein–protein interactions or indirectly through close genomic proximity, we treated our lysates with either micrococcal nuclease (MNase), capable of cutting only in nucleosome-free regions, or Benzonase nuclease, which can cleave DNA regardless of nucleosome occupancy. Although micrococcal nuclease had minimal impact, Benzonase nuclease significantly reduced the intensity of a subset of protein–protein interactions with HDAC3 (Fig. 1c and Supplementary Data 2). This suggested that these proteins are not in direct contact with HDAC3, and thus these protein–protein associations are likely facilitated by DNA. In addition, interactors associated with HDAC3 exhibiting lower enrichments tended to be more susceptible to Benzonase treatment, implying that proteins displaying greater abundance were, in general, more likely to occur through protein–protein interactions. The group of proteins maintained upon nuclease treatment included PROX1, indicating that this robust HDAC3 interactor was likely more directly bound and not dependent on DNA bridging for its interaction.
The interaction of PROX1 and HDAC3 in liver was confirmed by coimmunoprecipitation experiments using the reversible cross-linker DSP from livers expressing HA-tagged HDAC3 or endogenously from wild-type livers (Fig. 1d). PROX1 and HDAC3 interaction likely occurs in the context of the NCoR complex, as interaction between PROX1 and NCOR1 was observed in cell culture, and also requires multiple surfaces in the N terminus of PROX1 for binding (Supplementary Fig. 1a, b). We next sought to better understand the HDAC3 interactome from a more global functional perspective. Classifying the interactors and their biological links and annotating them using information from the STRING database revealed several interesting clusters of proteins associating with HDAC3 (Fig. 1e). In addition to the NCoR complex, HDAC3 was found to associate with several other transcriptionally important complexes including SWI/SNF, Integrator, Cohesin, and components of the NuRD and CoREST repressor complexes. We also observed a number of sequence-specific DNA-binding transcription factors of functional relevance to liver physiology such as C/EBPs, FXR, RXRα, and HNF4α. We also utilized cytoHUBBA, a tool to define network topology38, and identified members of the NCoR complex, NuRD/CoREST complex, Integrator complex, and nuclear receptors as important nodes in the liver HDAC3 interactome (Supplementary Fig. 2). These results indicate that HDAC3 likely has an important role in regulating the expression of liver gene programs through interaction with an array of transcription factors and suggest that PROX1, as a strong interactor in liver, may be of more general importance to the functionality of HDAC3.
PROX1 and HDAC3 co-localize extensively at the genome
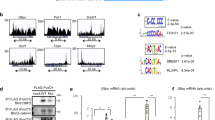
To further investigate the functional interplay between PROX1 and HDAC3, we performed ChIP-seq on PROX1 and HDAC3 in mouse liver. Bioinformatic analysis of the peaks discovered in the two data sets revealed a strong overlap between the binding sites of the HDAC3 and PROX1 cistromes (Fig. 2a). Visual inspection of individual ChIP-seq tracks confirmed a striking correlation (Fig. 2b). We next performed ChIP–reChIP analysis, which demonstrated that both HDAC3 and PROX1 were co-bound to the same DNA fragments (Fig. 2c). Analysis of overlapping peaks by pathway analysis (Reactome) revealed that co-bound sites were enriched at genes involved in a variety of important metabolic pathways, including those that regulate lipid metabolism (Fig. 2d). As expected peaks bound by HDAC3 selectively showed enrichment both for liver metabolic genes and those involved in circadian rhythm, indicating that HDAC3 likely has functions with Rev-erbα independent of its interaction with PROX1 (Supplementary Fig. 3a). HOMER motif analysis of the overlapping peaks displayed a strong enrichment for the HNF4 and C/EBP motifs, two transcription factors that are known to have important roles in liver development and adult liver function (Fig. 2e). Upon closer examination of overlapping, HDAC3-only, and PROX1-only peaks, we observed that the HNF4α motif was significantly reduced at HDAC3-only peaks, whereas the Rev-erbα (DR2) motif was significantly enriched at these sites (Fig. 2f). The C/EBP motif followed a similar pattern observed for HNF4α indicating that it may also have a role in coordinating these sites but not those involved in circadian function (Supplementary Fig. 3b). These results suggest that HDAC3 is recruited to at least two subsets of sites, ones that contain only HDAC3, which are recruited by Rev-erbα, and those that have both PROX1 and HDAC3, and are recruited by HNF4α.
HDAC3 and PROX1 exhibit extensive co-binding and reveal a metabolic signature. a Venn diagram displaying overlap of peaks identified in PROX1 (2 rpm cutoff) and HDAC3 (1.5 rpm cutoff) ChIP-seq. Peaks required 50% overlap and have a minimum 1 rpm signal for the other factor. b Representative browser tracks of HDAC3 and PROX1 ChIP-seq. Scale is reads per ten million (RPTM). c Co-occupancy of PROX1 and HDAC3 as indicated by ChIP–reChIP (n = 3) from liver. Legend indicates reChIP antibody following primary PROX1 ChIP elution. Data are presented as mean ± s.d., one-tailed unpaired Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001, ns not significant. d Reactome analysis of the nearest genes within 100 kb from the top 1000 overlapping HDAC3 and PROX1 peaks. e HOMER motif analysis of co-bound peaks displaying over-represented sequences. f HOMER motif enrichment analysis of the indicated motifs (HNF4α and Rev-erbα DR2) at overlapping and non-overlapping peaks determined in (a). Numbers above brackets indicate P-values, χ 2 test
HNF4α is required for recruitment of the HDAC3–PROX1 module
Given our observation that the HNF4α motif was found prominently at co-occupied sites, we hypothesized that this important liver transcription factor may have a role in the recruitment of HDAC3 and PROX1. ChIP-seq analysis of HNF4α in control liver demonstrated HNF4α binding at >60% of HDAC3–PROX1 co-bound peaks (Fig. 3a). In stark contrast, HDAC3-only peaks displayed significantly reduced HNF4α binding (~14%). Motif analysis of the triply bound peaks showed an increased enrichment of the HNF4 motif and a mild de-enrichment of the C/EBP motif when compared to all HDAC3–PROX1 co-bound peaks (Fig. 3b). To address whether HNF4α was required for the recruitment of HDAC3–PROX1 at the co-bound sites we injected Hnf4α fl/fl mice with AAV8 TBG Cre to delete Hnf4α in hepatocytes (Supplementary Fig. 4a). ChIP-seq analysis of HNF4α in these mice revealed near complete loss of HNF4α binding at the genome (Fig. 3c and Supplementary Fig. 4b–d). Remarkably, a significant portion of both HDAC3 and PROX1 peaks were strongly reduced upon loss of HNF4α (Fig. 3d). Moreover, the HDAC3 and PROX1 sites that were affected were almost exclusively the same sites (Supplementary Fig. 5a).
The HDAC3–PROX1 module is recruited by HNF4α in liver. a Comparison of HNF4α binding (>2 rpm, filtered on the HNF4α KO, 50% minimum overlap) at overlapping and non-overlapping peaks in the HDAC3 and PROX1 cistromes from Fig. 2a. Numbers above brackets indicate P-values, χ 2 test. b HOMER motif analysis of peaks co-bound by HDAC3, PROX1, and HNF4α displaying over-represented sequences. c Representative browser tracks of HNF4α, PROX1, and HDAC3 ChIP-seq in Hnf4α fl/fl livers infected with AAV8 TBG Egfp (Ctl) or Cre (Hnf4α KO). Indicated scales are in RPTM. d Scatter plots of PROX1 (left) and HDAC3 (right) ChIP-seq in control versus Hnf4α KO. Red and blue shaded regions indicate a twofold decrease in peak intensity upon loss of HNF4α. e, f Co-occupancy of HDAC3–PROX1 and HNF4α as indicated by ChIP–reChIP (n = 3) from liver. Legend indicates reChIP antibody following primary HDAC3 or PROX1 ChIP elution. Data are presented as mean ± s.d, one-tailed unpaired Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001, ns not significant
Co-binding of both PROX1 and HDAC3 with HNF4α nearby several lipid-related genes were confirmed by ChIP–reChIP (Fig. 3e, f). In addition, we observed a decreased interaction of PROX1 and HDAC3 in the Hnf4α knockout mice by Co-IP western blot, suggesting HNF4α may be required to stabilize the HDAC3–PROX1 interaction in vivo (Supplementary Fig. 5b). We compared the binding of both HNF4α and C/EBPβ at the co-bound sites that were lost in Hnf4α knockout versus those that were unaffected and observed a positive relationship between the strength of HNF4α binding and the dependency of HDAC3/PROX1 on HNF4α (Supplementary Fig. 5c). C/EBP, serving as a control comparison, did not display a similar correlation. These results strongly suggest that HNF4α recruits the HDAC3–PROX1 module to a substantial subset of binding sites, whereas other factors, like C/EBP, may be more important elsewhere in the genome.
Liver depletion of PROX1 increases hepatic triglycerides
To determine the function of PROX1 in liver, Prox1 was knocked down in adult hepatocytes of wild-type male mice using an AAV-based shRNA delivery vector (AAV8 TBG shProx1), which led to a ~70% depletion in PROX1 protein 3 weeks following injection (Fig. 4a). Importantly, no significant change in HDAC3 protein levels were observed upon knockdown of PROX1 (Supplementary Fig. 6a). Hepatic triglycerides (TG) were significantly elevated following 3 weeks of PROX1 knockdown, with a more striking difference in TG levels observed at 6 weeks post infection (Fig. 4b). Oil red O staining confirmed an increase in neutral lipid in livers of male mice infected with Prox1 shRNAs (Fig. 4c). Thus, similar to HDAC3 knockout, depletion of PROX1 in liver results in increased triglyceride content. Of note, hepatic PROX1 levels were not significantly changed after 12 weeks of high fat diet (Supplementary Fig. 6b). Moreover, knockdown of Prox1 in female livers led to a modest but non-significant increase in TG, potentially indicating a gender-specific response (Supplementary Fig. 6c).
The HDAC3–PROX1 module suppresses steatosis by controlling a hepatic lipid metabolism gene program. a Western blot of liver samples from mice treated for 3 weeks with AAV8 TBG shLuciferase (Prox1 Ctl) or shPROX1 (Prox1 KD). Densitometry of the western blot shown above (n = 3). Data are presented as mean ± s.d. b Hepatic triglyceride assay of livers infected for 3 weeks (n = 3) or 6 weeks (n = 6) with indicated virus. Data are presented as mean ± s.d. c Oil red O staining of livers infected for 6 weeks as in (b). Scale bar is 50 µm. d RNA-seq analysis of Hdac3 fl/fl mice infected for 2 weeks with AAV8 TBG Egfp (n = 2) or Cre (Hdac3 KO, n = 3) vs. wild-type mice infected with AAV8 TBG shLuciferase (n = 3) or shPROX1 (Prox1 KD, n = 3) for 3 weeks. Heatmap displays coregulated genes (1.7-fold change, P < 0.05) grouped by expression correlation (135 genes) or anti-correlation (50 genes) upon ablation of HDAC3 or PROX1. Scale bar represents log2(fold change). Inset shows the P-value and corresponding gene list for the highest ranking Reactome pathway in the co-upregulated cluster. e Heatmap displays co-upregulated lipid-related genes upon ablation of HDAC3 or PROX1 and the corresponding binding strength of adjacent HDAC3–PROX1 co-bound peaks (−50kb upstream of the transcription start site, TSS through +2 kb from the transcription end site, TES) in the HNF4α KO liver relative to control. Scale bar represents log2(fold change) for RNA-seq and fold-change for ChIP-seq. f Example ChIP-seq and GRO-seq browser tracks at the G0s2 locus. Red boxes indicate location of putative G0s2 enhancers. g Luciferase assay (n = 3) indicating transcriptional response to co-expression of HNF4α, PROX1 and C/EBPα at G0s2 enhancers as identified in (f). Data are presented as mean ± s.d. Two-tailed unpaired Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001, ns not significant
HDAC3 and PROX1 coregulate a lipid gene expression program
To determine if transcriptional mechanisms of coregulation by HDAC3 and PROX1 had a role in regulating this striking liver lipid phenotype, we performed RNA-seq on Hdac3 knockouts (Supplementary Fig. 6d) and 3 week Prox1 shRNA livers. We observed a significant overlap of genes whose expression was altered by loss of either factor, with a striking over-representation of genes whose directionality of change was correlated and a strong preference for upregulated genes, consistent with the function of both HDAC3 and PROX1 as transcriptional repressors (Fig. 4d and Supplementary Data 3). Pathway analysis (Reactome) revealed the group of transcripts co-upregulated by HDAC3 and PROX1 loss to be highly enriched for regulation of lipid metabolism, including a number of important mediators of lipid synthesis and lipolysis (Fig. 4d). Correlation analysis comparing the frequency of HNF4α-dependent HDAC3–PROX1 binding sites at unchanged and HDAC3–PROX1 coregulated genes revealed a statistically significant relationship (chi-square with Yates correction, P < 0.0001). This association was consistent at lipid-related genes (Fig. 4e), and point to a strong correlation between co-upregulated lipid-related gene expression and the co-occupancy of HNF4α-dependent, HDAC3–PROX1 co-bound sites.
Transcriptional coregulation of a number of genes whose expression are critical to maintenance of lipid homeostasis, such as G0s2 39, Elovl6 40, Mfsd2a 41, and Cidec 42 were confirmed by qPCR following Hdac3 knockout or Prox1 knockdown (Supplementary Fig. 7). Upon examination, a number of these loci exhibited a strong overlap of HDAC3, PROX1, and HNF4α ChIP-seq signals, consistent with our genome-wide analysis (Fig. 4f and Supplementary Fig. 8a–d). In addition, we observed upstream co-bound regions that exhibited high H3K27 acetylation, suggesting that these sites may be putative enhancers. Indeed, upon loss of HDAC3 or PROX1, we observed an increase in HNF4α-dependent H3K27 acetylation at these putative enhancer regions. Moreover, analysis of nascent transcription, utilizing an existing global run-on sequencing (GRO-seq) data set in liver43, revealed bi-directional enhancer RNAs (eRNAs) at a number of co-bound regions. To determine whether these binding sites act as enhancers, we cloned the G0s2 −17 kb and −1.7 kb binding sites into the pGL4 Luciferase reporter. Co-transfection of the enhancer reporter plasmid with an HNF4α expression vector revealed strong transcriptional induction by HNF4α at both the −17 kb and −1.7 kb sites, whereas mutation of the HNF4α binding site severely disrupted activation by HNF4α (Fig. 4g and Supplementary Fig. 9). Interestingly, expression of PROX1 significantly repressed HNF4α-mediated transcriptional activation, whereas it was unable to repress trans-activation induced by C/EBPα expression. These data indicate that the −17 kb, and to a lesser extent the −1.7 kb, binding sites act as HNF4α-controlled enhancers for the expression of G0s2 and that HDAC3 and PROX1 act as direct repressors of these enhancers. Overall, our results indicate that the interaction between HDAC3 and PROX1 is critical for regulating liver metabolic gene expression and maintenance of hepatic lipid homeostasis.
Discussion
In this study, we determined in vivo protein interactors of the class I histone deacetylase HDAC3 in mouse liver. This interactome revealed a number of binding partners in addition to confirming the well-established role of HDAC3 as a component of the NCoR complex16, 17, 20. Among these binding partners, we explored the functional interaction between HDAC3 and the Prospero-related homeobox protein PROX1. We determined that HDAC3 and PROX1 co-occupy a high number of genomic binding sites and that these binding sites correlate strongly with both the DNA-binding motif and the cistrome of HNF4α. Moreover, upon loss of HNF4α, a significant portion of HDAC3 and PROX1 co-occupied sites exhibit a profound loss of genomic binding. Depletion of PROX1 specifically in mature adult liver resulted in a marked increase in liver triglyceride content similar to that observed upon loss of HDAC312, likely due to the dysregulation of a coregulated gene expression program important for lipid synthesis and lipolysis. Furthermore, we defined a liver enhancer responsible for controlling the expression of the lipolysis inhibitor G0S2, and showed that its transcriptional activation by HNF4α can be specifically repressed by PROX1. Overall, these results strongly suggest HDAC3 and PROX1 function in liver to corepress gene transcription important for maintenance of lipid homeostasis.
Protein–protein interactions are critical for determining biological functionality, including the control of chromatin function and gene expression, and can be strongly influenced when a binding partner is restricted to a specific cell type or tissue. Previous studies defining interacting partners for HDAC3 by mass spectrometry have not provided significant depth or elucidated tissue-specific factors bound with HDAC3 in vivo15, 18, 44, 45. Our strategy for identifying HDAC3 interactors in liver differs from methods that have been successful in cultured cells46 in two important ways. First, through co-expression of fusion-tagged HDAC3 and Cre recombinase in Hdac3 fl/f animals via adeno-associated viral vectors (AAV8) under the control of the thyroxine binding globulin promoter (TBG), we were able to express our bait protein in vivo in hepatocytes lacking endogenous HDAC3. Second, we found that cross-linking isolated nuclei rather than whole liver depleted significant cytoplasmic and mitochondrial liver contaminants leading to reduced sample complexity and increased depth and sensitivity of our interacting protein mass spectrometric identifications. Nuclear isolation prior to functional assays, co-IP, and ChIP has been widely utilized in the transcriptional field47, most recently being employed for techniques such as GRO-seq48 and INTACT49. In addition, we have compared the cistromes of HDAC3 from standard ChIP-seq and those produced with our protocol and found them to be highly correlated (Pearson correlation, R = 0.781), suggesting that the nuclear isolation step is not having a major effect on the biological landscape that we are attempting to elucidate.
Similar to cross-linking methods developed for cultured cells46, the present method is improved in its ability to capture more transient interactions, including those at the genome. Of note, although previous work has shown the circadian nuclear receptor Rev-erbα and other nuclear receptors to be interacting partners for the HDAC3-containing NCoR complex6, 21, these proteins were not found in a previous HDAC3 proteomic screen performed in CEM T cells45. By contrast, the current study revealed several important HDAC3-bound nuclear receptors in liver including HNF4α, RXRα, Rev-erbα, and FXR, implicating HDAC3 in their functions. Our screen also revealed several non-nuclear receptor sequence-specific transcription factors such as C/EBPα, ChREBP, CREB1, and ETV6, possibly pointing to a broader role of HDAC3/NCoR in corepressing non-nuclear receptor transcription factors in liver. Alternatively, it is conceivable that our cross-linking-based sample preparation is capable of capturing targets of HDAC3 enzymatic activity. Indeed, a number of the interactors elucidated in our screen have been shown to be regulated by lysine acetylation including C/EBPα50, ChREBP51, CREB152, and others, and it is therefore interesting to speculate that HDAC3 may regulate their functions through this mechanism.
Interaction of HDAC3 was also observed with several important nuclear complexes such as Integrator53, 54, Cohesin55, SWI/SNF56, and NuRD57, which have been implicated in the regulation of transcription through binding and functional control of gene-specific enhancers. Previous studies suggested that HDAC3 genomic binding correlates primarily with regions of active transcription58, which our results strongly support and extend to suggest that these components of the transcriptional machinery may be involved. It will be interesting to determine whether HDAC3 has a role as a bona fide member of these complexes or rather regulates their function through enzymatic or non-enzymatic mechanisms. The processes by which transcriptional activators and repressors act together at these enhancers to specifically tune gene expression remains an important unanswered question.
Although PROX1 has been shown to be critical for the development and specification of several tissues25,26,27,28,29, 59, its functions in the physiology of adult liver have not been determined. Our study provides evidence that PROX1 in adult liver helps maintain proper lipid homeostasis, as its depletion results in metabolic gene transcriptional changes and a strong upregulation of hepatic triglycerides. A previous study suggested that haploinsufficiency of Prox1 in mice results in obesity60, and these animals exhibited an increase in liver lipids, which was suggested to be secondary to obesity. Our results suggest that the haploinsuffficiency of Prox1 in hepatocytes might also contribute to the hepatosteatotic phenotype.
Unlike PROX1, which displays significant tissue specificity31, HDAC3 expression is relatively ubiquitous. Even so, PROX1 and HDAC3 are implicated in the control of several overlapping tissues such as pancreas, heart, central nervous system, and liver. Thus it is possible that, in these biological contexts, PROX1 provides tissue-specific functionality to HDAC3 through protein–protein interaction. HDAC3 has been implicated in beta cell function and the regulation of glucose homeostasis61. Interestingly, genome-wide association studies have revealed a CC variant SNP of Prox1 (rs340841) that contributes to the control of fasting glucose levels and the development of diabetes62, 63. Perhaps in pancreatic beta cells, as in liver, HDAC3 and PROX1 form a complex to regulate gene expression involved in the control of organismal metabolism.
NAFLD is a disorder whose prevalence is increasing alongside metabolic syndrome, obesity, and type 2 diabetes and affects as high as 30% of the adult population in developed countries1, 2. As a major risk factor for liver inflammation (NASH) and scarring/fibrosis3, understanding the molecular mechanisms of liver function is of critical importance to development of therapies for this unmet medical need. Here, we have defined an interaction between HDAC3 and PROX1 that is nucleated at the genome by HNF4α, and elucidated the role of this repressor module in controlling hepatic triglyceride content by modulating lipid synthesis and lipolytic gene expression. These results highlight the importance tissue-specific corepressor interactions in maintaining liver metabolism and illustrate new pathways for therapeutic intervention.
Methods
Animal studies
Mice were housed in a temperature-controlled specific-pathogen-free facility under 12 h light/dark cycles (lights on at 0700 hours, off at 1900 hours). Adult male mice between the ages of 10–16 weeks old were used in all experiments unless otherwise indicated. AAV8 TBG Cre was intravenously injected at 1.5 × 1011 GC per mouse to induce hepatocyte-specific gene knockout, using AAV8 TBG GFP as a negative control. For MS experiments AAV8 TBG HDAC3-HA or control AAV8 TBG HA-EGFP vectors were injected at 5 × 1010 GC per mouse in combination with either AAV8 TBG Cre or AAV TBG GFP, respectively. All mice in an experiment received equal total dosages of AAV vectors by supplementing with AAV8 TBG empty or AAV8 TBG GFP vectors where appropriate. All mice were killed by CO2 inhalation followed by tissue harvest at 2 weeks after viral injection at ZT10 except indicated otherwise. All animal procedures followed the guidelines of the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Constructs and viral vectors
AAV8 TBG viral constructs for the expression of HA-EGFP and HDAC3-HA were generated by PCR cloning and Gibson Assembly. All constructs were sequence verified by Sanger sequencing at the Penn Genomic Analysis Core. Adeno-associated viruses were produced and purified by the University of Pennsylvania Vector Core. AAV-based knockdown vectors were generated through Gibson cloning of Egfp and the UltramiR mir-30 scaffold64 into a modified AAV8 TBG vector containing a downstream Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element (WPRE). Two target sequences used for Prox1 knockdown, determined using the shERWOOD algorithm64, were: (TTCAGAGCAGGATGTTGAATA) and (GAGAAGGCAGCAACAAAGAAA). The control shRNA sequence targeting Luciferase was (CGCTGAGTACTTCGAAATGTC). For in vivo knockdown experiments mice received 1 × 1012 GC per mouse of each Prox1 targeting virus or 2 × 1012 GC per mouse of the control shLuciferase virus. For enhancer luciferase reporter vectors, enhancers for G0s2 were selected by comparing H3K27Ac and GRO-seq tracks (GEO Accession number GSM1437738)43 with binding sites for HDAC3, PROX1, and HNF4α. Enhancer regions of 200 bp were cloned into the MCS of the pGL4 luciferase reporter system (Promega). pRL-SV40 renilla was used as a control for firefly luciferase expression. Primers used in this study can be found in Supplementary Data 4.
Liver triglyceride measurement and oil red O staining
For measuring triglyceride, livers were lysed in lysis buffer (140 mM NaCl, 50 mM Tris, and 1% Triton X-100, pH 8) followed by triglyceride assay using LiquiColor kit (Stanbio). Oil red O staining was performed by the University of Pennsylvania Center for Molecular Studies in Digestive and Liver Disease Molecular Pathology & Imaging Core.
Cell culture and luciferase assay
HEK 293T cells purchased from ATCC were maintained in DMEM medium containing 10% FBS and 100 µg ml−1 Penicillin/streptomycin at 5% CO2 and 37 °C. Transfections of plasmids for the expression of proteins and luciferase reporter constructs were performed using Fugene 6 (Roche). For luciferase assays, after 18–24 h of transfection, cells were washed once with PBS followed by lysis in passive lysis buffer (Promega). Lysates were analyzed for firefly and control renilla luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions and read on a microplate reader equipped with a dual-injection system (BioTek).
Immunoprecipitation and western blotting
For western blot analysis of total lysates, samples were lysed in a TissueLyser (Qiagen) in radioimmunoprecipitation assay buffer (RIPA buffer) supplemented with complete EDTA-free protease inhibitor (Roche) and 1 mM PMSF. Samples were resolved by Tris-glycine SDS-PAGE (Biorad), transferred to nitrocellulose membrane (Biorad), and blotted with the indicated antibodies. Antibodies for western blotting were anti-PROX1 (Millipore, 07-537, 1:1000), anti-HDAC3 (GeneTex, GTX113303, 1:1000), anti-HNF4α (Santa Cruz, sc-8987, 1:500), anti-HA 3F10 High Affinity (Roche, 12013819001, 1:5000), anti-FLAG M2 HRP (Sigma, A8592, 1:10,000), anti-Actin HRP (Santa Cruz, sc-1616 HRP, 1:5000), anti-HSP90 (Cell Signaling, 4874, 1:1000), and anti-rabbit HRP (Cell Signaling, 7074, 1:10,000). For immunoprecipitation followed by western blot livers were cross-linked with 2 mM dithiobis(succinimidyl propionate), DSP (Pierce) in PBS at room temperature for 30 min, quenched with glycine, washed with ice cold PBS, and subsequently lysed in RIPA buffer containing protease inhibitors and PMSF. Samples were pre-cleared with protein A sepharose CL-4B (GE Healthcare), and incubated with either anti-HA agarose (Sigma, A2095) or with anti-HDAC3 (GeneTex, GTX113303, 10 μg) or anti-PROX1 (Millipore, 07-537, 10 μg) antibodies and captured with TrueBlot anti-Rabbit IgG agarose (Rockland). Immunoprecipitates were washed five times with RIPA and eluted with SDS loading dye. For in vitro coimmunoprecipitation analysis, HEK 293T cells were transfected with pcDNA FLAG-NCOR1 or PROX1 vectors with Fugene 6 (Promega). For domain mapping of PROX1 interaction with HDAC3, pcDNA FLAG-tagged PROX1 mutants, HDAC3-HA and EGFP were transfected as indicated. At 72 h after transfection, cells were washed with PBS and lysed in IP buffer (50 mM Tris 7.4, 150 mM NaCl, and 0.5% NP-40) containing protease inhibitors and 1 mM PMSF. Samples were pre-cleared with protein A sepharose CL-4B (GE Healthcare), and incubated with anti-FLAG M2 resin (Sigma, A2220). Immunoprecipitates were washed five times with IPLS and eluted with SDS loading dye for western blot analysis. Uncropped scans of western blots are shown in Supplementary Fig. 10.
Mass spectrometry
Livers from Hdac3 fl/fl animals transduced with AAV8 TBG HDAC3-HA and AAV8 TBG Cre or AAV8 TBG HA-EGFP and AAV8 TBG EGFP were harvested after 2 weeks of infection. A 250 mg sample of liver was dissected, dounced 10 strokes with pestle A in 15 ml swelling buffer (10 mM Tris-HCl pH 7.4, 2 mM MgCl2, 3 mM CaCl2) supplemented with 1 mM PMSF, and incubated for 20 min on ice. Swelled cells were dounced 15 strokes with pestle B, an additional 15 ml swelling buffer with PMSF was added, filtered through a 100 µm cell strainer, and spun at 400×g for 10 min to collect cells. Cells were resuspended in 5 ml swelling buffer with 10% glycerol and PMSF and an additional 5 ml of swelling buffer supplemented with 10% glycerol, 1% NP-40, and 1 mM PMSF was added slowly while vortexing. Following lysis, nuclei were washed 1 time with PBS and subsequently fixed with 1% formaldehyde diluted in PBS for 15 min at room temperature. Cross-linked samples were quenched with glycine, washed once with PBS, and resuspended in 1 ml NCB (20 mM Tris-HCl pH 8, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 1 mM DTT) supplemented with complete EDTA-free protease inhibitor tablet (Roche) and 1 mM PMSF. Lysis was allowed to proceed for 1 h followed by sonication with a probe sonifier (Branson). Cross-linked chromatin extracts were cleared by centrifugation and where indicated samples were treated with 1000 Kunitz units of either micrococcal nuclease (NEB) or Benzonase nuclease (Sigma) for 30 min at 30 °C. Extracts were immunoprecipitated with anti-HA agarose (Sigma, A2095) overnight, washed three times in NCB supplemented with protease inhibitors and 1 mM PMSF, washed two times in NCB, washed two times in HPLC-grade water, eluted with 10% ammonium hydroxide diluted in HPLC-grade water, and dried to completion in a SpeedVac (Eppendorf).
Samples were prepared for MS65. Briefly, after reduction/alkylation, samples were digested with LysC (Wako) for 2 h followed by Trypsin (Promega) at 37 °C overnight. Samples were resuspended in 1% acetic acid, and desalted with C18 stage tips (Empore, 2215). EASY-nanoLC (Thermo Fisher Scientific) was configured with a 75 µm ID × 17 cm Reprosil-Pur C18-AQ (3 µm; Dr Maisch GmbH, Germany) nano-column and coupled with an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). Full scan MS spectrum (m/z 360−1600) was performed in the Orbitrap with a resolution of 120,000 (at 200 m/z). Fragmentation was performed with higher energy collisional dissociation (HCD) and a maximum injection time of 120 ms. MS/MS data were collected in centroid mode in the ion trap mass analyzer. Peptides were identified using MaxQuant (v1.5.3.30) using the Mus Musculus UniProt FASTA database (March 2016, Q921N8 added) and MaxQuant contaminants FASTA with an FDR < 1% at the peptide spectrum match and protein levels. Protein abundance was deconvoluted from peptide intensity using the intensity-based absolute quantification (iBAQ)24 algorithm, followed by log2 transformation, normalization by the median intensity, and missing values imputed (width 0.25, downshift 2.0) using Perseus (v1.5.5.3) (Supplementary Data 1). Significance was estimated using a two-tailed homoscedastic t-test (P-value < 0.05).
RT-qPCR and RNA-seq
Total RNA was extracted using TRIzol (Invitrogen) and RNeasy mini kit (Qiagen). Contaminating DNA was removed using the RNase-Free DNase Set (Qiagen) on-column digestion protocol per manufacturer’s instructions. RT-qPCR was performed with the High Capacity RT kit (Thermo Fisher Scientific), Power SYBR Green PCR Master Mix (Thermo Fisher Scientific), and a QuantStudio™ 6 Flex Real-Time PCR System (Thermo Fisher Scientific) using absolute quantification method with standard curves. 36b4 (Arbp) was used as housekeeping control. RNA-seq libraries were generated using the Tru-seq kit (Illumina). Raw reads were aligned to the mm9 reference genome using Tophat version 2.1.9 using the parameters recommended by the original author. Differential expression analysis was performed using CuffDiff 2 using default parameters.
ChIP-qPCR, ChIP–reChiP, and ChIP-seq
For ChIP and ChIP-seq6, 66, livers were mildly dissociated by dounce with pestle A for six strokes in PBS containing 1% formaldehyde and rocked for 15 min, quenched with glycine, washed with PBS, and sonicated with a probe-type sonifier (Branson) in RIPA supplemented with protease inhibitors and PMSF. Sonicated extracts were immunoprecipitated with antibodies for HDAC3 (Abcam, ab7030, 10 μg), PROX1 (Millipore, 07-537, 10 μg), HNF4α (Santa Cruz, sc-8987, 10 μg), or H3K27Ac (Abcam, ab4729, 2 μg), and captured with bovine serum albumin blocked CL-4B protein-A sepharose beads (GE). PROX1 ChIP-seq was validated by comparing results of ChIP-seq with two different PROX1 antibodies raised against separate epitopes (Millipore, 07-537 and Proteintech, 51043-1-AP), which showed a highly significant Pearson correlation (R = 0.96). The HNF4α and HDAC3 antibodies used for ChIP in this study have been validated in the literature67, 68.
ChIP–reChIP from three biological replicate livers was performed essentially as standard ChIP, except following the first IP chromatin-protein complexes were eluted in 1% SDS with 10 mM DTT for 15 min at 65 °C. Subsequent to elution of the first ChIP, complexes were re-diluted in 10 volumes RIPA supplemented with protease inhibitors (Roche), PMSF (Sigma), 5 mg ml−1 BSA, and 2 μg Lambda DNA/HindIII (NEB), followed by a second ChIP with the either anti-rabbit IgG (Cell Signaling Technologies, #2729) or the indicated antibody.
For ChIP-seq, ChIP was performed independently on livers from different mice (n = 3). The precipitated DNA samples were barcoded and amplified according Illumina guide protocols, followed by deep sequencing on a HiSeq 2000 (Illumina). Sequencing reads of biological replicates for PROX1 and HDAC3 were aligned to the mm9 genome using Bowtie v0.12.7. Duplicate reads were removed and replicates were pooled using HOMER v4.7. Genome browser tracks were generated and peaks were called using HOMER with default parameters and IgG as input. Peaks more than threefold over input and >2 rpm in PROX1 and >1.5 rpm in HDAC3 were used for further downstream analyses. Venn diagram was generated using bedtools v2.26.0 and Vennerable R package with peaks overlapping at least 50% and having at least 1 rpm in the other ChIP-seq. Motif analyses were performed with HOMER using 200 bp peak windows. Gene ontology was performed using Reactome 2016. Track visualization was performed using the Integrated Genomics Viewer (Broad Institute).
Sequencing reads of biological replicates for PROX1, HDAC3, and HNF4α in Hnf4α fl/fl and Hnf4α conditional liver knockout animals were processed similarly. Previously published C/EBPα ChIP-seq (GEO Accession numbers GSM1816821 and GSM1816822, replicates combined)69 and C/EBPβ ChIP-seq (GEO Accession number GSM1446070)66 were re-processed using the same parameters. Peaks >1 rpm for PROX1 and HDAC3 and >2 rpm for HNF4α were used for further downstream analyses. Scatter plots and box plots were generated using the HOMER annotatePeaks command and R.
Statistical methods
Data are presented as mean ± s.d. unless otherwise stated. Microsoft Excel software, GraphPad Prism 7, MathWorks MATLAB, or R was utilized for all graphing and statistical tests. For comparison between two groups, two-tailed unpaired Student’s t-test were utilized unless otherwise stated, where P < 0.05 was considered statistically significant and the significance is marked by *P < 0.05, **P < 0.01, and ***P < 0.001 unless otherwise noted. Wilcoxon–Mann–Whitney test was performed for statistical comparison between HNF4α binding and C/EBPα or C/EBPβ binding in the HDAC3–PROX1 down peaks in Hnf4α liver KO vs.HDAC3–PROX1 unchanged peaks in Hnf4α liver KO (Supplementary Fig. 5c) and exact P-values were reported. For correlation analysis of HDAC3–PROX1 coregulated genes with HNF4α-dependent binding sites, a χ2 contingency table with Yates correction was used. All other statistical comparisons for two groups of peaks were done using Pearson’s χ2 contingency table tests and P-values were reported for each pair. All statistical tests are fully described in figure legends. The required sample size was calculated based on similar experiments and analyses carried out previously. The number of animals in each experiment is stated in the respective figure legends.
Data availability
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus70 and are accessible through GEO Series accession number GSE90533 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90533). All mass spectrometry data reported here have been deposited in Chorus under ID number 1251. All other data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Browning, J. D. & Horton, J. D. Molecular mediators of hepatic steatosis and liver injury. J. Clin. Invest. 114, 147–152 (2004).
Tilg, H., Moschen, A. R. & Roden, M. NAFLD and diabetes mellitus. Nat. Rev. Gastroenterol. Hepatol. 14, 32–42 (2016).
Cohen, J. C., Horton, J. D. & Hobbs, H. H. Human fatty liver disease: old questions and new insights. Science 332, 1519–1523 (2011).
Sladek, F. M., Zhong, W. M., Lai, E. & Darnell, J. E. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 4, 2353–2365 (1990).
Hayhurst, G. P., Lee, Y. H., Lambert, G., Ward, J. M. & Gonzalez, F. J. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell Biol. 21, 1393–1403 (2001).
Feng, D. et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 (2011).
Calkin, A. C. & Tontonoz, P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 13, 213–224 (2012).
Gross, B., Pawlak, M., Lefebvre, P. & Staels, B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 13, 36–49 (2016).
Xu, X., So, J.-S., Park, J.-G. & Lee, A.-H. Transcriptional control of hepatic lipid metabolism by SREBP and ChREBP. Semin. Liver Dis. 33, 301–311 (2013).
Wang, Y., Viscarra, J., Kim, S.-J. & Sul, H. S. Transcriptional regulation of hepatic lipogenesis. Nat. Rev. Mol. Cell Biol. 16, 678–689 (2015).
Knutson, S. K. et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 27, 1017–1028 (2008).
Sun, Z. et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 18, 934–942 (2012).
Hörlein, A. J. et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377, 397–404 (1995).
Chen, J. D. & Evans, R. M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377, 454–457 (1995).
Wen, Y. D. et al. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl Acad. Sci. USA 97, 7202–7207 (2000).
Guenther, M. G., Barak, O. R. R. & Lazar, M. A. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell Biol. 21, 6091–6101 (2001).
You, S.-H. et al. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat. Struct. Mol. Biol. 20, 182–187 (2013).
Guenther, M. G. et al. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14, 1048–1057 (2000).
Zhang, J., Kalkum, M., Chait, B. T. & Roeder, R. G. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9, 611–623 (2002).
Lazar, M. A. Nuclear receptor corepressors. Nucl. Recept. Signal. 1, e001 (2003).
Ishizuka, T. & Lazar, M. A. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell Biol. 23, 5122–5131 (2003).
Kulozik, P. et al. Hepatic deficiency in transcriptional cofactor TBL1 promotes liver steatosis and hypertriglyceridemia. Cell Metab. 13, 389–400 (2011).
Shimizu, H. et al. NCoR1 and SMRT play unique roles in thyroid hormone action in vivo. Mol. Cell Biol. 35, 555–65 (2015).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Wigle, J. T. & Oliver, G. Prox1 function is required for the development of the murine lymphatic system. Cell 98, 769–778 (1999).
Wigle, J. T., Chowdhury, K., Gruss, P. & Oliver, G. Prox1 function is crucial for mouse lens-fibre elongation. Nat. Genet. 21, 318–322 (1999).
Sosa-Pineda, B., Wigle, J. T. & Oliver, G. Hepatocyte migration during liver development requires Prox1. Nat. Genet. 25, 254–255 (2000).
Burke, Z. & Oliver, G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech. Dev. 118, 147–155 (2002).
Risebro, C. A. et al. Prox1 maintains muscle structure and growth in the developing heart. Development 136, 495–505 (2009).
Kivelä, R. et al. The transcription factor Prox1 is essential for satellite cell differentiation and muscle fibre-type regulation. Nat. Commun. 7, 13124 (2016).
Steffensen, K. R. et al. Functional conservation of interactions between a homeodomain cofactor and a mammalian FTZ-F1 homologue. EMBO Rep. 5, 613–619 (2004).
Qin, J. et al. Prospero-related homeobox (Prox1) is a corepressor of human liver receptor homolog-1 and suppresses the transcription of the cholesterol 7-alpha-hydroxylase gene. Mol. Endocrinol. 18, 2424–2439 (2004).
Song, K.-H., Li, T. & Chiang, J. Y. L. A Prospero-related homeodomain protein is a novel co-regulator of hepatocyte nuclear factor 4alpha that regulates the cholesterol 7alpha-hydroxylase gene. J. Biol. Chem. 281, 10081–10088 (2006).
Charest-Marcotte, A. et al. The homeobox protein Prox1 is a negative modulator of ERR{alpha}/PGC-1{alpha} bioenergetic functions. Genes Dev. 24, 537–542 (2010).
Dufour, C. R. et al. Genomic convergence among ERRα, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 7, e1002143 (2011).
Takeda, Y. & Jetten, A. M. Prospero-related homeobox 1 (Prox1) functions as a novel modulator of retinoic acid-related orphan receptors α- and γ-mediated transactivation. Nucleic Acids Res. 41, 6992–7008 (2013).
Lengler, J. et al. Antagonistic action of Six3 and Prox1 at the gamma-crystallin promoter. Nucleic Acids Res. 29, 515–526 (2001).
Chin, C.-H. et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8, S11 (2014).
Wang, Y. et al. The g0/g1 switch gene 2 is an important regulator of hepatic triglyceride metabolism. PLoS ONE 8, e72315 (2013).
Matsuzaka, T. et al. Elovl6 promotes nonalcoholic steatohepatitis. Hepatology 56, 2199–2208 (2012).
Berger, J. H., Charron, M. J. & Silver, D. L. Major facilitator superfamily domain-containing protein 2a (MFSD2A) has roles in body growth, motor function, and lipid metabolism. PLoS ONE 7, e50629 (2012).
Matsusue, K. et al. Hepatic steatosis in leptin-deficient mice is promoted by the PPARγ Target Gene Fsp27. Cell Metab. 7, 302–311 (2008).
Fang, B. et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159, 1140–1152 (2014).
Bantscheff, M. et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 29, 255–265 (2011).
Joshi, P. et al. The functional interactome landscape of the human histone deacetylase family. Mol. Syst. Biol. 9, 1–21 (2013).
Mohammed, H. et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 3, 342–349 (2013).
Chaya, D. & Zaret, K. S. Sequential chromatin immunoprecipitation from animal tissues. Methods Enzymol. 376, 361–372 (2003).
Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Bararia, D. et al. Acetylation of C/EBPα inhibits its granulopoietic function. Nat. Commun. 7, 10968 (2016).
Bricambert, J. et al. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Invest. 120, 4316–4331 (2010).
Lu, Q., Hutchins, A. E., Doyle, C. M., Lundblad, J. R. & Kwok, R. P. S. Acetylation of cAMP-responsive element-binding protein (CREB) by CREB-binding protein enhances CREB-dependent transcription. J. Biol. Chem. 278, 15727–15734 (2003).
Lai, F., Gardini, A., Zhang, A. & Shiekhattar, R. Integrator mediates the biogenesis of enhancer RNAs. Nature 525, 399–403 (2015).
Gardini, A. et al. Integrator regulates transcriptional initiation and pause release following activation. Mol. Cell 56, 128–139 (2014).
Kulemzina, I. et al. A reversible association between smc coiled coils is regulated by lysine acetylation and is required for cohesin association with the DNA. Mol. Cell 63, 1044–1054 (2016).
Euskirchen, G. M. et al. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7, e1002008 (2011).
Whyte, W. A. et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221 (2012).
Wang, Z. et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 (2009).
Dyer, M. A., Livesey, F. J., Cepko, C. L. & Oliver, G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat. Genet. 34, 53–58 (2003).
Harvey, N. L. et al. Lymphatic vascular defects promoted by Prox1 haploinsufficiency cause adult-onset obesity. Nat. Genet. 37, 1072–1081 (2005).
Remsberg, J. R. et al. Deletion of histone deacetylase 3 in Adult beta cells improves glucose tolerance via increased insulin secretion. Mol. Metab. 6, 30–37 (2016).
Dupuis, J. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42, 105–116 (2010).
Lecompte, S. et al. Genetic and molecular insights into the role of PROX1 in glucose metabolism. Diabetes 62, 1738–1745 (2013).
Knott, S. R. V. et al. A computational algorithm to predict shRNA potency. Mol. Cell 56, 796–807 (2014).
Armour, S. M. et al. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol. Cell Biol. 33, 1487–1502 (2013).
Lim, H. W. et al. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 25, 836–844 (2015).
Odom, D. T. et al. Control of pancreas and liver gene expression by HNF transcription factors. Science 303, 1378–1381 (2004).
Papazyan, R. et al. Physiological suppression of lipotoxic liver damage by complementary actions of HDAC3 and SCAP/SREBP. Cell Metab. 24, 863–874 (2016).
Bauer, R. C. et al. Tribbles-1 regulates hepatic lipogenesis through posttranscriptional regulation of C/EBPα. J. Clin. Invest. 125, 3809–3818 (2015).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Acknowledgements
We thank K. Kaestner (University of Pennsylvania) for providing the Hnf4α fl/fl mice. We acknowledge the Functional Genomics Core and the Viral Vector Core of the Penn Diabetes Research Center (P30DK19525) for next-generation sequencing and virus production. We thank the Molecular Pathology and Imaging Core of the University of Pennsylvania (P30DK050306) for histology. We thank C.L. Lanzillotta, X.J. Cao, Z. Sun, M. Emmett, K. Kulej, P.M. Titchenell, D. Steger, and D. Cohen for technical support and insightful discussions on the project. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants F32DK102284 (to S.M.A.) and R37DK43806 (to M.A.L.), and National Institute of General Medical Sciences Grants T32GM008275 (to J.R.R.) and R01GM110174 (to B.A.G.).
Author information
Authors and Affiliations
Contributions
S.M.A. and J.R.R. planned and conducted the experiments. M.D. and Z.L. provided additional bioinformatics analyses. W.Y.H. performed additional experiments and provided technical support. S.S. and B.A.G. provided mass spectrometry instrumentation and support. S.M.A., J.R.R., and M.A.L. designed the study, analyzed and interpreted the data, and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
M.A.L. is a member of the Scientific Advisory Boards of KDAC, Eli Lilly and Company, and Pfizer, Inc. The remaining authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Armour, S.M., Remsberg, J.R., Damle, M. et al. An HDAC3-PROX1 corepressor module acts on HNF4α to control hepatic triglycerides. Nat Commun 8, 549 (2017). https://doi.org/10.1038/s41467-017-00772-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-017-00772-5
This article is cited by
-
Nuclear receptor corepressors non-canonically drive glucocorticoid receptor-dependent activation of hepatic gluconeogenesis
Nature Metabolism (2024)
-
Caloric restriction remodels the hepatic chromatin landscape and bile acid metabolism by modulating the gut microbiota
Genome Biology (2023)
-
The role of HDAC3 and its inhibitors in regulation of oxidative stress and chronic diseases
Cell Death Discovery (2023)
-
Coordinated transcriptional upregulation of oxidative metabolism proteins in long-lived endocrine mutant mice
GeroScience (2023)
-
Transcriptional control of energy metabolism by nuclear receptors
Nature Reviews Molecular Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.