Abstract
Therapies based on immune cells have been applied for diseases ranging from cancer to diabetes. However, the viral and electroporation methods used to create cytoreagents are complex and expensive. Consequently, we develop targeted mRNA nanocarriers that are simply mixed with cells to reprogram them via transient expression. Here, we describe three examples to establish that the approach is simple and generalizable. First, we demonstrate that nanocarriers delivering mRNA encoding a genome-editing agent can efficiently knock-out selected genes in anti-cancer T-cells. Second, we imprint a long-lived phenotype exhibiting improved antitumor activities into T-cells by transfecting them with mRNAs that encode a key transcription factor of memory formation. Third, we show how mRNA nanocarriers can program hematopoietic stem cells with improved self-renewal properties. The simplicity of the approach contrasts with the complex protocols currently used to program therapeutic cells, so our methods will likely facilitate manufacturing of cytoreagents.
Similar content being viewed by others
Introduction
Therapeutic methods based on immune cells have experienced a substantial metamorphosis from interventions involving straightforward blood transfusions and bone marrow transplants into a nascent healthcare industry. Currently, over 500 companies are involved in the development and commercialization of cell-based therapeutic products1, and hematopoietic stem cell (HSC) transplants have evolved into the standard-of-care for treating leukemia and other bone and blood cancers (with over one million transplants performed worldwide to date2). But also, different sorts of cell therapy products are undergoing clinical evaluation for treating a variety of diseases, including tissue degeneration, chronic inflammation, autoimmunity, genetic disorders, cancer, and infections3,4,5,6,7,8. It has become possible to focus immune responses towards these diseases by genetically engineering T-cells to express targeted chimeric antigen receptors (CARs) or T cell receptors (TCRs), and this approach has presented positive clinical responses in cancer patients who have no other curative options9, 10. Thanks to a strong clinical presence, the expanding array of cell therapy products has catalyzed the field of cellular bioengineering with the goal of maximizing the therapeutic performance of these cytoreagents in patients11, 12.
Some gene therapy applications require chronic expression systems that stably integrate the engineered transgene into the patient’s DNA. One example is the expression of cancer-specific receptor genes by T-cells, which converts them into ‘living drugs’ that can increase in number while they serially destroy tumor cells9, 10. Another is the introduction of gamma-globin genes into transplanted HSCs as a way to reverse beta thalassemia13. Despite the time and cost required for their production, as well as restrictions on the size and number of genes that they can package, viral vectors are currently the most effective means to stably express these transgenes14, 15.
It is also possible to elicit phenotypic changes via transient expression of macromolecules, designed to accomplish “hit-and-run” genetic programming. In most of these kinds of applications, permanent expression of the therapeutic transgene is undesirable and potentially dangerous16. Examples include the use of transcription factors to control cell differentiation17, 18, and the expression of sequence-specific nucleases to engineer genomes19.
Although there is a growing number of applications where transient gene therapy could improve the curative potential of engineered cells, currently available methods (which, like the chronic expression methods described above, are mostly based on viral vectors) are complicated by the time and expense involved in the elaborate protocols required for their implementation20. Electroporation is an alternative transfection method, but physical permeabilization of plasma membranes compromises cell viability, which means these approaches are not suited for scale-up applications. Besides, like virus-based methods, electroporation cannot selectively transfect specific cell types from a heterogeneous pool, so it must be preceded by a cell purification process.
Here, we describe a nanoreagent that, via a comparatively simple process, produces transient gene expression in cultured cells. We demonstrate that an appropriately designed messenger RNA (mRNA) nanocarrier can accomplish dose-controlled delivery of functional macromolecules to lymphocytes or HSCs simply by mixing the reagent with the cells in vitro (Fig. 1a). These nanoparticles (NPs) can be designed to target particular cell subtypes and, upon binding to them, stimulate receptor-mediated endocytosis, thereby introducing the synthetic mRNA they carry which the cells can now express. Because nuclear transport and transcription of the transgene are not required, this process is fast and efficient. Here, we illustrate in three examples how this new platform can be implemented to manufacture effective cell products for clinical use.
Creating mRNA nanoparticles to program therapeutic T-cells. a Schematic explaining how cultured T-cells can be programmed to express therapeutically relevant transgenes carried by polymeric nanoparticles (NPs). These particles are coated with ligands that target them to specific cell types, enabling them to introduce their mRNA cargoes and cause the targeted cells to express selected proteins (like transcription factors or genome-editing agents). b Design of targeted mRNA-carrying NPs. The inset shows a transmission electron micrograph of a representative NP; scale bar, 50 nm. Also depicted is the synthetic mRNA encapsulated in the NP, which is engineered to encode therapeutically relevant proteins
In the first case, we used mRNA nanocarriers to edit the genome of T-cells and established that targeted delivery of mRNA encoding a rare-cleaving megaTAL nuclease21 into lymphocytes can efficiently disrupt their expression of T cell receptors. In the second application, we transiently expressed Foxo1 to reprogram the differentiation of effector cells into functionally competent memory cells22, 23. Our results demonstrate that engineered NPs can bias CAR-T-cells toward a central memory phenotype.
To assure that our transient gene delivery platform can easily be integrated into existing protocols for manufacturing other therapeutic cell types, we engineered our third example in HSCs. For many decades, these have been successfully used for treating hematological and immune diseases2. However, their limited number, especially when isolated from umbilical cord, prevents broader application of HSC-based therapies. Attempts to propagate these cells in vitro have generally failed, primarily because self-renewal and in vivo regenerative capacity are rapidly lost in culture24, 25. We used our nanotechnology platform to deliver transgenes into cultured HSCs, and demonstrated that transient expression of a key “stemness” regulator (Musashi-2 protein26) increases their capacity for self-renewal and maintains therapeutically desirable properties in the stem cells.
The most significant benefit of our system is its simplicity in achieving genetic modifications of therapeutic cells at a clinical scale: all that is required is mixing the appropriate NP reagent with the cultured cells. Our approach patently contrasts with those currently used to transiently deliver genetic materials, which are less effective and involve many expensive and proprietary procedures that limit their availability. Beyond the T-cells and HSCs tested in our experiments, the technology described here could be adapted to improve the curative potentials of other cell types used in the clinic to treat disease (e.g., natural killer cells, regulatory T-cells, dendritic cells, or mesenchymal stem cells) without increasing handling time, risk, or complexity.
Results
Designing mRNA nanocarriers to choreograph gene expression
To create a reagent that can genetically modify primary T lymphocytes (which are notoriously refractory to non-viral transfection methods) simply by contact, we bioengineered polymeric NPs comprised of four functional components (Fig. 1b): (i) surface-anchored targeting ligands that selectively bind the NPs to T-cells and initiate rapid receptor-induced endocytosis to internalize them. In our experiments we used anti-CD3 and anti-CD8 antibodies; (ii) a negatively charged coating that shields the NPs to minimize off-target binding by reducing the surface charge of the NPs. Because it is already widely used in drug delivery platforms, we selected polyglutamic acid (PGA) to accomplish this; (iii) a carrier matrix that condenses and protects the nucleic acids from enzymatic degradation while they are in the endosome, but releases them once the particles are transported into the cytoplasm, thereby enabling transcription of the encoded protein. For this, we used a biodegradable poly(β-amino ester) (PBAE) polymer formulation that has a half-life between 1 and 7 h in aqueous conditions; and (iv) nucleic acids that are encapsulated within the carrier and produce gene editing or transient expression of proteins that can permanently alter the phenotype of the T-cell. mRNA is an ideal platform for transient therapeutic protein expression, because it has no potential for genomic integration and does not require nuclear localization for expression. However, unmodified mRNA can activate intracellular toll-like receptors, limiting protein expression and leading to toxicity27. To improve the stability and reduce the immunogenic potential of the mRNA we deliver, we used synthetic versions that incorporate modified nucleotides. For example, substitution of uridine and cytidine with the engineered bases pseudouridine and 5-methyl-cytidine synergistically blocks identification by innate pattern recognition receptors and increases mRNA translation28.
The NPs were manufactured utilizing a two-step, charge-driven self-assembly process. First, the synthetic mRNA was complexed with a positively-charged PBAE polymer, which condenses the mRNA into nano-sized complexes (Supplementary Fig. 1a, b). This step was followed by the addition of antibody-functionalized PGA, which shields the positive charge of the PBAE-mRNA particles and confers lymphocyte-targeting. The resulting mRNA nanocarriers had a size of 109.6 ± SE/26.6 nm and an almost neutral surface charge (1.1 ± SE/5.3 mV zeta potential, Supplementary Fig. 1c).
T-cell viability following mRNA nanocarrier transfection
Our goal is to streamline the manufacture of cell-based therapies, so we first tested whether simply adding targeted mRNA nanocarriers to an established culture of human lymphocytes is sufficient to choreograph robust transfection in them. We found that when CD3-targeted NPs carrying mRNA encoding a reporter (enhanced green fluorescent protein, eGFP) are incubated with these cells, they not only bind to them but also stimulate receptor-mediated endocytosis, providing entry for the genes they carry (Fig. 2a). Following a single NP application (NP:T cell ratio = 2 × 104:1), we routinely transfected >80% of these primary T-cells (Fig. 2b, c), with transgene expression observed as early as 5 h post-transfection (Supplementary Fig. 2a–c). Thus, this process is fast and efficient. Importantly, it was not necessary to prepare mRNA NPs freshly for each application—it was possible to lyophilize them before use with no change in properties or efficacy (Supplementary Fig. 2d). We also found that CD3-targeted nanoparticles selectively bind T lymphocytes, as their interactions with off-target cells were low (Fig. 2b, c; Supplementary Fig. 3). T cell proliferation did not impair NP-uptake/transfection, as (i) gene transfer into naive versus effector T-cells was comparable (Supplementary Fig. 4a, b), and (ii) addition of nanoparticles to stimulated T-cells at the peak of their expansion (day 17) yielded similar transfection relative to adding nanoparticles to freshly stimulated cells (day 5; Supplementary Fig. 4c).
mRNA nanoparticle transfection choreographs robust transgene expression by lymphocytes. a Primary T-cells were mixed with CD3-targeted polymeric nanoparticles (NPs) carrying Cy5-labeled mRNA. Confocal microscopy establishes that these particles are rapidly internalized from the cell surface. The images are representative of 15 randomly chosen fields. Scale bars, 2 μm. b Flow cytometry of preactivated PBMCs 24 h after incubation with CD3-targeted or isotype control antibody-targeted nanoparticles bearing eGFP-encoding mRNA. c Bar graph summarizing transfection efficiencies from three independent experiments conducted in duplicate. d, e Comparison of the effects electroporation and NP gene delivery have on cell expansion. Left panels show the workflow for transfection with NPs (top) and electroporation (bottom). Right panels show the -fold expansion of PBMC cultures from three independent donors treated with stimulatory beads on days 0 and 12. Matched cultures from each donor were not treated, or transfected using CD3/CD28-targeted NPs (d, right) or electroporation (e, right) on days 5 and 17. Every line represents one donor and each dot reflects the -fold T-cell expansion. Pairwise differences between groups were analyzed with the unpaired, two-tailed Student’s t test; n.s., non-significant; *, significant, n = 3). f Relative viability of NP-transfected and electroporated T-cells. Samples of 2 × 106 activated T-cells per condition were untreated, transfected with NPs, or electroporated. 18 h after treatment, cells were labeled with fluorescent dyes to assess viability. Results from three separate experiments conducted in duplicate are summarized in the bar graph shown in g. Statistical analysis between groups was performed using the unpaired, two-tailed Student’s t Test. *P < 0.0001
We next assessed the impact of targeted mRNA−carrying NPs on T-cell expansion. Because malignancies often progress quickly, it is important that engineered T-cells can be expanded to clinically relevant scales equally quickly. One widely used approach to multiply polyclonal lymphocytes is to incubate them with beads that are coated with antibodies against TCR/CD3 and co-stimulatory CD28 receptors. We found that even repeated transfections with CD3-targeted NPs did not interfere with T-cell expansion stimulated by these coated beads (Fig. 2d). These results contrast sharply with T-cell electroporation, which we tested side-by-side. Not only did electroporation add complex handling steps (Fig. 2e), it compromised viability of the lymphocytes (Fig. 2f, g) and reduced their yield by 60-fold (Fig. 2e, right panel).
Nanoparticle methods integrate into CAR-T cell manufacture
To test our approach in a clinically relevant application, we incorporated NP-mediated mRNA transfection methods into the manufacture of leukemia-specific 19-41BBζ CAR T-cells (Fig. 3a). CD19-targeted receptors are the most investigated CAR-T cell product today, with nearly 30 ongoing clinical trials internationally29. Our ability to perform genome engineering offers the potential to improve the safety and efficacy of CAR-T-cells. For example, we can inhibit expression of endogenous TCRs to avoid graft-versus-host disease, or selectively delete immune checkpoint genes in these cells to strengthen their anti-cancer activity in the suppressive tumor milieu30, 31. Here, we tested the ability of NPs to deliver gene-editing agents by preparing particles carrying mRNA encoding megaTAL nuclease, which targets the constant region (TRAC) of the TCR alpha gene. Taking advantage of the flexibility offered by our NP formulation methods, we included mRNA for the DNA repair endonuclease TREX2 to improve knockout efficiency32, along with eGFP mRNA so we could track transfection. Control particles were loaded with eGFP mRNA only. We found that, in contrast to eGFP-transfection (which did not impact TCR expression; Fig. 3b, top row), the addition of TCRα-megaTAL-carrying particles to the T-cell culture efficiently disrupted TCR expression by day 5, an effect that was maintained after loss of the mRNA by day 12 (Fig. 3b, bottom row). Average TCR knockout efficiency was 60.8% ( ± SE/17.7%; Fig. 3c), which corresponds with the percentage of indel frequencies (a measure of targeting efficiency) determined using the Surveyor assay (Fig. 3d). Importantly, the presence of mRNA-carrying NPs did not affect virus-mediated gene transfer of the tumor-specific CAR, as we achieved equal transduction efficiencies with a lentiviral vector encoding 19-41BBζ CAR in NP-transfected and non-transfected T-cells (Fig. 3e, f). Following NP-mediated genome editing and lentiviral transduction, CAR-programmed T-cells fully maintained their capacities to proliferate, secrete cytokines, and eliminate leukemia target cells (Fig. 3g–i). In summary, these findings establish that lymphocyte-targeted mRNA nanocarriers can mediate efficient genome editing of CAR-T-cells without compromising their function.
Nanoparticles can knockout T cell receptors in CAR-programmed lymphocytes. a Integration of nanoparticle (NP) transfection into normal manufacturing of CAR-T-cells. After stimulation with anti-CD3/CD28-coated beads (day 0), CD8-targeted mRNA NPs were introduced on days 1 and 2, then lentiviral transduction with a vector encoding the leukemia-specific 19-41BBz CAR was performed on day 3. We added either NPs carrying mRNAs encoding megaTAL nuclease plus eGFP, or control particles loaded with eGFP mRNA alone. b Flow cytometry of NP transfection efficiencies (based on eGFP signals) correlated with surface expression levels of TCRs (based on CD3 signals) by T-cells following NP treatments. c Summary plot showing editing efficiency as measured by loss of CD3 surface expression at day 14 (n = 6). d Surveyor assay confirming TCRα chain gene locus disruption. e Flow cytometry of lentiviral transduction in genome-edited versus control T-cells. f Bar graph showing mean viral transductions and SE of three independent experiments conducted in duplicate; n.s., not significant g, h Proliferation and cytolytic activity of TCR+ (FACS sorted TCR-positive, unedited 19-41BBz CAR-T-cells) and TCR- (FACS sorted TCR-negative, genome edited) 19-41BBz CAR-T-cells. To measure proliferation, T-cells were co-cultured on irradiated TM-LCL leukemia cells. Cytolytic assays were performed with CD19-expressing K562 target cells. i T cell IFN-γ release was measured with ELISA 48 h after stimulation on CD19+ TM-LCL leukemia cells or control LNCaP C4-2 prostate adenocarcinoma cells. Data from two experiments run in triplicate are shown
Nanoparticle programming for Foxo1 enhances CAR-T-cells
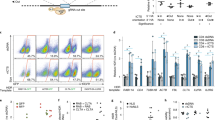
We next examined whether lymphocyte-targeted NPs can improve the therapeutic activity of CAR-T-cells by delivering mRNAs that program them toward a favorable phenotype. Clinical testing has already established that lymphocytes derived from CD62L+ central memory T-cells (TCM) present improved engraftment and function in animal models, and high fractions of CD62L+ TCM cells in infused products are linked to successful CAR treatment33, 34. However, to develop into therapeutically relevant numbers, these cells must undergo multiple rounds of in vitro stimulation/expansion—a process that drives cells away from the TCM lineage and toward terminal differentiation and senescence35. To address this problem, we manufactured T-cell-targeted NPs loaded with mRNA encoding the forkhead family transcription factor Foxo1, which controls the effector-to-memory transition in CD8 T-cells22, 23. During in vitro stimulation/expansion, TCR and cytokine signaling activate AKT kinase, which phosphorylates Foxo1 and thereby causes its cytoplasmic segregation and blocks its transcriptional activity. To maintain active Foxo1 levels in cultured T-cells, we used an AKT-insensitive nuclear retaining Foxo13A variant in which three key phosphorylated residues are mutated to alanine (Supplementary Fig. 5)36. We hypothesized that addition of Foxo13A-containing NPs to T cell culture medium during ex vivo expansion would promote the development of CD62L + TCM cells that have improved therapeutic potential. The effect transcription factors have on reprogramming is sensitive to the magnitude and duration of their expression. To determine these values after Foxo13A-NP addition, we measured Foxo1 protein and mRNA expression in the treated cells. In the Jurkat T-cell line, endogenous expression of Foxo1 is low and Foxo13A-NP treatment led to large increases in total expression of the factor, as measured by intracellular labeling (Fig. 4a, left panel). In primary T-cells, expression levels of Foxo1 protein is already high, and Foxo13A-NPs only led to modest increases. These findings indicate that NP treatment can maintain near-physiological levels of the active transcription factor (Fig. 4a, right panel). To examine mRNA dynamics after it was delivered into activated, proliferating T-cells by NPs, we measured the expression of Foxo13A using real-time quantitative PCR specific for the engineered mRNA. We found mRNA expression was maximal at day 1, and was close to baseline by 8 days post-transfection (Fig. 4b). Treatment with Foxo13A-NPs after T-cell priming in vitro rapidly increased expression of CD62L (Fig. 4c), which is the primary surface marker that distinguishes TCM from effector and effector memory populations37. To determine if transient expression of Foxo13A can lead to persistent alterations in CD8+ T-cell differentiation, cells from three independent donors were treated with CD8-targeted Foxo13A-eGFP NPs, sorted based on eGFP expression, and maintained in vitro. An increased frequency of CD62L+ cells was observed by 24 h after transfection, and this was maintained even 8 and 20 days after NP addition (Fig. 4d). Foxo13A overexpression did not induce T-memory stem cells, as the lymphocytes were universally CD45RO+ (Supplementary Fig. 6).
Nanoparticles can induce markers and transcriptional patterns characteristic of memory T-cells. a Expression of total Foxo1 protein measured by intracellular labeling in Jurkat and primary T-cells treated with CD3-targeted control (GFP+) or Foxo13A-GFP NPs. Mean Foxo1 fluorescence intensities (MFI) for cells transfected with control NPs (shown in blue) compared to Foxo13A-NPs (shown in red) are indicated above the respective histograms. b qPCR measurements of relative Foxo13A mRNA expression over time after cells were exposed to Foxo13A-eGFP nanoparticles (NPs). Data are representative of two independent experiments. Graphs show mean ± SE. c Effect of CD8-targeted Foxo13A-GFP NPs on CD62L expression after 24 h of particle treatment. d Percentage of CD62L+ cells in sorted CD8+ eGFP+ cells treated with CD8-targeting control or Foxo13A/eGFP-encoding NPs at 1, 8, and 20 days of culture after the particles were introduced. These results are from three independent donors. *P < 0.05; **P < 0.01 between the indicated conditions as calculated from a ratio-paired t-test. e, f Heat maps of TCM signature gene expression in TCM, naive, and control cells 8 days after treatment. g Volcano plot of differential gene expression in Foxo13A-NP-treated cells after 8 days. TCM signature genes and selected memory phenotype genes are indicated. P value of overlap between Foxo13A and the TCM signature gene set was determined by GSEA (via analysis shown in h)
To understand the genetic regulatory network induced by Foxo13A and its connections to the TCM lineage, we performed RNASeq on ex vivo-isolated naive CD8, TCM CD8, and in vitro-cultured CD8 T-cells that were treated with Foxo13A-encoding NPs or control particles. We identified a TCM signature consisting of the most frequent 500 genes that have lower (Fig. 4e) or higher (Fig. 4f) expression in TCM versus average CD8 T-cells. The majority of these genes are coordinately regulated in naive CD8 T-cells, which is consistent with the close transcriptional relationship we observed between naive and TCM cells38. Foxo13A-encoding NP treatments led to differential expression of many genes. As expected, these included those encoding the key memory transcriptional effector KLF2, and the surface molecules SELL (CD62L), CD28, and S1PR1, all crucial mediators of CD8 TCM trafficking and function (Fig. 4g)39, 40. Overlaying the TCM gene signature onto the Foxo13A volcano plot reveals a strong concordance of transcripts: this means TCM signature genes were up-regulated in Foxo13A-programmed CD8 cells. Gene set enrichment analysis confirmed the strong connection between Foxo13A-regulated transcripts and TCM-associated gene expression (Fig. 4h).
To test whether genetically imprinting a TCM phenotype in CAR-T-cells translates into improved antitumor efficacy, we studied the effect of Foxo13A-NP transfection on the in vivo activity of 19-41BBζ CAR-modified T-cells in a mouse model of B-cell lymphoma. Immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were inoculated with 5 × 105 CD19 + Raji cells expressing firefly luciferase. After 7 days, one treatment group received a single low-dose infusion of 2.5 × 106 CD8 + 19-41BBζ CAR T-cells, the second group was injected with the same dose of Foxo13A-NP transfected CAR T-cells, and controls received no treatment. After adoptive immunotherapy, we used bioluminescence imaging to serially quantify tumor growth and monitored overall survival. We found that low-dose treatments with conventional 19-41BBζ CAR-T-cells produced only a temporary delay in disease progression (29 compared with 23 days median overall survival in the untreated control group; Fig. 5a–c). By contrast, mice treated with Foxo13A-transfected 19-41BBζ CAR-T-cells showed substantial disease regression, with an average 11.3-fold reduced tumor burden compared to 19-41BBζ CAR-T-cells 2 weeks after therapy (Fig. 5b), which translated into a median 14-day improvement in survival (Fig. 5c).
Foxo13A-NP-transfection improves the anti-cancer activities of CAR T-cells. NSG mice were inoculated with CD19 + Raji-luc tumor cells. After 7 days the mice were injected with luciferin and imaged on an IVIS before being randomly sorted into groups (n = 9) with representative tumor burden. Next 2.5 × 106 CD8+ 19-41BBζ CAR + T-cells (transfected with NPs loaded either with Foxo13A mRNA or GFP mRNA) were infused intravenously. Control mice received no treatment. a Representative IVIS imaging depicting five mice per cohort. b Quantified tumor burden (as mean radiance from luciferase activity from each mouse from a ± SE). Pairwise differences between groups were analyzed with the unpaired, two-tailed Student’s t Test; n.s., non-significant; *, significant. c Kaplan–Meier survival curves for treated and untreated control mice. Shown are nine mice per treatment group pooled from two independent experiments. ms, median survival. Statistical analysis between the treated experimental and the untreated control group was performed using the Log-rank test; P < 0.05 was considered significant
In summary, these results indicate that Foxo13A-encoding NPs induce persistent alterations in surface markers, and transcriptional programming towards a TCM-like phenotype with improved antitumor function.
Nanoparticle-mediated transfection can promote HSC expansion
We further examined the generality of our approach by genetically engineering stem cells, focusing on the problem that integration of HSCs into clinical use is limited by an inability to expand them ex vivo without inducing differentiation24, 25. Several stem cell self-renewal genes have been identified, including homeobox B4 (HOXB4) and Musashi-2 (MSI2)26, 41. Unfortunately, it is unsafe to permanently integrate these transgenes into the genome of human HSCs, as the factors they encode can promote malignant progression42.
We first examined whether mRNA nanocarriers can perform specific transfection of CD34+ human HSCs without affecting viability or triggering differentiation. To accomplish this, we incubated CD34+ cells (that had been purified from cytokine-mobilized peripheral blood) with GFP mRNA-loaded NPs coated with a nonspecific antibody (control Ab-eGFP NPs) or anti-CD105 (α-CD105 NPs). We chose to target CD105 because this surface marker is specific for the most immature, long-term repopulating HSCs43, 44, and because it can mediate the internalization of bound material45. We found that α-CD105-coated NPs produced efficient transfection levels (mean 49% ± SE/6.9%; Fig. 6a, b; Supplementary Fig. 7), which were similar to those described for transductions of these cells using highly purified lentiviral vectors45. In contrast, transgene expression in HSCs treated with control NPs coated with nonspecific antibodies was low (mean 4.9% ± SE/0.9%). Importantly, NP-transfection did not affect viability or proliferative activity of expanded HSCs (Fig. 6c, d). We also could not measure phenotypic differences between NP-transfected and unmodified HSCs (Fig. 6e, f), which indicates that receptor-mediated internalization of bound NPs from the cell surface does not trigger undesirable differentiation of these hematopoietic progenitor cells. In line with these results, colony forming unit (CFU) assays revealed no difference in the ability of NP-transfected HSCs to form multi-lineage colonies when compared with unmodified HSCs (Fig. 6g, h).
Transfection of stem cells with mRNA nanoparticles can promote their expansion and self-renewal. a Targeting of CD105 enables specific transfection of HSC CD34+ cells. Cells were left untreated, or transfected with eGFP-encoding mRNA in nanoparticles (NPs) coated with PGA coupled to a control antibody or anti-CD105. Transfection efficiency was assayed by flow cytometry after 24 h. b NP transfection efficiency in CD34+ samples from three independent donors. Viability is shown in c. d Expansion of CD34 + PBSCs after NP-transfection. e Phenotypical characterization of PBSC-derived CD34+ subpopulations after 2 days in culture. Cells were either transfected with eGFP NPs on day 1 or left unmodified. Gating is indicated in brackets on top of each column. f Summary bar graph showing mean frequencies and SE of primitive Hematopoietic Stem Cells (HSCs), Multipotent Progenitors (MPPs), Lymphoid-primed Multipotent Progenitors (LMPs), and Early Myeloid Progenitors (EMPs). PBSCs from four independent donors were analyzed. Error bars represent mean ± SE. g Colony output of sort-purified GFP-NP transfected versus unmodified CD34+ cells from day 7 cultures (n = 3 cultures from independent donors); n.s., non-significant. Arising colonies were identified as colony forming unit (CFU) granulocyte (CFU-G), macrophage (CFU-M), granulocyte-macrophage (CFU-GM) and burst forming unit-erythrocyte (BFU-E). Colonies consisting of erythroid and myeloid cells were scored as CFU-MIX; n.s., non-significant. Error bars represent mean ± SE. h Representative images of CFU-MIX colonies from untransfected and GFP-NP-transfected CD34+ cells (×4-magnification; scale bar 1000 µm). i qPCR measurements of NP-delivered Musashi-2 (MSI2) mRNA expression over time. Error bars represent mean ± SE. j Comparison of CD133 and CD34 expression in HSCs transfected with control GFP mRNA-NPs versus MSI2 mRNA-NPs, assessed by flow cytometry 8 days after NP exposure and cell expansion. Data represent two independent experiments conducted in triplicate. k Cellular fold expansion of CD34− (differentiated) and CD34+ CD133+ (progenitor) cells. Bar graphs show mean and SE of three independent experiments. Data represent two independent experiments conducted in triplicate. l Colony forming unit outputs of untransfected versus MSI2-NP-transfected HSCs (n = 3 cultures from independent donors); Pairwise differences between groups were analyzed with the unpaired, two-tailed Student’s t Test. *P = 0.049, **P = 0.012, ***P = 0.011; n.s., non-significant. Error bars represent mean ± SE. m Representative images of colonies from untransfected and MSI2-NP-transfected CD34+ cells (scale bar, 300 µm)
To determine if transient expression of self-renewal genes directed by mRNA-carrying NPs can facilitate stem cell expansion, we loaded α-CD105-targeted NPs with synthetic mRNA encoding MSI2 and added them to cultures of HSCs. Using real-time quantitative PCR specific for the engineered (codon-optimized) MSI2 mRNA, we found that mRNA expression in HSCs was maximal at day 1, and was close to baseline by 7 days post-transfection (Fig. 6i). This transient NP-mediated upregulation of MSI2 increased the frequency and total numbers of CD34+ CD133+ cells by 2.6-fold and 2.3-fold, respectively, after 8 days of culture (Fig. 6j, k). At the same time, the outgrowth of differentiated CD34− HSC populations was reduced by two-fold (Fig. 6k). To confirm that MSI2 NP-transfection enhances the regenerative potential of human HSCs, we examined their clonogenic potential in colony forming cell (CFC) assays. NP-mediated expression of MSI2 increased the overall ability of HSCs to form multi-lineage colonies by an average 30% (Figs. 6l, m), principally due to a 67% increase in erythroid burst-forming units (BFU-E) and a 41% increase in the most primitive CFU-granulocyte erythrocyte monocyte megakaryocyte (CFU-MIX) colony types.
In summary, these results demonstrate that mRNA NPs that induce key regulators of self-renewal can accelerate in vitro growth and regenerative potential of primitive HSCs.
Discussion
As research findings produce new insights into the molecular mechanisms and master regulators that control cell fate and function, the list of genes potentially useful for rationally designing powerful disease-fighting cytoreagents is growing longer46. Unfortunately, the available gene therapy toolbox, which includes virus-mediated transduction approaches and physical electroporation methods, does not allow clinicians to apply the potential of genetic engineering without adding specialized protocols to an already complex and costly manufacturing process.
Here, we demonstrate using two therapeutic cell-based products–CAR-programmed T-cells and stem cells–that appropriately designed mRNA nanocarriers can transiently program gene expression to improve their therapeutic potential. We show how cell function and/or differentiation can be permanently reprogrammed by the simple addition of bioengineered NPs to cultures of cells used for therapy. This nanotechnology platform does not require special cell handling, so it can be easily integrated into established protocols for the manufacture of therapeutic cells without changing the workflow, or the equipment used in the process. Compared to RNA electroporation, which is currently the method of choice for ‘hit-and-run’ gene therapy in cell-based products47,48,49, this could provide a significant advantage in manufacturing. As depicted in Fig. 2, in addition to the expensive equipment involved, electroporation requires many culture medium exchanges, centrifugation steps, and washing cycles. Each of these procedures is prone to error, and increases the risk of contamination and compromising the final cell product. Even state-of-the-art flow electroporation devices reduce cell viability, and hence product yield and quality50, 51. Our approach does not rely on mechanical permeabilization of cell membranes to deliver transgenes. Instead, engineered NPs bind to target cells and stimulate receptor-mediated endocytosis – a physiological process that provides entry for the RNA they carry without compromising cell viability (Fig. 2).
Cell-penetrating peptides (CPPs), which are small proteins that facilitate cellular uptake of various molecular cargos, have also been used to transport therapeutics into cells52. Even large proteins that harbor CPP domains can be introduced into the cytoplasm using this approach. However, specific targeting of selected cell types is not possible with CPPs, and protein transfer is relatively inefficient53, 54. The duration of therapeutic impact also strongly depends on the half-life of the transferred protein. By contrast, nanocarriers loaded with synthetic mRNA are targeted to specified cells, and every RNA molecule they deliver serves as a template for the translation of multiple protein copies.
The surface molecules we targeted to deliver the NPs are just examples of the many antigens that could be used to selectively shuttle mRNA into cytoreagents. For instance, to selectively modify only defined T cell subsets, such as antigen-experienced lymphocytes, activation markers (e.g., CD25, 4-1BB, OX40, or CD40L) could be targeted. Similarly, antibodies recognizing CD34, CD133, or CD46 could be explored as targeting ligands for human HSCs. Also, the choices for the core polymer and the charge-negating coating material are flexible, and will likely be optimized before production in a clinical setting comes about. In terms of the former, our group tested a panel of cationic matrices, including hyperbranched STAR polymer, polyethylene glycol-grafted polyethylenimine, and mesoporous silica nanoparticles, and elected to use PBAE 447 based on its superior transfection efficacy and low cytotoxicity in primary cells. This compatibility is the result of the high biodegradability of this formulation, which has a half-life between 1 and 7 h in aqueous conditions55. That time frame is ideal for gene therapy, as the polymer condenses and protects the mRNA against degradation while it is encapsulated in the endosome, but releases it soon after transfer into the cytoplasm, thus enabling transcription of the encoded protein. Importantly, in all NP designs we tested, a negatively-charged coating was required to shield the positive charge of RNA/PBAE polyplexes and prevent off-target binding.
In summary, our project demonstrates that exposing pharmaceutical cells to an NP reagent can improve their therapeutic value by accomplishing ‘hit-and-run’ gene modification. This platform does not add complexity to manufacturing because it involves no special equipment or training. Thus, it can substantially streamline the manufacture of cell-based therapies at clinical scales, which means that treating patients with genetically engineered cells could become less expensive, and more effective as a disease-fighting intervention.
Methods
PBAE 447 synthesis
This polymer was synthesized using a method similar to that described by Mangraviti et al55. 1,4-butanediol diacrylate was combined with 4-amino-1-butanol in a 1.1:1 molar ratio of diacrylate to amine monomer. The mixture was heated to 90 °C with stirring for 24 h to produce acrylate-terminated poly(4-amino-1-butanol-co-1,4-butanediol diacrylate). 2.3 g of this polymer was dissolved in 2 ml tetrahydrofuran (THF). To form the piperazine-capped 447 polymer, 786 mg of 1-(3-aminopropyl)-4-methylpiperazine dissolved in 13 ml THF was added to the polymer/THF solution. The resulting mixture was stirred at RT for 2 h, then the capped polymer was precipitated with 5 volumes of diethyl ether. After the solvent was decanted, the polymer was washed with 2 volumes of fresh ether, then the residue was dried under vacuum for 2 days before it was formed into a stock of 100 mg/ml in DMSO, which was stored at −20 °C.
PGA-antibody conjugation
15 kD poly-glutamic acid (from Alamanda Polymers) was dissolved in water to form 20 mg/ml and sonicated for 10 min. An equal volume of 4 mg/ml 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (Thermo Fisher) in water was added, and the solution was mixed for 5 min at RT. The resulting activated PGA was then combined with antibodies at a 4:1 molar ratio in phosphate buffered saline (PBS) and mixed for 6 h at RT. To remove unlinked PGA, the solution was exchanged 3 times against PBS across a 50,000 NMWCO membrane (Millipore). Antibody concentrations were determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific). The antibodies we used for T cell experiments were anti-CD3 (clone OKT3, BioXCell; Cat# BE0001-2), anti-CD4 (clone OKT4, BioXCell; Cat# BE0003-2), anti-CD8 (clone OKT8, BioXCell; Cat# BE0004-2), and anti-CD28 (clone 9.3, BioXCell; Cat# BE0248). Clone C1.18.4 was used as a control antibody (BioXCell; Cat# BE0085). For HSC transduction, we used polyclonal goat anti-human CD105 (R&D Systems; Cat# AF1097) and non-specific polyclonal goat IgG antibodies (Biolegend; Cat# 400102). There were no dilutions for all above antibodies. We coupled them to polyglutamic acid at a 4:1 molar ratio.
mRNA synthesis
Codon-optimized mRNA for eGFP, Foxo1, Trex2, TRAC-megaTAL, and Musashi-2, which were fully substituted with the modified ribonucleotides pseudouridine (Ψ) and 5-methylcytidine (m5C) and capped with ARCA, were produced by TriLink Biotechnologies. We conjugated Ψ and m5C-modified eGFP mRNA with cy5 (also from TriLink) to track delivery of these transcripts.
Nanoparticle preparation
mRNA stocks were diluted to 100 µg/ml in 25 mM nuclease-free sodium acetate buffer, pH 5.2 (NaOAc). PBAE-447 polymer in DMSO was diluted to 6 mg/ml in NaOAc, and added to mRNA at a 60:1 (w:w) ratio. After the resulting mixture was vortexed for 15 s at medium speed, it was incubated for 5 min at room temperature so NPs could form. To add targeting elements to the NPs, PGA-linked antibodies were diluted to 250 µg/ml in NaOAc and added at a 2.5:1 (w:w) ratio to the mRNA. The resulting mixture was vortexed for 15 s at medium speed, and then incubated for 5 min at room temperature to permit binding of PGA-Ab to the NPs. The NPs were lyophilized by mixing them with 60 mg/ml D-sucrose as a cryoprotectant, and flash-freezing them in liquid nitrogen, before processing them in a FreeZone 2.5L Freeze Dry System (Labconco). The lyophilized NPs were stored at −80 °C until use. For application, lyophilized NPs were re-suspended in a volume of sterile water to restore their original concentration.
Nanoparticle characterization
The hydrodynamic radius of the particles we created was measured with a Nanosite (Malvern), and their zeta potential was determined using dynamic light scattering detected with a Zetapals instrument (Brookhaven Instrument Corporation). The particles were diluted 1:400 (v/v) in PBS (pH 7.4) for size measurements, and 1:40 for zeta potential quantitation. For transmission electron microscopy, a 25-μl sample of NPs was applied to a glow discharge-activated 200 mesh carbon/formvar-coated copper grid. After 30 s, grids were touched sequentially to a drop of ½ Karnovsky’s fixative, a drop of 0.1 M cacodylate buffer, 8 drops of dH2O, and then a drop of 1% (w/v) filtered uranyl acetate. These samples were examined using a JEOL JEM-1400 transmission electron microscope (JEOL USA).
Cell lines and culture media
K562-CD19 and control K562 cells were provided by Dr. Stanley Riddell (Fred Hutchinson Cancer Research Center). TM-LCL is a CD19+ EBV-transformed lymphoblastoid cell line that has been optimized to use as a feeder for T cell expansion56. The Jurkat-E6 T cell line was obtained from the American Type Culture Collection (Cat# TIB-152). These cells were cultured in T cell medium (TCM): RPMI-1640 containing 10% fetal bovine serum, 0.8 mM l-glutamine, 25 mM HEPES buffer, and 1% penicillin-streptomycin. Raji tumor cells (American Type Culture Collection; Cat# CCL-86), lentivirally transduced with firefly luciferase (Raji/ffluc cells34) were also cultured in TCM.
Cell lines were screened for the presence of mycoplasma on a monthly basis. Primary human peripheral blood mononuclear cells (PBMC) and T-cells were cultured in TCM supplemented with 50 IU IL-2/ml (Preprotech), or in ImmunoCult-XF T Cell Expansion Medium (XFSFM) (Stemcell) as indicated.
mRNA transfection of T-cells
Cryopreserved PBMC from normal donors were thawed by drop-wise addition of warm TCM, followed by centrifugation (450 × g for 10 min). Where indicated, CD8 T-cells were isolated by negative selection (Stemcell). Cells were cultured in TCM + IL-2 at 106 cells/ml and stimulated with CD3/CD28 beads (Dynabeads, Life Technologies) at a 1:1 bead:cell ratio. For experiments involving αCD3-targeted NP transfection, these beads were removed 24 h before NP addition.
For NP-mediated transfections, the T-cells were resuspended in XFSFM to a concentration of 2 × 106/ml. Antibody-targeted NPs containing 2.5 µg of mRNA/106 cells were mixed into this suspension for an exposure of 2 h at 37 °C before diluting it four-times by adding T cell medium supplemented with 50 IU IL-2/ml. Control NPs contained eGFP mRNA. TCRα gene editing NPs contained TRAC-megaTAL, Trex2, and eGFP mRNAs at a 42:42:16 w:w:w ratio. Foxo13A-NPs contained Foxo13A and eGFP mRNAs at an 84:16 w:w ratio.
For electroporation, 2 × 106 T-cells were washed twice with PBS containing 0.5% bovine serum albumin (BSA), resuspended in 100 µl of T-cell electroporation medium (Lonza) containing 3 µg of eGFP mRNA, transferred to an electroporation cuvette, and treated in a Nucleofector (Lonza) instrument using program T-20. The electroporated cells were transferred into a plate containing 2 ml TCM + IL-2 without antibiotics.
In all T-cell genome editing experiments, cells were incubated at 30 °C for 40 h following nanoparticle transfection. This transient cold shock was previously reported to enhance genome editing in mammalian cells57.
Nanoparticle transfection of CD34+ cells
CD34+ cells purified from peripheral blood stem cells (PBSC) previously mobilized from normal donors were obtained from the Hematopoietic Cell Processing and Repository Core at the Fred Hutchinson Cancer Research Center. After thawing, the cells were counted then cultured overnight at a concentration of 106/ml in HSC medium: StemSpan SFEMII serum-free medium supplemented with 50 ng/ml human Stem Cell Factor (Scf), 50 ng/ml murine Flt3/Flk-2 ligand, and 25 ng/ml human thrombopoietin (Stemcell Technologies). The next day the cells were harvested, counted, and resuspended in 100 µl HSC without cytokines at 2.5 × 104 cells/well in a 96-well tissue culture plate (Costar). The cells were treated with CD105-targeted or control NPs containing 1 µg eGFP or Musashi-2 mRNA per well for 1 h, then washed twice with 1 ml HSC medium without cytokines. Washed cells were then transferred into 500 µl complete HSC medium in 24-well tissue culture plates; 48 h later, the cells were labeled for CD34, CD133, and CD105 (BioLegend) for analysis by flow cytometry.
PCR amplification and detection of indels for TCRα
Indel detection was performed with a Geneart Genomic Cleavage Detection Kit (Invitrogen) according to the manufacturer’s instructions. Briefly, T-cells were lysed, and genomic DNA flanking the TCRα MegaTAL target site was amplified by PCR using these primers: TRAC-Forward CCCGTGTCATTCTCTGGACT, and TRAC-Reverse ATCACGAGCAGCTGGTTTCT. The PCR product was denatured, re-annealed, and treated with the detection enzyme. Indel formation was assessed by comparing gel band density for germline vs specifically cleaved bands.
Lentiviral transduction and expansion of T-Cells using 19-41BBζ CAR
Human anti-CD19 CAR construct containing 41BB and CD3ζ signaling domains (19-41BBζ) was modified with a single StrepTag as described58 and transferred into the epHIV7 lentiviral vector. VSVG pseudotyped lentivirus was produced via calcium phosphate transfection (Invitrogen) of Lenti-X 293 T-cells (Clontech) with epHIV7 lentiviral vector and the viral packaging plasmids pCMVdR8.91 and pMD2.G. For lentiviral transduction, T-cells were transferred to retronectin-coated plates (Takara) with 8 µg/ml polybrene and 19-41BBζ-CAR encoding lentivirus at a MOI of 5:1, then spin infected for 1 h at 800 × g at 34 °C. For selective expansion of 19-41BBζ-transduced cells, the lymphocytes were stimulated with irradiated (7000 rads) CD19+ TM-LCL cells at a 1:7 ratio in TCM + IL-2.
To prepare tumor-targeting cells for in vivo therapy, CD8 T-cells were activated and transfected with α-CD8-targeted NPs containing eGFP or Foxo13A + eGFP mRNA on day 1 and 2 after activation. On day 3, GFP + cells were sorted from each treatment group, and transduced with 19-41BBζ-CAR-encoding lentivirus, followed by expansion for 5–7 days in TCM + IL-2. CAR T-cells were expanded with irradiated TM-LCLs. To maintain a differentiated state during this secondary expansion, T-cells were transfected with α-CD8-targeted NPs containing eGFP or Foxo13A + eGFP mRNA 1 day before and 1 day after addition of TM-LCL cells.
Cell sorting and flow cytometry
Data were acquired using a BD LSRFortessa or FacsCanto II cell analyzer running FACSDIVA software, sorted on the BD FACS ARIA-II, and analyzed with FlowJo v10.1. Antibodies used in flow cytometry are listed in Supplementary Table 1.
Cytokine secretion assays
T-cell IFN-γ release was measured with ELISA (R&D Systems) 48 h after stimulation by irradiated TM-LCL (a CD19+ EBV-transformed lymphoblastoid cell line that has been optimized for use as a feeder cell for T-cell culture)59 or C42 prostate control tumor cells as a negative control.
Intracellular staining for foxo1
106 Jurkat T-cells were transfected with anti-CD3-targeted NPs containing 3 µg eGFP mRNA, or 2.5 µg Foxo13A and 0.5 µg eGFP mRNA. 24 h later, cells were fixed with 4% paraformaldehyde in PBS, washed once, and permeabilized with 90% ice-cold methanol for 30 min. These samples were blocked with 0.5% BSA in PBS at room temperature, then labeled with rabbit anti-Foxo1 (clone C29H4) or isotype (clone DAE1), followed by anti-rabbit IgG F(aB′)2 Alexa-647 (Cell Signaling).
CAR T cell killing assay
Specific cytolysis of CAR target cells was assayed by flow cytometry. Target K562-CD19 cells were labeled with low (0.4 µM), and control K562 with high (4.0 µM) carboxyfluorescein succinimidyl ester (CFSE) for 15 min at 37 °C. Both samples were washed in complete medium containing serum, mixed at a ratio of 1:1, then co-cultured with 19-41BBζ−transduced CAR T-cells at the indicated effector:target ratios. To assess specific cytolysis, each condition was stained with anti-CD8 mAbs (BioLegend; Cat# 301035, used at 1:200 dilution) to identify T-cells and with 7AAD to exclude dead cells, and analyzed by flow cytometry. Specific cell killing was assessed by measuring the ratio of viable CD19+ target cells (low CFSE) to control CD19- K562 cells (high CFSE).
Microscopy
106 T-cells in 400 µl of XFSFM were treated with anti-CD3-targeted NPs containing 3 µg cy5-labeled eGFP mRNA for 1 h at 4 °C for surface binding, followed by a 2-h incubation at 37 °C for internalization. Following these treatments, the cells were washed 3 times with cold PBS, and loaded onto poly-l-lysine (Sigma)-coated slides for 30 min at 4 °C. The samples were fixed in 2% paraformaldehyde, mounted in ProLong Gold Antifade reagent (Invitrogen), and imaged with a Zeiss LSM 780 NLO laser scanning confocal microscope.
RNA isolation, qPCR, sequencing, and bioinformatic analysis
After cells (T-cells in Fig. 4b, HSCs in Fig. 6g) were lysed in Trizol reagent (Ambion), total RNA was isolated using a DirectZol kit (Zymo) with on-column DNA digestion following the manufacturer’s instructions. For real-time quantitative PCR (qPCR), cDNA was prepared with a high capacity cDNA kit (Applied Biosystems). Expression levels of codon-optimized (nanoparticle-delivered) FOXO13A (Fig. 4b) and codon-optimized MSI2 (Fig. 6i) relative to the housekeeping gene B2M were measured using PrimeTime qPCR assays (Integrated DNA technology) and a QuantStudio5 machine (Applied Biosystems). The following primers were used: Foxo13A Forward: GGACAGCCTAGAAAGAGCAG; Foxo13A Probe: AGGTCGGCGTAGCTCAGATTGC; Foxo13A Reverse: CTCTTGACCATCCACTCGTAG; MSI2 (codon optimized) Forward: CTGCTAGACCTGGCGGATT; MSI2 (codon optimized) Reverse: CCGTACAGATCGGCCACT. For RNAseq analysis, RNA samples were isolated from in vitro-cultured control NP- and Foxo13A-NP-treated CD8+ cells after 3 and 8 days, and compared with sorted reference naive (CD8+ CD45RA+ CD62L+ CCR7+) and TCM (CD8+ CD45RA− CD62L+ CCR7+) cells from two independent donor-matched cryopreserved PBMC samples.
RNASeq libraries were prepared using the TruSeq sample preparation kit (Illumina) according to the manufacturer’s instructions. Libraries were sequenced for 50 cycles (paired end) with a HiSeq platform (Illumina). We aligned results that passed Illumina’s base call and quality filters to the human hg38 genome using TopHat v2.1.0. Counts were generated for each gene with htseq-count (v0.6.1p1), implemented in the “intersection-strict” overlap mode. We used the GLM method in edgeR for data normalization and differential expression analysis. TCM signature gene sets were defined as the top 500 genes ranked by statistical significance with higher (TCM Up) or lower (TCM Down) expression in CD8+ TCM versus donor-matched control NP-treated CD8+ T-cells at day 8. Gene set enrichment was analyzed with GSEAPreranked software using gene lists ranked by the sign of the fold change × 1/(P value)58. Raw and processed data from RNAseq analysis have been deposited in NCBI’s Gene Expression Omnibus, GEO series accession number GSE8913460.
Mice and in vivo tumor models
Animals were housed in the animal facility of Fred Hutchinson Cancer Research Center, and used in the context of an animal protocol approved by their Institutional Animal Care and Use Committee. Four- to 6-week-old female NOD.Cg-Prkdc scid Il2rg tm1Wjl/SzJ (NSG) mice were bred in house and engrafted via tail vein with 5 × 105 Raji cells expressing firefly luciferase. One week later, tumor burden was determined by bioluminescent imaging, and mice with detectable tumor were sorted into groups with matching tumor burden. Groups were then randomly assigned to treatment conditions, receiving no therapy, or intravenous injections of 2.5 × 106 CD8+ 19-41BBζ CAR+ cells transfected with eGFP or Foxo13A mRNA encapsulating nanoparticles. Based on pilot studies, 5 mice per group were used for data analysis to provide an 80% power to attain a P-value of 0.02. The investigators were not blinded to group allocation.
In vivo bioluminescence and fluorescence imaging
We used D-Luciferin (Xenogen) in PBS (15 mg/ml) as a substrate for F-luc (imaging of Raji-luc lymphoma cells). Bioluminescence images were collected with a Xenogen IVIS Spectrum Imaging System (Xenogen, Alameda, CA). Living Image software version 4.4 (Caliper Life Sciences) was used to acquire (and later quantitate) the data 10 min after intraperitoneal injection of D-luciferin into animals anesthetized with 150 mg/kg of 2% isoflurane (Forane, Baxter Healthcare). Acquisition times ranged from 10 s to 5 min.
Colony-forming cell assay
For Colony-forming Cell (CFC) assays 400 sorted HSCs cells were seeded into 1 ml MethoCult H4435 (StemCell Technologies). Hematopoietic colonies were scored after 12–14 days. Arising colonies were identified as colony forming unit- (CFU-) granulocyte (CFU-G), macrophage (CFU-M), granulocyte-macrophage (CFU-GM) and burst forming unit-erythrocyte (BFU-E). Colonies consisting of erythroid and myeloid cells were scored as CFU-MIX.
Statistical analysis
The statistical significance of the differences we measured in T-cell expansion (Fig. 2d, e), viability (Fig. 2g), lentiviral gene transfer (Fig. 3f), bioluminescent tumor signal (Fig. 5b), the viability of HSCs (Fig. 6d), and the HSCs differentiation (Fig. 6f, g, l) was analyzed with the unpaired, two-tailed Student’s t Test. The P value of overlap between Foxo13A and the TCM signature gene set (Fig. 4e–g) was determined by GSEA (via analysis shown in Fig. 4h. A P value < 0.05 was considered significant. We characterized survival data (Fig. 5c) using the Log-rank test. All statistical analyses were performed using GraphPad Prism software version 6.0.
Study approval
Blood samples were obtained from healthy donors. Donors provided written informed consent for research protocols approved by the Institutional Review Board of the FHCRC. The FHCRC Institutional Care and Use Committee approved all mouse experiments.
Data availability
Raw and processed data from RNAseq analysis have been deposited in NCBI’s Gene Expression Omnibus, GEO series accession number GSE89134. Step-by-step protocols describing nanoparticle synthesis, as well as T-cell transfection were uploaded to Protocol Exchange (“Stephan Lab-FHCRC”). All other relevant data can be requested from Dr M. Stephan by others both within and outside the scientific community.
References
Jain, K. K. Cell Therapy - Technologies, Markets and Companies, ID: 39071, 1–1144 (Jain PharmaBiotech, 2017).
Gratwohl, A. et al. One million haemopoietic stem-cell transplants: a retrospective observational study. Lancet Haematol. 2, e91–e100 (2015).
Mao, A. S. & Mooney, D. J. Regenerative medicine: Current therapies and future directions. Proc. Natl Acad. Sci. USA 112, 14452–14459 (2015).
Freitag, J. et al. Mesenchymal stem cell therapy in the treatment of osteoarthritis: reparative pathways, safety and efficacy–a review. BMC Musculoskelet. Disord. 17, 230 (2016).
Ellebrecht, C. T. et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science 353, 179–184 (2016).
DeWitt, M. A. et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 8, 360ra134 (2016).
Fesnak, A. D., June, C. H. & Levine, B. L. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat. Rev. Cancer. 16, 566–581 (2016).
Arasaratnam, R. J. & Leen, A. M. Adoptive T cell therapy for the treatment of viral infections. Ann. Transl. Med. 3, 278 (2015).
Brentjens, R. J. et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med. 5, 177ra138 (2013).
Porter, D. L. et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med. 7, 303ra139 (2015).
Maeder, M. L. & Gersbach, C. A. Genome-editing technologies for gene and cell therapy. Mol. Ther. 24, 430–446 (2016).
Burga, R. A. et al. Improving efficacy of cancer immunotherapy by genetic modification of natural killer cells. Cytotherapy. 18, 1410–1421 (2016).
Negre, O. et al. Preclinical evaluation of efficacy and safety of an improved lentiviral vector for the treatment of beta-thalassemia and sickle cell disease. Curr. Gene. Ther. 15, 64–81 (2015).
Zhang, L. et al. MicroRNA-31 negatively regulates peripherally derived regulatory T-cell generation by repressing retinoic acid-inducible protein 3. Nat. Commun. 6, 7639 (2015).
Cribbs, A. P., Kennedy, A., Gregory, B. & Brennan, F. M. Simplified production and concentration of lentiviral vectors to achieve high transduction in primary human T cells. BMC Biotechnol. 13, 98 (2013).
Wurm, M. et al. Ectopic expression of HOXC6 blocks myeloid differentiation and predisposes to malignant transformation. Exp. Hematol. 42, 114–125.e114 (2014).
Themeli, M. et al. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat. Biotechnol. 31, 928–933 (2013).
Costa, A. et al. Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development 142, 1948–1959 (2015).
Cox, D. B., Platt, R. J. & Zhang, F. Therapeutic genome editing: prospects and challenges. Nat. Med. 21, 121–131 (2015).
Nightingale, S. J. et al. Transient gene expression by nonintegrating lentiviral vectors. Mol. Ther. 13, 1121–1132 (2006).
Boissel, S. & Scharenberg, A. M. Assembly and characterization of megaTALs for hyperspecific genome engineering applications. Methods. Mol. Biol. 1239, 171–196 (2015).
Tejera, M. M., Kim, E. H., Sullivan, J. A., Plisch, E. H. & Suresh, M. FoxO1 controls effector-to-memory transition and maintenance of functional CD8 T cell memory. J. Immunol. 191, 187–199 (2013).
Kim, M. V., Ouyang, W., Liao, W., Zhang, M. Q. & Li, M. O. The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity 39, 286–297 (2013).
Delaney, C. et al. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat. Med. 16, 232–236 (2010).
Pineault, N. & Abu-Khader, A. Advances in umbilical cord blood stem cell expansion and clinical translation. Exp. Hematol. 43, 498–513 (2015).
Rentas, S. et al. Musashi-2 attenuates AHR signalling to expand human haematopoietic stem cells. Nature 532, 508–511 (2016).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Quabius, E. S. & Krupp, G. Synthetic mRNAs for manipulating cellular phenotypes: an overview. Nat. Biotechnol. 32, 229–235 (2015).
Sadelain, M. CAR therapy: the CD19 paradigm. J. Clin. Invest. 125, 3392–3400 (2015).
Menger, L. et al. TALEN-mediated inactivation of PD-1 in tumor-reactive lymphocytes promotes intratumoral T-cell persistence and rejection of established tumors. Cancer Res. 76, 2087–2093 (2016).
Torikai, H. et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood 119, 5697–5705 (2012).
Certo, M. T. et al. Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat. Methods 9, 973–975 (2012).
Louis, C. U. et al. Antitumor activity and long-term fate of chimeric antigen receptor-positive T cells in patients with neuroblastoma. Blood 118, 6050–6056 (2011).
Sommermeyer, D. et al. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia 30, 492–500 (2016).
Wang, X. et al. Phenotypic and functional attributes of lentivirus-modified CD19-specific human CD8+ central memory T cells manufactured at clinical scale. J. Immunother. 35, 689–701 (2012).
Kodama, S., Koike, C., Negishi, M. & Yamamoto, Y. Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol. Cell Biol. 24, 7931–7940 (2004).
Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999).
Kaech, S. M. & Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012).
Skon, C. N. et al. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 14, 1285–1293 (2013).
Boesteanu, A. C. & Katsikis, P. D. Memory T cells need CD28 costimulation to remember. Semin. Immunol. 21, 69–77 (2009).
Lee, J. et al. Improved ex vivo expansion of adult hematopoietic stem cells by overcoming CUL4-mediated degradation of HOXB4. Blood 121, 4082–4089 (2013).
Zhang, X. B. et al. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J. Clin. Invest. 118, 1502–1510 (2008).
Pierelli, L. et al. CD105 (endoglin) expression on hematopoietic stem/progenitor cells. Leuk. Lymphoma 42, 1195–1206 (2001).
Pierelli, L. et al. CD34+/CD105+ cells are enriched in primitive circulating progenitors residing in the G0 phase of the cell cycle and contain all bone marrow and cord blood CD34+ /CD38low/− precursors. Br. J. Haematol. 108, 610–620 (2000).
Kays, S. K. et al. CD105 is a surface marker for receptor-targeted gene transfer into human long-term repopulating hematopoietic stem cells. Stem Cells. Dev. 24, 714–723 (2015).
Darcy, P. K. & Neeson, P. J. Adoptive immunotherapy: a new era for the treatment of cancer. Immunotherapy 7, 469–471 (2015).
Schumann, K. et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl Acad. Sci. USA 112, 10437–10442 (2015).
Wang, J. et al. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res. 44, e30 (2016).
Bai, Y. et al. Enhancement of the in vivo persistence and antitumor efficacy of CD19 chimeric antigen receptor T cells through the delivery of modified TERT mRNA. Cell Discov. 1, 15040 (2015).
Koh, S. et al. A practical approach to immunotherapy of hepatocellular carcinoma using T cells redirected against hepatitis B virus. Mol. Ther. Nucleic Acids 2, e114 (2013).
Liu, X. et al. Affinity-tuned ErbB2 or EGFR chimeric antigen receptor T cells exhibit an increased therapeutic index against tumors in mice. Cancer Res. 75, 3596–3607 (2015).
Copolovici, D. M., Langel, K., Eriste, E. & Langel, U. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano 8, 1972–1994 (2014).
Liu, J., Gaj, T., Patterson, J. T., Sirk, S. J. & Barbas, C. F. 3rd Cell-penetrating peptide-mediated delivery of TALEN proteins via bioconjugation for genome engineering. PLoS ONE 9, e85755 (2014).
Liu, J., Gaj, T., Wallen, M. C. & Barbas, C. F. 3rd Improved cell-penetrating zinc-finger nuclease proteins for precision genome engineering. Mol. Ther. Nucleic Acids 4, e232 (2015).
Mangraviti, A. et al. Polymeric nanoparticles for nonviral gene therapy extend brain tumor survival in vivo. ACS Nano 9, 1236–1249 (2015).
Riddell, S. R. & Greenberg, P. D. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J. Immunol. Methods 128, 189–201 (1990).
Doyon, Y. et al. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat. Methods 7, 459–460 (2010).
Liu, L. et al. Inclusion of Strep-tag II in design of antigen receptors for T-cell immunotherapy. Nat. Biotechnol. 34, 430–434 (2016).
Terakura, S. et al. Generation of CD19-chimeric antigen receptor modified CD8+ T cells derived from virus-specific central memory T cells. Blood 119, 72–82 (2012).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
We thank Dr Liu and Dr Riddell for the gift of cell lines and StrepTagII antibody. We thank the Fred Hutchinson Genomics Core staff for assistance in the preparation and analysis of RNAseq data. This work was supported in part by the Fred Hutchinson Cancer Research Center’s Immunotherapy Initiative with funds provided by the Bezos Family, a FHCRC/UW Cancer Consortium Cancer Center Support Grant of the National Institutes of Health under Award Number P30 CA015704, a Prostate Cancer Research Program - Synergistic Idea Development Award from the Department of Defense (PC140519P1), and by the National Science Foundation (CAREER Award #1452492 and EAGER Award #1644363). M. Stephan was also supported by a Research Scholar Grant, RSG-16-110-01—LIB, from the American Cancer Society.
Author information
Authors and Affiliations
Contributions
H.F.M. conceived the study, designed and performed the experiments, and wrote the manuscript. M.E.C. and L.M. designed and performed the experiments. S.R. performed all HSC colony forming assays. S.B.S. produced polymers and antibody conjugates. A.L. and B.L.S. provided reagents, gave advice on experimental design, and critiqued the manuscript. H.P.K. provided reagents and gave advice on experiments related to HSCs. M.T.S. conceived the study, designed the experiments, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The Fred Hutchinson Cancer Center, H.F.M. and M.T.S. have filed a patent pertaining to mRNA delivery via targeted polymeric nanoparticles. The remaining authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Moffett, H.F., Coon, M.E., Radtke, S. et al. Hit-and-run programming of therapeutic cytoreagents using mRNA nanocarriers. Nat Commun 8, 389 (2017). https://doi.org/10.1038/s41467-017-00505-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-017-00505-8
This article is cited by
-
The quest for nanoparticle-powered vaccines in cancer immunotherapy
Journal of Nanobiotechnology (2024)
-
Biomaterials to enhance adoptive cell therapy
Nature Reviews Bioengineering (2024)
-
Drug delivery methods for cancer immunotherapy
Drug Delivery and Translational Research (2024)
-
FOXO1 is a master regulator of memory programming in CAR T cells
Nature (2024)
-
mRNA-based vaccines and therapeutics: an in-depth survey of current and upcoming clinical applications
Journal of Biomedical Science (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.