Abstract
Cross-ethnic genetic studies can leverage power from differences in disease epidemiology and population-specific genetic architecture. In particular, the differences in linkage disequilibrium and allele frequency patterns across ethnic groups may increase gene-mapping resolution. Here we use cross-ethnic genetic data in sporadic amyotrophic lateral sclerosis (ALS), an adult-onset, rapidly progressing neurodegenerative disease. We report analyses of novel genome-wide association study data of 1,234 ALS cases and 2,850 controls. We find a significant association of rs10463311 spanning GPX3-TNIP1 with ALS (p = 1.3 × 10−8), with replication support from two independent Australian samples (combined 576 cases and 683 controls, p = 1.7 × 10−3). Both GPX3 and TNIP1 interact with other known ALS genes (SOD1 and OPTN, respectively). In addition, GGNBP2 was identified using gene-based analysis and summary statistics-based Mendelian randomization analysis, although further replication is needed to confirm this result. Our results increase our understanding of genetic aetiology of ALS.
Similar content being viewed by others
Introduction
For people of European ancestry, the lifetime risk of amyotrophic lateral sclerosis (ALS) is 0.3–0.5%1, 2, with peak age of onset of 58–63 years3, and median survival of 2–4 years4. Investigations of families with multiple affected individuals have led to the identification of mutations that segregate with disease in a number of genes, including SOD1, C9orf72, TARDBP, FUS and TBK1 5, 6. However, about 90% of cases5 (‘sporadic ALS’ (sALS)) present with sparse or no family history. Nonetheless, genome-wide association studies (GWAS) have provided direct evidence of a genetic contribution to sALS, with estimates that ~8.5%7 of variance in liability is tagged by common single-nucleotide polymorphisms (SNPs). Currently, only a small proportion of this variation (0.2% of variance in liability)7 is accounted for by the six common loci (C9orf72, UNC13A, SARM1, MOBP, SCFD1, C21orf2) identified as significant based on association analysis of 12,577 cases and 23,475 controls7. The SNP-heritability estimate implies that more risk loci will be detected with increasing sample size, as found for other complex genetic diseases8. Whole-exome sequencing (WES) studies, designed to identify genes enriched for rare variants, have also been conducted for sALS. The largest study, comprising 2,874 cases and 6,405 controls, identified TBK1 as a novel ALS risk gene6, with GWAS support for association of common loci (p = 6.6 × 10−8)7. Rare variant burden analysis in a WES of 1,022 index familial cases identified p.Arg261His in NEK1 as an ALS associated variant, and follow-up in large samples suggest that this variant together with NEK1 loss of function mutations account for ~3% of ALS cases9.
To date, the largest genetic studies for ALS are in the subjects of European ancestry, but common variants associated with disease are likely to be ancient and shared across ethnicities. Given sufficient power, cross-ethnic genetic studies can aid fine mapping of disease loci, exploiting differences in allele frequency and linkage disequilibrium (LD). In China, the lifetime risk of ALS is estimated to be lower (0.1%)1 and its mean age of onset is estimated to be a few years earlier than in Europe4, 10. High penetrance mutations in known ALS genes identified in Europeans have been detected in Chinese cases11, but the frequency of the C9orf72 expansion is much lower (0.3%)12 than in Europeans (frequency 7%)5, and it may have arisen on a different haplotype background12.
In a cross-ethnic meta-analysis of the largest GWAS for ALS in Europeans7, together with a new Chinese data set, we identify the GPX3-TNIP1 locus to be significantly associated with ALS (p = 1.3 × 10−8). This association is replicated in two independent Australian cohorts with a combined p-value of 1.7 × 10−3. Previous studies indicate functional relevance of both GPX3 and TNIP1 13,14,15,16,17,18. The identification of this locus contributes to a better understanding of the genetic aetiology of ALS.
Results
Genome-wide association analysis
We conduct a genome-wide (GW) association analysis in a Chinese sample of 1,234 sALS cases and 2,850 controls (Supplementary Table 1 and Supplementary Figs 1−3). The genomic inflation factor λ GC of 1.02 and λ 1000 of 1.01 showed no evidence for inflation in test statistics. The combined effects of all common genetic variants on ALS liability (SNP-heritability) estimated from the Chinese GWAS data is 15.1% (SE): 4%; p = 9.5 × 10−5) using GCTA-GREML19 and 15.0% (SE: 3.5%) using LD score regression20 (intercept 1.0, which also shows no evidence of population stratification). Given the SE, these estimates are not different from the estimate of 8.5% (SE 0.5%) from European data7. Partitioning of the SNP-heritability by chromosome showed a significant positive correlation with chromosome length (Supplementary Fig. 4a) consistent with a polygenic architecture. Based on minor allele frequency (MAF) bin, the SNP-heritability was attributed to SNPs across the MAF range, but SEs per bin were large (Supplementary Fig. 4b); similar analyses based on European data suggested that less common SNPs tagged more variation than other MAF classes7.
No individual SNPs passed the GW significant p value threshold of 5 × 10−8, and none of the significant SNPs reported in the European7 GWAS replicated in our samples (p > 0.05). We also checked for the associations of two GW significant SNPs in previous GWAS of Chinese cohort of ALS patients21. However, we could not replicate the association in that study. We note that despite evidence for population stratification, principal components derived from SNP data of the previous study were not included as covariates in their association analysis. The p values for rs6703183 and rs8141797 are 0.07 and 0.12 in our Chinese samples and 0.66 and 0.94 in European GWAS results, respectively. Direction of effect sign tests (Supplementary Table 2) and polygenic risk scoring analyses (Supplementary Fig. 5) provided no conclusive evidence of shared risk loci (Nagelkerke’s R 2 = 0.002; p = 0.01). These results are not unexpected given the size of our sample and effect sizes estimated in Europeans. The Chinese GWAS sample had 80% power to identify common genetic variants of genotype relative risk of 1.4 and 1.8 for risk allele frequency of 0.2 and 0.05, respectively, at the GW threshold of significance p = 5 × 10−8.
Meta-analysis
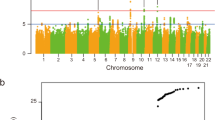
Meta-analysis of our results with those of the European7 GWAS identified a new GW significant locus at chromosome 5p33.1 (rs10463311, risk allele C, odds ratio (OR) 1.11 95% confidence interval (CI): 1.06–1.14, p logistic = 2.9 × 10−8; p lmm = 1.3 × 10−8) spanning the genes GPX3 and TNIP1 (Figs. 1 and 2; Table 1; Supplementary Data 1) for which the risk allele is more common in Chinese than in Europeans (0.46 vs. 0.25). The association result was replicated in an independent Australian sample (145 cases, 116 controls, OR = 1.66; 95% CI: 1.16–2.38; p = 5.8 × 10−3) and had the same direction of effect in a second Australian sample (431 cases, 567 controls, OR = 1.22; 95% CI: 1.00–1.48; p = 6.2 × 10−2), giving a combined replication OR of 1.32 (95% CI: 1.11–1.58; p = 1.7 × 10−3) (Table 1).
Regional ALS association plot of the GPX3-TNIP1 locus from the meta-analysis results created using LocusZoom44. From the meta-analysis, rs10463311 is the SNP with the strongest association with ALS (p = 1.3 × 10−8). This SNP is replicated in two independent Australian cohorts with combined p = 1.7 × 10−3
Functional relevance of GPX3 and TNIP1
Both GPX3 and TNIP1 are genes that could have functional relevance for ALS. The protein glutathione peroxidase 3 (GPX3), is an antioxidant molecule functionally related to superoxide dismutase 1 (SOD1)13; many SOD1 single-nucleotide variants are pathogenic for ALS. In a mass spectrometric screen of sera of SOD1H46R rats compared to their wild-type (WT) controls in the presymptomatic stage (12 weeks of age) of ALS, Gpx3 was detected as one of the two significant results (1.3-fold increase in expression)14. In the same study, Gpx3 expression was significantly lower (0.74 fold, p = 0.009) compared to WT controls by disease end stage, a finding which was replicated in blood sera of sporadic ALS cases (n = 18) and controls (n = 35) (GPX3 0.41-fold lower, p = 0.008)14. Both GPX3 and TNIP1 are functionally associated with NF-κB, the master regulator of inflammation17, 19, with upregulation of NF-κB associated with death of motor neurones15. Protein–protein interaction analysis18 links GPX3 to SOD1 and TNIP1 to OPTN, and OPTN also harbours mutations associated with familial ALS5. TNIP1 is associated with a wide range of immune disorders22, 23, although our most associated SNP (rs10463311) is not in LD with specific SNPs associated with these disorders24. We investigated differential expression of GPX3 and TNIP1 between ALS patients and controls, but given small sample sizes, the results were not conclusive (Supplementary Note 1, Supplementary Table 3, Supplementary Fig. 6). In a pleiotropy informed analysis25 applied to the European GWAS summary statistics7, rs10463311 was identified as an ALS-associated SNP, providing additional, albeit not fully independent, support for this locus.
Gene-based association analysis
No genes were significantly associated with ALS from gene-based association analysis implemented in fastBAT26 of Chinese data (based on Bonferroni correction for ~18,000 autosomal genes, significance declared at 2.8 × 10−6), but meta-analysed results (Supplementary Table 4) identified multiple genes (reflecting LD and overlapping gene boundaries) at the previously reported chromosome 5, 9, 14 and 17 GWAS loci. Two new loci on Chromosome 17 (17q12 and 17q21.2) were also significant (minimum genic p = 3.3 × 10−7 and 1.2 × 10−7, respectively). The former locus was also supported by summary statistic-based Mendelian randomization (SMR) analysis27 that combines the disease–SNP association with gene expression–SNP association results and has GW significance threshold of p SMR < 8.4 × 10−6) (Supplementary Fig. 7; Supplementary Data 2), with most significant association for GGNBP2 (European only p SMR = 4.6 × 10−6; meta-analysis p SMR = 9.8 × 10−6). The two replication samples did not provide support for the GGNBP2 SNP implicated from the SMR analysis (Supplementary Table 5); larger sample sizes are needed to confirm the association and to provide evidence to exclude ZNHIT3 (p SMR = 3.1 × 10−5) or MYO19 (p SMR = 2.2 × 10−4) as contributing to the association in this region. Gene-set pathway analysis implemented in MAGMA and applied to the Chinese/European meta-analysis results did not find any ALS significant pathways that passed multiple testing correction (Supplementary Table 6).
Discussion
In summary, using a cross-ethnic design we identify association of the GPX3-TNIP1 locus with ALS. This locus was identified by combining GWAS results from our Chinese data with the largest European GWAS data7 and replicated in independent Australian samples. In addition, GGNBP2 was identified using gene-based analysis and SMR analysis, although further replication is needed to confirm this result. The discovery of a novel risk locus significantly advances our understanding of ALS aetiology.
Methods
Chinese ALS cases and controls
The samples comprised 1,324 ALS cases and 3,115 controls. ALS cases were recruited from the Department of Neurology, the Peking University Third Hospital (Beijing, China) from 2003 to 2013. The cases were diagnosed by a neurologist specializing in ALS using the revised El Escorial criteria28. The controls are individuals who attended the Peking University Third Hospital, Peking University Sixth Hospital or Shanghai Changzheng Hospital (Shanghai) with no medical or family history of neurological disorders. All cases and controls are of Chinese origin from Mainland China and provided written informed consent for the study. The sample collections were approved by the ethics committees at the respective hospitals12. The study is compliant with the Guidance of the Ministry of Science and Technology (MOST) for the Review and Approval of Human Genetic Resources. Analyses conducted at the University of Queensland were approved by the University human research ethics committee.
Australian replication cohort 1
ALS cases were recruited from the Royal Brisbane & Women’s Hospital (RBWH), Brisbane, Queensland and the Macquarie University Multidisciplinary Motor Neurone Disease Clinic29, New South Wales. The cases (N = 159) were diagnosed using the revised El Escorial criteria10. The controls are healthy individuals (N = 132), sourced from 4 different sites, RBWH (27 individuals), Neurology at Macquarie University, Sydney (25 individuals), the Older Australian Twin Study (OATS)30 comprising 90 monozygotic (MZ) twin pairs recruited in Brisbane (QIMR Berghofer Medical Research Institute (QIMR)) and Sydney (University of New South Wales (UNSW)) and Melbourne (University of Melbourne (UM)). The OATS study recruits MZ twins aged ≥65 years and were chosen for this study because the Discovery sample controls were younger than Discovery sample cases. Twin pair data helped in quality control checks but only one twin from each pair was used in analyses. The subjects provided written informed consent for the study. The study was approved by the RBWH31, QIMR, UNSW, UM, University of Queensland and Macquarie University Research Ethics Committees.
Australian replication cohort 2
Patients and controls were ascertained from Macquarie University Multidisciplinary Motor Neurone Disease and Neurology Clinics, Sydney and from the Australian MND DNA bank. Patients were diagnosed with definite or probable ALS according to the revised El Escorial criteria. Patients with a family history for ALS were excluded. Control subjects were healthy individuals free of neuromuscular diseases. DNA from 471 cases and 586 controls were available for genotyping. The subjects provided written informed consent for the study. The study was approved by Macquarie University Research Ethics Committee.
DNA extraction
In the Chinese cohort, genomic DNA was extracted from whole blood using the DNA Extraction Kit (Beijing Aide Lai Biotechnology Co. Ltd., Beijing, China). In the Australian replication cohorts, the majority of DNA was extracted from fresh whole blood using manual extraction protocols, except for 90% (118 out of 131) of UNSW/UM control samples, where DNA was extracted from frozen whole blood or lymphocytes using an automated purification system, Qiagen Autopure LS (Qiagen, Valencia, CA, USA).
Genome-wide association study
We performed GW genotyping in the discovery cohort using the Illumina HumanOmni ZhongHua-8 v1.0 and v1.1 arrays. These arrays contain 900,015 (v1.0) and 894,517 (v1.1) variants, respectively. Before testing for the association between each variant and disease status, we carried out quality control (QC) steps to identify and exclude poor quality samples and genetic variants. We excluded individuals based on the following QC filters: (i) genotyping call rate <99% (134 individuals); (ii) sex mismatch between genotype and clinical information (6 individuals); (iii) ancestry outliers (6 SDs from HapMap-CHB means of PC1 and PC2; 30 individuals); and (iv) duplicated or related individuals (genetic relationship matrix >0.05; 195 individuals). We excluded genetic variants based on the following criteria: (i) low genotype call rate <99%; (ii) MAF <1%; (iii) deviation from Hardy–Weinberg equilibrium p < 10−6; and (iv) differential missingness in genotypes between cases and controls (p < 10−6). After these QC steps, 1,234 cases and 2,850 controls with genotypic information from 753,038 markers remained for the subsequent analyses.
We imputed unobserved genotypes into the 1000 Genomes Project Phase 1 v3 (all ethnicities) using samples and markers that passed QC. We implemented a two-step process, i.e., haplotyping using HAPI-UR32 and imputation using IMPUTE33. We imputed 38,033,906 SNPs, but after QC (i.e., excluding markers with MAF <0.01, imputation quality score <0.80 and HWE p < 10−6), 6,613,544 SNPs were available for analysis.
Validation sample genotyping
The first validation sample was genotyped on the Illumina Human Core Exome Array. QC and imputation followed the same pipeline as for the Chinese samples. After QC, 145 cases and 116 controls were available for analysis. For the second validation sample, SNPs were genotyped via Taqman assay such that the reaction mix included 1.0 μl of genomic DNA (10 ng/μl), 0.25 μl Custom TaqMan genotyping assay 20× (Life Technologies), 2.5 μl TaqMan SNP genotyping MasterMix 2X (Life Technologies) and 6.25 μl MilliQ. The thermocycler program included 30 s at 60 °C, 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C and a final step of 30 s at 60 °C. Fluorescent signals were analysed on a Viia7 Real-Time PCR System and genotypes were determined by allelic discrimination using the Viia7 Real-Time PCR System Software (Life Technologies). Genotype calling rates were 94% for rs4958872 (LD r 2 = 1 proxy for rs10463311) and 91% for rs9906189. After QC, 431 cases and 567 controls were available for analysis.
Genetic association analysis
The association analysis between genetic variants and disease was conducted using a linear mixed model framework implemented in GCTA (mlma-loco)34. To compare the results, we also used a logistic regression model by fitting five principal components as covariates. Genomic inflation factor was calculated as the median of Chi-square test statistics divided by its expected value (0.455).
Gene-based analysis
To test for the association between a set of variants within a gene (±50 kb) and ALS, we used GCTA-fastBAT26 with SNP association analysis p values as input. This test complements SNP–disease association analysis, identifying genes that may show evidence for independent associations that individually have not achieved association significance. For Chinese data analysis, we used our own GWAS data as the reference to calculate LD and ARIC samples (dbGAP accession phs000090.v1.p1) for the European sample.
Whole-genome estimation analysis
Genomic relationship matrix (GRM) restricted maximum likelihood (GREML) analysis using GCTA19, 35, 36 was used to estimate the total contribution of common genetic variants on the liability of ALS or SNP-heritability. This analysis fits all SNPs simultaneously in a mixed model linear framework to estimate the proportion of variance in disease liability explained by all SNPs. To avoid bias, for example, due to common environmental factors, we excluded related individuals based on GRM values >0.05. Lifetime disease risk of 0.002 was used in the conversion of the estimate to the liability scale37 (compared to 0.0025 used in the European conversion, although the results are robust to these choices). LD-score regression20 was applied to GWAS summary statistics as an alternative method to estimate the contribution of common genetic variants to variation in the liability of ALS.
Genetic overlap analysis
We considered estimation of the genetic correlation between ALS risk in Europeans and Chinese, using popcorn38 (the cross-ethnicity LDscore regression method), but calculated39 that the relatively small sample size for the Chinese cohort would generate an unacceptably large SE. Instead we used polygenic risk scoring (PRS) to investigate the genetic relationship between ALS in the two ethnicities. PRS were estimated for all Chinese cases and controls as the sum of risk alleles weighted by the log OR of association estimated in the European GWAS. Eight PRS were constructed for each individual using independent SNPs (based on SNPs pruned (r 2 < 0.25 in 200-kb window) that are significant at p value thresholds of 0.001, 0.005, 0.01, 0.05, 0.10, 0.25, 0.5 and 1. We also constructed a PRS using all SNPs without pruning for LD because of the difference in allele frequencies and LD between ethnicities. Association between the case–control status and PRS was evaluated by logistic regression. Binomial sign tests were also used to evaluate evidence of overlap in signal between Chinese and European association statistics.
Meta-analysis
Inverse variance meta-analysis was conducted between the largest GWAS for ALS in European7 and our Chinese GWAS results using METAL40.
In silico functional analyses
To help interpret biological function of the SNP– and gene–ALS associations, gene-set pathway analyses were performed using MAGMA41; this method was selected based on results of a method comparison study42. Gene-set pathway analyses aim to identify sets of biological pathways that are relevant to disease based on a set of disease-associated variants42.We also conducted SMR analysis27 that combines the GWAS summary statistics with gene expression association results. Here we used gene expression from blood43 as this is currently the largest gene expression quantitative trait loci data set. The SMR test identifies pleiotropic association of a variant that affects both the expression level of a gene and the trait. The SMR-HEIDI test attempts to determine whether the effect of the disease-associated gene on gene expression reflects a single causal variant, thus prioritizing loci for functional follow-up studies.
Data availability
GWAS summary statistics results and gene expression data are available from http://cnsgenomics.com/data/benyamin_et_al_2017_nc/BenyaminEtAl_NatComm_Data.zip.
References
Chiò, A. et al. Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology 41, 118–30 (2013).
Johnston, C. A. et al. Amyotrophic lateral sclerosis in an urban setting: a population based study of inner city London. J. Neurol. 253, 1642–3 (2006).
Kiernan, M. C. et al. Amyotrophic lateral sclerosis. Lancet 377, 942–955 (2011).
Chio, A. et al. Prognostic factors in ALS: A critical review. Amyotroph. Lateral Scler. 10, 310–323 (2009).
Renton, A. E. et al. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 17, 17–23 (2014).
Cirulli, E. T. et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347, 1436–1441 (2015).
van Rheenen, W. et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 48, 1043–1048 (2016).
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Kenna, K. P. et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 48, 1037–1042 (2016).
Liu, M. S., Cui, L. Y. & Fan, D. S., Chinese ALS Association. Age at onset of amyotrophic lateral sclerosis in China. Acta Neurol. Scand. 129, 163–7 (2014).
Liu, Q. et al. Mutation spectrum of Chinese patients with familial and sporadic amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 87, 1272–1274 (2016).
He, J. et al. C9orf72 hexanucleotide repeat expansions in Chinese sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 36, 2660.e1–2660.e8 (2015).
Chi, L., Ke, Y., Luo, C., Gozal, D. & Liu, R. Depletion of reduced glutathione enhances motor neuron degeneration in vitro and in vivo. Neuroscience 144, 991–1003 (2007).
Tanaka, H. et al. ITIH4 and Gpx3 are potential biomarkers for amyotrophic lateral sclerosis. J. Neurol. 260, 1782–97 (2013).
Frakes, A. E. et al. Microglia induce motor neuron death via the classical NF-κB pathway in amyotrophic lateral sclerosis. Neuron 81, 1009–23 (2014).
Oliveira-Marques, V., Marinho, H. S., Cyrne, L. & Antunes, F. Role of hydrogen peroxide in NF-kappaB activation: from inducer to modulator. Antioxid. Redox Signal. 11, 2223–43 (2009).
Rahighi, S. et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136, 1098–109 (2009).
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–9 (2010).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–5 (2015).
Deng, M. et al. Genome-wide association analyses in Han Chinese identify two new susceptibility loci for amyotrophic lateral sclerosis. Nat. Genet. 45, 697–700 (2013).
Gateva, V. et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 41, 1228–33 (2009).
Nair, R. P. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 41, 199–204 (2009).
Staley, J. R. et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 32, 3207–3209 (2016).
McLaughlin, R. L. et al. Genetic correlation between amyotrophic lateral sclerosis and schizophrenia. Nat. Commun. 8, 14774 (2017).
Bakshi, A. et al. Fast set-based association analysis using summary data from GWAS identifies novel gene loci for human complex traits. Sci. Rep. 6, 32894 (2016).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–7 (2016).
Brooks, B. R., Miller, R. G., Swash, M. & Munsat, T. L. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Motor Neuron Disord 1, 293–9 (2000).
Williams, K. L. et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat. Commun. 7, 11253 (2016).
Sachdev, P. S. et al. A comprehensive neuropsychiatric study of elderly twins: the Older Australian Twins Study. Twin Res. Hum. Genet. 12, 573–82 (2009).
Devine, M. S., Kiernan, M. C., Heggie, S., McCombe, P. A. & Henderson, R. D. Study of motor asymmetry in ALS indicates an effect of limb dominance on onset and spread of weakness, and an important role for upper motor neurons. Amyotroph. Lateral Scler. Frontotemporal Degener 15, 481–7 (2014).
Williams, A. L., Patterson, N., Glessner, J., Hakonarson, H. & Reich, D. Phasing of many thousands of genotyped samples. Am. J. Hum. Genet. 91, 238–51 (2012).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–13 (2007).
Yang, J., Zaitlen, N. A., Goddard, M. E., Visscher, P. M. & Price, A. L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 46, 100–6 (2014).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Yang, J. et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43, 519–25 (2011).
Lee, S. H. et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 44, 247–250 (2012).
Brown, B. C., Ye, C. J., Price, A. L. & Zaitlen, N. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet. 99, 76–88 (2016).
Visscher, P. M. et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 10, e1004269 (2014).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
de Leeuw, C. A., Neale, B. M., Heskes, T. & Posthuma, D. The statistical properties of gene-set analysis. Nat. Rev. Genet. 17, 353–364 (2016).
Westra, H.-J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–43 (2013).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 27, 2336–2337 (2011).
Acknowledgements
We thank all the participants for their generous contribution to this study. We also thank staff and researchers at the Peking University Third Hospital (Beijing, China), the Royal Brisbane and Women Hospital (Brisbane, Australia) and Multidisciplinary Motor Neurone Disease Clinic (Sydney, Australia). This work was funded by the Australian Research Council (ARC) Linkage Grant (M.A.B., P.F.B., P.M.V., H.X., R.J.W., B.J.M., D.C.R. and M.M), the Peter Goodenough Foundation, the National Natural Science Foundation of China (grants to D.S.F.: 81030019, and J.H.: 81601105 and Z.B.J.: 81522014), the National Health and Medical Research Council (NHMRC) (N.R.W.: 1078901, 1083187, B.B.: 1084417, 1079583, P.M.V.: 1078037, I.P.B.: 1095215, K.L.W.: 1092023), MNDRIA (P.G.N.: Grant-in-Aid 2013, B.B.: Mick Roger Benalla Grant, F.G.: Bill Gole Fellowship, I.P.B.: MND Australia Leadership Grant, E.P.M. and J.A.F.: scholarship top ups), Ross Maclean Senior Research Fellowships (M.M, R.J.W.), the Sylvia & Charles Viertel Charitable Foundation (J.Y.). O.A.T.S. was supported by an NHMRC/ARC Strategic Award (Grant 401162) and the Project Grant (104532) and was facilitated through access to the Australian Twin Registry, which is funded by an NHMRC Enabling Grant (310667).
Author information
Authors and Affiliations
Contributions
M.A.B., D.F., R.H.W., H.X. and P.F.B. conceived the study. M.A.B., P.F.B., P.M.V., H.X., R.J.W., D.C.R., B.J.M., M.M., R.H.W., N.R.W., B.B., F.G. and P.G.N. obtained funding. B.B., J. He, Q.Z., J.G., P.L., Z.L., Z.-H.Z., P.M.V. and N.R.W. performed statistical analysis. B.B., F.G., Z.L., M.M., S. Shah, J.Y. and N.R.W. performed follow-up analysis. D.F., J. He, L.T., L.C., X.L. and M.A.B. provided Chinese ALS and control samples. H.X., D.Z., W.Y., X.W., T.L., M.L. and M.A.B. provided Chinese GWAS control samples. J.W., Z.-B.J., Z. Li, A.K.H., J.Y., P.G.N., R.L.J., M.D., S.F., S.T.N. and T.J.B. provided data. H.-W.D., Y.L., S.R., Y.-Y.H., L.J.T. and X.-D.C. provided HNU WES controls. P.A.M., R.D.H., R.P., D.B.R., I.P.B., P.S., K.L.W., A.K.H., E.P.M., J.A.F. and N.R.W. provided Australian ALS and control samples. L.A., K.C., J.E., J. Harris, S. Song, A.K.H., L.W., H.X. and M.A.B. performed genotyping. J.H.V., W.v.R., A.A.-C. and L.H.v.d.B. provided European GWAS data. B.B., Q.Z., J.G., F.G. and N.R.W. wrote the first draft of the manuscript. All authors contributed to revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Benyamin, B., He, J., Zhao, Q. et al. Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat Commun 8, 611 (2017). https://doi.org/10.1038/s41467-017-00471-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-017-00471-1
This article is cited by
-
Abnormal Brain Protein Abundance and Cross-tissue mRNA Expression in Amyotrophic Lateral Sclerosis
Molecular Neurobiology (2024)
-
Discovery and Exploration of Lipid-Modifying Drug Targets for ALS by Mendelian Randomization
Molecular Neurobiology (2024)
-
Cross-ancestry genetic architecture and prediction for cholesterol traits
Human Genetics (2024)
-
Genetic control of DNA methylation is largely shared across European and East Asian populations
Nature Communications (2024)
-
Genome-wide analyses identify NEAT1 as genetic modifier of age at onset of amyotrophic lateral sclerosis
Molecular Neurodegeneration (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.