Abstract
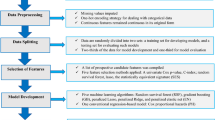
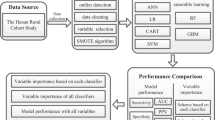
Current studies have shown the controversial effect of genetic risk scores (GRSs) in hypertension prediction. Machine learning methods are used extensively in the medical field but rarely in the mining of genetic information. This study aims to determine whether genetic information can improve the prediction of incident hypertension using machine learning approaches in a prospective study. The study recruited 4592 subjects without hypertension at baseline from a cohort study conducted in rural China. A polygenic risk score (PGGRS) was calculated using 13 SNPs. According to a ratio of 7:3, subjects were randomly allocated to the train and test datasets. Models with and without the PGGRS were established using the train dataset with Cox regression, artificial neural network (ANN), random forest (RF), and gradient boosting machine (GBM) methods. The discrimination and reclassification of models were estimated using the test dataset. The PGGRS showed a significant association with the risk of incident hypertension (HR (95% CI), 1.046 (1.004, 1.090), P = 0.031) irrespective of baseline blood pressure. Models that did not include the PGGRS achieved AUCs (95% CI) of 0.785 (0.763, 0.807), 0.790 (0.768, 0.811), 0.838 (0.817, 0.857), and 0.854 (0.835, 0.873) for the Cox, ANN, RF, and GBM methods, respectively. The addition of the PGGRS led to the improvement of the AUC by 0.001, 0.008, 0.023, and 0.017; IDI by 1.39%, 2.86%, 4.73%, and 4.68%; and NRI by 25.05%, 13.01%, 44.87%, and 22.94%, respectively. Incident hypertension risk was better predicted by the traditional+PGGRS model, especially when machine learning approaches were used, suggesting that genetic information may have the potential to identify new hypertension cases using machine learning methods in resource-limited areas.
Clinical trial registration
The Henan Rural Cohort Study has been registered at the Chinese Clinical Trial Register (Registration number: ChiCTR-OOC-15006699). http://www.chictr.org.cn/showproj.aspx?proj=11375.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Mills KT, Bundy JD, Kelly TN, Reed JE, Kearney PM, Reynolds K, et al. Global disparities of hypertension prevalence and control: a systematic analysis of population-based studies from 90 countries. Circulation. 2016;134:441–50.
Lawes CMM, Vander Hoorn S, Rodgers A, Hypertens IS. Global burden of blood-pressure-related disease, 2001. Lancet. 2008;371:1513–8.
Rapsomaniki E, Timmis A, George J, Pujades-Rodriguez M, Shah AD, Denaxas S, et al. Blood pressure and incidence of twelve cardiovascular diseases: Lifetime risks, healthy life-years lost, and age-specific associations in 1·25 million people. Lancet. 2014;383:1899–911.
Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–87.
Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–25.
International Consortium for Blood Pressure Genome-Wide Association S, Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9.
Li C, Sun D, Liu J, Li M, Zhang B, Liu Y, et al. A prediction model of essential hypertension based on genetic and environmental risk factors in Northern Han Chinese. Int J Med Sci. 2019;16:793–9.
Fava C, Sjogren M, Montagnana M, Danese E, Almgren P, Engstrom G, et al. Prediction of blood pressure changes over time and incidence of hypertension by a genetic risk score in Swedes. Hypertension. 2013;61:319–26.
Lim NK, Lee JY, Lee JY, Park HY, Cho MC. The role of genetic risk score in predicting the risk of hypertension in the Korean population: Korean genome and epidemiology study. PLoS One. 2015;10:e0131603.
Lu X, Huang J, Wang L, Chen S, Yang X, Li J, et al. Genetic predisposition to higher blood pressure increases risk of incident hypertension and cardiovascular diseases in Chinese. Hypertension. 2015;66:786–92.
Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–30.
Zhang L, Wang Y, Niu M, Wang C, Wang Z. Machine learning for characterizing risk of type 2 diabetes mellitus in a rural Chinese population: the Henan Rural Cohort Study. Sci Rep. 2020;10:4406.
Heo J, Yoon JG, Park H, Kim YD, Nam HS, Heo JH. Machine learning-based model for prediction of outcomes in acute. Stroke Stroke. 2019;50:1263–5.
Zhang L, Wang Y, Niu M, Wang C, Wang Z. Nonlaboratory-based risk assessment model for type 2 diabetes mellitus screening in Chinese rural population: a joint bagging-boosting model. IEEE J Biomed Health Inform. 2021. https://doi.org/10.1109/JBHI.2021.3077114. Online ahead of print.
Liu X, Mao Z, Li Y, Wu W, Zhang X, Huo W, et al. Cohort Profile: The Henan Rural Cohort: a prospective study of chronic non-communicable diseases. Int J Epidemiol. 2019;48:1756–1756j.
Liu LS, Hypertension WGoCGftMo. 2010 Chinese guidelines for the management of hypertension. Chin J Cardiol. 2011;39:579–615.
Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–8.
Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–93.
Lu X, Wang L, Lin X, Huang J, Charles Gu C, He M, et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum Mol Genet. 2015;24:865–74.
Hong KW, Go MJ, Jin HS, Lim JE, Lee JY, Han BG, et al. Genetic variations in ATP2B1, CSK, ARSG and CSMD1 loci are related to blood pressure and/or hypertension in two Korean cohorts. J Hum Hypertens. 2010;24:367–72.
He J, Kelly TN, Zhao Q, Li H, Huang J, Wang L, et al. Genome-wide association study identifies 8 novel loci associated with blood pressure responses to interventions in Han Chinese. Circ Cardiovasc Genet. 2013;6:598–607.
Cox DR. Regression models and life-tables. J R Stat Soc Ser. 1972;34:187–202.
Bishop CM. Neural networks for pattern recognition. Oxford; Oxford University: 1996.
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
Friedman J. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001;29:1189–232.
Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594.
Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36.
Pencina MJ, D’Agostino RB Sr., D’Agostino RB Jr., Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–72.
Pencina MJ, D’Agostino RB Sr., Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21.
Kerr KF, Brown MD, Zhu K, Janes H. Assessing the clinical impact of risk prediction models with decision curves: guidance for correct interpretation and appropriate use. J Clin Oncol. 2016;34:2534–40.
Forman JP, Stampfer MJ, Curhan GC. Diet and lifestyle risk factors associated with incident hypertension in women. JAMA. 2009;302:401–11.
Kaur P, Rao SR, Radhakrishnan E, Rajasekar D, Gupte MD. Prevalence, awareness, treatment, control and risk factors for hypertension in a rural population in South India. Int J Public Health. 2012;57:87–94.
Liu MW, Yu HJ, Yuan S, Song Y, Tang BW, Cao ZK, et al. Association between fruit and vegetable intake and the risk of hypertension among Chinese adults: a longitudinal study. Eur J Nutr. 2018;57:2639–47.
Holtermann A, Schnohr P, Nordestgaard BG, Marott JL. The physical activity paradox in cardiovascular disease and all-cause mortality: the contemporary Copenhagen General Population Study with 104 046 adults. Eur Heart J. 2021;42:1499–511.
Niiranen TJ, Havulinna AS, Langen VL, Salomaa V, Jula AM. Prediction of blood pressure and blood pressure change with a genetic risk score. J Clin Hypertens. 2016;18:181–6.
Oikonen M, Tikkanen E, Juhola J, Tuovinen T, Seppala I, Juonala M, et al. Genetic variants and blood pressure in a population-based cohort: the Cardiovascular Risk in Young Finns study. Hypertension. 2011;58:1079–85.
Taal HR, Verwoert GC, Demirkan A, Janssens AC, Rice K, Ehret G, et al. Genome-wide profiling of blood pressure in adults and children. Hypertension. 2012;59:241–7.
Kullo IJ, Jouni H, Austin EE, Brown SA, Kruisselbrink TM, Isseh IN, et al. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES clinical trial). Circulation. 2016;133:1181–8.
Taylor RA, Moore CL, Cheung KH, Brandt C. Predicting urinary tract infections in the emergency department with machine learning. PLoS One. 2018;13:e0194085.
Sato M, Morimoto K, Kajihara S, Tateishi R, Shiina S, Koike K, et al. Machine-learning approach for the development of a novel predictive model for the diagnosis of hepatocellular carcinoma. Sci Rep. 2019;9:7704.
Kruse C, Eiken P, Vestergaard P. Machine learning principles can improve hip fracture prediction. Calcif Tissue Int. 2017;100:348–60.
Acknowledgements
The authors thank all the participants, coordinators, and administrators for their support and help during the research.
Funding
This research was supported by the Henan Natural Science Foundation of China (Grant No: 182300410293), Foundation of National Key Program of Research and Development of China (Grant no: 2016YFC0900803, 2019YFC1710002), National Natural Science Foundation of China (Grant no: 81573243, 81602925), Foundation of Medical Science and Technology of Henan Province (No: 201702367, 2017T02098), and the Discipline Key Research and Development Program of Zhengzhou University (Grant no: XKZDQY202008, XKZDQY202002). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Niu, M., Wang, Y., Zhang, L. et al. Identifying the predictive effectiveness of a genetic risk score for incident hypertension using machine learning methods among populations in rural China. Hypertens Res 44, 1483–1491 (2021). https://doi.org/10.1038/s41440-021-00738-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41440-021-00738-7
Keywords
This article is cited by
-
Development of risk models of incident hypertension using machine learning on the HUNT study data
Scientific Reports (2024)