Abstract
The human Major Histocompatibility Complex (MHC) or Human Leukocyte Antigen (HLA) super-locus is a highly polymorphic genomic region that encodes more than 140 coding genes including the transplantation and immune regulatory molecules. It receives special attention for genetic investigation because of its important role in the regulation of innate and adaptive immune responses and its strong association with numerous infectious and/or autoimmune diseases. In recent years, MHC genotyping and haplotyping using Sanger sequencing and next-generation sequencing (NGS) methods have produced many hundreds of genomic sequences of the HLA super-locus for comparative studies of the genetic architecture and diversity between the same and different haplotypes. In this special issue on ‘The Current Landscape of HLA Genomics and Genetics’, we provide a short review of some of the recent analytical developments used to investigate the SNP polymorphisms, structural variants (indels), transcription and haplotypes of the HLA super-locus. This review highlights the importance of using reference cell-lines, population studies, and NGS methods to improve and update our understanding of the mechanisms, architectural structures and combinations of human MHC genomic alleles (SNPs and indels) that better define and characterise haplotypes and their association with various phenotypes and diseases.
Similar content being viewed by others
Introduction
The human Major Histocompatibility Complex (MHC) on the short arm of chromosome 6 (band p21.3) is a Human Leukocyte Antigen (HLA) super-locus composed of clusters of many tightly linked supergenes involved with various phenotypic functions, mostly in connection with the immune response1,2,3,4. The MHC genes are defined as supergenes on the basis that they are clusters of tightly linked functional genetic elements spanning hundreds of kilobases that control complex balanced phenotypes and are inherited as a unit [haplotype] owing to reduced or absent recombination within them5, and because many have evolved by genomic duplications, deletions and inversions6. Although the most common mechanism of supergene formation is considered to be by inversion7,8, in which single crossovers between heterozygotes may lead to unbalanced gametes, the MHC genomic organisation reveals a variety of haplotypes with segmental duplications9,10,11, and structurally variant loci such as C4 and DRB12, and a variety of duplicated repeat elements6,13,14, that exist possibly due to balancing selection15,16. These duplicated and inverted homologues probably generate recombinant haplotypes by varying rates of non-allelic and allelic homologous and nonhomologous recombinations and crossovers12,17. Thus, finding reliable phenotypic associations by genome-wide association studies (GWAS) is complicated and masked by the presence of hundreds of interlinked genes and regulatory elements in strong linkage disequilibrium (LD) within the super-locus18,19,20.
The HLA super-locus is characterised specifically by twelve classical class I and class II genes that encode antigen-presenting HLA proteins that present host (self) or foreign (nonself) peptides to interact with T-cell receptors in order to discriminate between self and nonself as part of the host immune response3,20,21,22,23. This is an important immunogenetic regulatory region24 of ~4 Mb in length with more than 120 non-HLA genes that together with the classical and non-classical HLA genes have been associated with more diseases than probably any other region of the human genome1,2,12,25. It is one of the most complex and diverse genomic regions with high levels of polymorphism, gene duplications, repeat elements, structural variations (indels), and long-range haplotype segments or blocks known as Conserved Extended Haplotypes (CEHs)18 or Ancestral Haplotypes (AHs)10. The diversity of the variable long-range haplotype segments within heterozygote individuals has provided problems and challenges for assigning SNPs to loci, and assembling structural variants of numerous duplicated genes particular in regard to associating them as genetic markers or causative agents for many of the immune-related phenotypes and diseases18. In recent years, more attention is being given to gaining a better understanding of MHC haplotypes by phased long-range sequencing as an extension of genotyping and identifying genic and non-genic alleles for associating them with disease, bone marrow transplantation, and for ascertaining the effects of immunotherapy26. Reliable MHC linkage mapping and haplotyping usually are dependent on pedigree studies of particular genotyped markers to evaluate their linkage or segregation in meiosis18 or on phased genomic sequences26, such as those that have been sequenced or genotyped using multilocus HLA-captured haplotype phasing27,28, de novo assembled trios29, MHC homozygous cell-lines11, sperm30 or single chromosomes31. Because of the complexity of the MHC as a HLA super-locus with a myriad of interconnected gene systems and sub-genomic regions, it is a gradual and continuing difficult process to build up the genetic, molecular and functional knowledge about the architectural and functional organisation of haplotypes in this region and their overall contribution to health and disease1,2,25,26,32,33.
In this brief review, we outline some of the recent analytical developments used to investigate the SNP polymorphisms, structural variants (indels), expression quantitative trait locus (eQTL) and haplotypes of the HLA super-locus. We highlight the importance of using reference cell-lines, population studies and next-generation sequencing (NGS) methods to overcome past problems and to improve and update our understanding of the mechanisms and architectural structures and combinations of human MHC genomic alleles (SNPs) that better define and characterise haplotypes, and their association with various phenotypes and diseases.
MHC genomic sequence and subdivisions of structural organisation
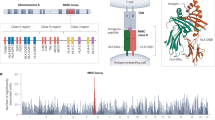
The first fully sequenced and gene annotated human genomic MHC was published in 1999 using the pioneering Sanger sequencing technology34. This primary sequence was a ‘virtual MHC’ composed of a mosaic of different human haplotypes rather than presenting any one particular haplotype. Subsequently, the first generation genomic sequences of eight human ancestral MHC haplotypes were published for a more precise comparative genomic analysis of the similarities and differences between different haplotypes35. Figure 1 shows the gene map of the HLA genomic region based on Genome Reference Consortium Human Build 38 patch release 14 (GRCh38.p14) in the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/genome/?term=human) and the MHC-PGF haplotype, one of the eight MHC haplotypes sequenced by the MHC Haplotype Consortium (Fig. 1A)35. The MHC genomic organisation has a high degree of evolutionary complexity with the remnants of many homologous segmental duplications6 as well as inversions (Fig. 1B); probably turned over and shuffled by many different ancestral hominoid haplotypes as a result of non-allelic and allelic homologous recombination, gene conversion (nonhomologous recombination) and sequence crossover between different homozygotes or heterozygotes (Fig. 1C).
A Gene map of the HLA genomic region that corresponds to the genomic coordinates of 29602228 (GABBR1) to 33410226 (KIFC1) on chromosome 6 in the human genome GRCh38.p13 primary assembly of the NCBI map viewer. The regions separated by arrows show the HLA sub-regions such as extended class I, class I, class III, classical class II and extended class II regions from telomere (left and top side) to centromere (right and bottom side). The red and blue double, horizontal arrows show the spans of alpha, kappa and beta blocks, and framework (FW), non-HLA gene blocks, FW1 and FW2, respectively. The Class III region is composed of non-HLA genes or FW genes, but is known traditionally as Class III. White or coloured (orange, red and blue) boxes, grey, and black boxes show protein-coding genes, non-coding RNAs (ncRNAs), and pseudogenes, respectively. Red, green and blue letters indicate HLA class I, MIC, and class II genes, respectively. Adapted from Shiina et al.1,2. B Cis and trans structural orientation of duplicated and inverted HLA class I and class II genes within their duplication blocks (alpha, kappa, beta, delta and epsilon) relative to the telomeric and centromeric ends (left to right, respectively) of the HLA super-locus. The duplicated MIC pseudogenes, MICE, MICG, MICF, MICD, MICC, and their locations in the alpha block (A) are not shown in B. All MIC pseudogenes and genes in the MHC genomic region are coded in the opposite direction to all the HLA class I genes and pseudogenes6. Solid arrows indicate the 5′ to 3′ direction of coding genes and dotted arrows indicate the 5′ to 3′ direction of pseudogenes (italicised). The structural variants for the HLA-DRB3, DRB4 and DRB5 genes in the class II region and the C4 genes in the class III region are indicated by the enclosed vertical boxes. The location and distribution of the duplicated genes are not shown to exact genomic scale. C Chromosomal or SNP density crossover (XO) junction in comparisons between two homozygous haplotype pairs (AB/AB and ab/ab) and two heterozygous recombinant (haplotype) pairs (AB/Ab and aB/ab). Chromosomal recombination is shown with a XO located between loci A and B within haplotype region ‘AB’ and between loci a and b within haplotype region ‘ab’ in a diploid cell during meiosis.
The HLA super-locus is divided into three regions related to the functions and distributions of the duplicated HLA genes and pseudogenes; the class I region located at the telomeric end and the class II region at the centromeric end, both separated from each other by an extended class III region of 61 protein-coding genes1,2. Whereas the HLA class I and class II genomic regions encode the highly polymorphic gene complex of the HLA class I and HLA class II genes, the class III region consists of many different non-HLA genes that are involved in stress response (HSPA1A, HSPA1B and HSPA1L), complement cascade (C4A, C4B, C2, CFB), immune regulation (NFKBIL1, FXBPL and DDX39B), inflammation (LTA, LTB, LST1, ABCF1, AIF1, NCR3 and TNF), leukocyte maturation (LY6G5B, LY6GSC, LY6G6D, LY6G6E and LY6G6C), and regulation of T cell development and differentiation (BTNL2)4,36. Recently, Zhou et al. showed that a quartet of MHC class III genes (NELF-E, SKIV2L, DXO and STK19) are involved with the metabolism and surveillance of RNA during the transcriptional and translational processes of gene expression37. The class II region also contains some proteosome-processing and peptide antigen transportation non-HLA genes such as PSMB8, PSMB9, TAP1, and TAP2. The TAP-binding protein, TAPBP, is in the extended class II region. The ‘Class I’ region (telomeric to centromeric ends) ranges from HLA-F to MICB, ‘Class III’ from PPIAP9 to BTNL2, and ‘Class II’ from HLA-DRA to HLA-DPA3. There also are sub-regions from the telomeric side of Class I and the centromeric side of Class II that are called the ‘Extended class I’ (telomeric side of HCG4P11) and ‘Extended class II’ (centromeric side of COL11A2) regions, respectively. The class I region has been divided into three genomic blocks, alpha, beta and kappa6,10,38, that include duplicated HLA genes on either side of two intervening blocks of framework (FW1 and FW2) genes (Fig. 1A) that include non-HLA genes39. HLA-A, -G and -F are in the alpha block, HLA-B and -C are in the beta block, and HLA-E is in the kappa block.
A total of 283 loci were identified and/or reclassified in the 3.78-Mb HLA genomic region of the PGF haplotype from GABBR1 located on the extended class I region to KIFC1 located on the extended class II region (Fig. 1A and Table 1). When all the loci of the HLA genomic region are grouped into four categories of gene types, then 144 loci are classified as a protein-coding gene, 53 loci are non-coding RNA (ncRNA), five loci are small nucleolar RNA (snoRNA) and 81 loci are pseudogenes (Table 1). Of the 283 loci, 15.5% (44 loci) are occupied by HLA and HLA-like genes (HLA class I, HLA class II and MHC class I polypeptide-related sequences or MIC genes). However, the genic and non-genic numbers in Table 1 are not absolute for the MHC genomic region because of haplotype differences that may involve structural variations due to duplications, deletions, and insertions.
Of the HLA and HLA-like genes, 18 HLA class I genes (six protein-coding genes and 12 pseudogenes) (Fig. 1B) and 7 MIC genes (two protein-coding genes and five pseudogenes) are located in the HLA class I region, and 18 HLA class II genes (13 protein-coding genes and five pseudogenes) are in the HLA class II region (Fig. 1A and Table 2). Also, one HLA class I 88-bp pseudogene (HLA-Z) is located within the ncRNA gene LOC100294145 close to the HLA-DMB gene in the HLA class II region. The classical HLA class I genes, HLA-A, -B and -C, and the classical HLA class II genes, HLA-DR, -DQ and -DP, are characterised by their extraordinary polymorphisms, whereas the non-classical HLA class I genes, HLA-E, -F and -G, are differentiated by their tissue-specific expression and limited polymorphism (Table 2).
Apart from the protein coding genes, pseudogenes, non-coding transcribed RNA loci, and small nucleolar transcribed RNAs (snoRNAs) loci, there are at least 8604 repeat elements including those known as transposable elements (TEs) and/or retroelements, and 723 simple repeats (microsatellites) in the MHC PGF haplotype sequence. Table 3 lists the main families of repeat elements identified and classified by RepeatMasker (http://www.repeatmasker.org) as a percentage of genomic sequence both within the intervening sub-regions, and within the entire MHC region from HLA-F to HLA-DPA3. The SINEs that congregated mainly in FW2 (26%) and class III (21%) regions were lowest in the alpha, kappa, beta, and class II blocks at <10%. The LINEs, mostly fragmented and of the mammalian L1M types, were found at highest percentage in the kappa block (31%), and within the beta block, FW1, and class II region, each at 26%. The ERVL subfamily of the LTR family were in the alpha and beta blocks at least at three to ten times higher percentage than within the other subregions. The LTR and ERVL were highest in the alpha block (25% and 13%, respectively) and lowest in the class III region (4% and 0.3%, respectively). Many of the LTR/HERVs form the building blocks of the transcriptional regulatory elements40, and their relatively high content in the alpha and beta blocks (Table 3) may reflect a role in the duplication of the HLA genes within the MHC6,41,42,43,44. The overall total percentage of the interspersed repeat elements (IREs) was highest in the beta (61%) and alpha (58%) blocks and lowest in the class III region (41%). On the other hand, the class III region and FW2 had the highest GC level percentage at 49% and 48%, respectively, possibly reflecting the greater density of coding genes within these two regions.
Homozygous cell-lines as MHC genomic sequence haplotype references
Haplotypes at the genomic sequence level are blocks of phased coding and non-coding nucleotide sequences of multiple loci that are in the same orientation (cis) as their mode of gene transcription and regulation26. The characterisation and understanding of MHC haplotypes in modern disease and population genetics began in 1967 with the introduction of the word ‘haplotype’ by Ruggero Ceppellini to describe alleles in the HLA system45, and expanded in the 1990s with the pedigree studies of the research groups of Alper9,18, and Dawkins10,46,47. Since then, the International Histocompatibility Workshop Group (IHWG) has provided at least a thousand commercially available cell-line samples from HLA heterozygous and homozygous donors, families, and diverse populations (https://www.fredhutch.org/en/research/institutes-networks-ircs/international-histocompatibility-working-group.html) that are important for research into MHC immunogenetics, comparative genomics, transcriptomics and haplomics11,18,28,35,46,47 These genotyped or fully sequenced MHC haplotypes provide standardised references to assist with the design and interpretation of HLA genotyped population studies and HLA-disease relationships. The genotyped cell-lines also provide excellent insights into the structural organisation of MHC phased haplotypes11, not previously available for detailed comparative analysis by just using blood or tissues samples collected from diploid heterozygous individuals. The first MHC genomic sequence variations in different haplotypes were produced by the Sanger Centre MHC Haplotype Project (SCMHP) using eight homozygous cell-lines35. These now are alternative reference sequences as part of the human reference genome GRCh3848. Initially, only two haplotypes were resolved completely at the base pair level (cell-lines PGF and COX); whereas the other six haplotypes were completed only at 51% (cell-line APD) to 93% (cell-line QBL) of the MHC genomic region. Seven of the SCMHP cell-lines were resequenced again as part of 95 near-complete haplotypes, using short-range and long-range NGS11,49. Overall, Norman et al. provided 137 genotyped loci for most of the 95 cell-lines that they sequenced11.
Table 4 shows the diversity of 68 different haplotypes at six HLA class I and class II loci for eight cell-lines sequenced by the SCMHP, and 82 IHWG reference cell-lines sequenced, genotyped, and annotated by Norman et al.11 whereas Norman et al.11 genotyped for polymorphisms at 139 MHC loci in the MHC class I, II and III regions, for simplicity, the haplotypes listed in Table 4 are shown only for the six HLA class I and class II loci of the classical genes, HLA-A, -C, -B, -DRB1, -DQA1 and -DQB1. Nevertheless, these 68 examples illustrate the segmental organisation of the haplotypes, whereby some blocks of consecutive loci are (1) the same or highly similar (homozygous, conserved, shared or matched), (2) different (heterozygous or diverse), or (3) a hybrid recombinant (mixed) composed of adjoining blocks of conserved and different sequences12,13,14,50. The AH/CEH nomenclature in Table 4 is taken from Dorak et al.47. The AH names use the B allele and if two or more AH carry the same B allele then sequential numbers are added to indicated the order of discovery, such as AH7.1 and AH7.247. In Table 4, four different cell-lines (PGF, SCHU, HO104, LD2B)11 have the haplotypic structure of AH7.147, which is a ‘homozygous’ or ‘conserved’ haplotype represented by the HLA lineage alleles A*03-C*07-B*07-DRB1*15-DQA1*01:02-DQB1*06. AH7.2 has C*07-B*07, but differs to AH7.1 at A*24-C*07-B*07-DRB1*01-DQA1*01:01-DQB1*0547. Similarly, AH8.147 is highly conserved in five different homozygous cell-lines (COX, STEINLIN, VAVY, L0541265, PF04015) with the HLA lineage alleles of A*01-C*07-B*08-DRB1*03-DQA1*05-DQB1*02 at six loci. These haplotype nomenclatures can be expanded from the one allelic set of digits up to four or six sets of digits. For example, the following AH8.147 is classified using 4 allelic digital numbers at five HLA loci: A*01:01-C*07:01-B*08:01-DRB1*03:01-DQA1*05:01-DQB1*02:01.
The allelic combinations of the BOLETH cell-line (AH62.1) and the MCF cell-line (A*02-C*03-B*15-DRB1*04-DQA1*03-DQB1*03) are totally different to those of the AH7.1 and AH8.1 cell-lines at the six MHC loci. The AH7.1 and AH8.1 allele lineages47 are different from each other at all the six loci except at HLA-C where they are both C*07; although they actually are different from each other at the two digital allelic level, C*07:02 and C*07:01, respectively. This two digital allelic difference represents the two amino acid difference between the HLA-C proteins for AH7.1 (PGF) and AH8.1 (COX) with K90N in exon 2 and S125Y in exon 3. Comparatively, most of the 68 haplotypes in the Norman et al.11 study are hybrids or recombinants that are different at one or more loci, but share the same alleles possibly at other loci. For example, the ten haplotypes with the allele A*01:01:01:01 at the HLA-A locus are different at one or more of the other five loci. However, some of these A*01 haplotypes have the same alleles at other loci. There are two haplotypes that are both A*01:01:01-C*07:01:01, but different from each other at the HLA-B, -DRB1, -DQA1 and -DQB1 loci. Similarly, there are two haplotypes that both have A*01:01:01-DRB1*11:01/02:01-DQA1*05:05:01, but differ from each other at the HLA-C and -B loci. This illustrates the considerable mixing and matching between different haplotypes in a process called shuffling50,51. Similarly, trends of loci shuffling are evident for the 21 haplotypes with A*02:01:01:01, and so on. Genomic sequence comparisons between MHC class I or between class II ‘hybrid’ haplotypes by Kulski et al.13,14 suggest that the haplotypic block or segmental SNP patterns with genomic sequence crossovers (Fig. 2) probably evolved ancestrally using recombination mechanisms17. Conserved and hybrid haplotypes are likely to have accumulated in interrelated populations or ethnic groups in relatively recent times, possibly over a few thousand generations or more52. These shuffling or recombination mechanisms are delineated also as SNP diversity plots in sequence alignments between two phased MHC genomic regions (Fig. 2).
The MHC gene markers and genomic distances (Mb) from left to right between the MOG and COL11A2 genes, and the regions of polymorphic frozen blocks known as alpha, kappa, beta, gamma, delta and epsilon (Dawkins et al.10, Shiina et al.2), are shown at the bottom of the Figure. The four SNP plots (A–D) are between the haplotype PGF: A*03:01-C*07:02-B*07:02-DRB1*15:01-DQA1*01:02-DQB1*06:02 and the haplotypes of A COX: A*01:01-C*07:01-B*08:01-DRB1*03:01-DQA1*05:01-DQB1*02:01:01, B LD2B: A*03:01-C*07:02-B*07:02-DRB1*15:01-DQA1*01:02-DQB1*06:02, C BM14: A*03:01-C*07:02-B*07:02-DRB1*04:01-DQA1*03:01-DQB1*03:02 and D MGAR: A*26:01-C*07:01-B*08:01-DRB1*15:01-DQA1*01:02-DQB1*06:02. The fifth SNP plot (E) is between the haplotype MGAR (see D) and YAR: A*26:01-C*12:03-B*38:01-DRB1*04:02-DQA1*03:01-DQB1*03:02. In F, the SNV plot is between the PGF reference sequence (CZUC02000001.1) and the chimpanzee (CHIMP) genomic reference sequence, GCF_002880755.1. The SNV regions label ‘del’ are genomic sequence regions absent from CHIMP sequence. The Chimpanzee MIC gene in the beta block is a hybrid of human MICA and MICB53,62. The Y-axis presents the number of SNP/kb (window size). The X-axis shows the SNP density positions (SNP/kb) across 3.6 Mb of genomic sequence between the MOG and COL11A2 genes. The red vertical lines along the X-axis that are above 100 on the Y axis are artifactual sequences or those representing sequence gaps, poor assembly, inversions or long runs of unspecified nucleotides. The yellow horizontal boxes labelled SNP POOR are regions of recombination (highly conserved nucleotide sequence with little or no SNPs between sequence alignments). In this context, the SNP POOR regions are those that are <1 SNP/kb, in contrast to the same regions that are SNP rich (>1 SNP/kb) in other haplotype sequence comparisons. The MHC class III and most FW genes in the class I region are always SNP poor, and consequently were not labelled as such in A, D or E. The ends of the ‘SNP POOR’ boxes represent regions of putative crossovers (vertical arrows) between SNP poor and SNP rich regions of different haplotypes in C–E. In B, PGF v LD2B shows the relative absence of SNPs across 3.6 Mb between two conserved (highly similar sequences) haplotypes. Extended genomic regions (>50 kb) with 1-50 SNP/kb are considered to be SNP rich regions, whereas extended regions of < 1 SNP/kb are SNP poor regions. The SNPs within SNP poor regions were easy to count manually because of small numbers (<0.1 SNP/kb), whereas SNP rich regions were difficult to count because of larger numbers at an average of 7 SNP/kb in the alpha block (320 kb), and up to 50 SNP/kb or greater in the delta block (185 kb) depending on haplotype comparisons. In the alpha block, the highest average SNP density between seven different haplotypes was 16 SNP/kb near HLA-A with the lowest density at ~2 SNP/kb near the HLA-J pseudogene13. The SNP count was <0.001 SNP/kb between the same HLA-A haplotypes in C and D. Spikes and peaks of SNPs above 100 SNP/kb were due mostly to nucleotide misalignments because of poor sequence assembly, structural variations, gaps, inversions or long runs of unspecified nucleotides. See recent SNP plots by Houwaart et al.49 for additional comparisons between MHC haplotypes.
Haplotype SNP diversity plots and crossover junctions
Figure 2 shows SNP diversity plots in nucleotide DNA comparisons between the same and different human MHC haplotypes as well as to that of a chimpanzee haplotype sequence. SNPs are the nucleotide sequence differences seen between two different phased haplotypes that have been aligned (Fig. 2A, E, F). Sequence alignments between different haplotypes (heterozygous sequences) reveal varying SNP densities (number of SNPs per kb) across the entire MHC with the greatest SNP densities occurring in the alpha block within the HLA-A gene region; the HLA-B and -C genes of the beta block; the delta block with HLA-DRB1, -DQA1 and -DQB1; and the epsilon block involving HLA-DPB1. Unsurprisingly, the highest SNP density peaks occur in the regions of the HLA classical class I and class II genes that correlate positively with the overall number of alleles detected for the different HLA gene loci (Table 2). In comparison, the SNP densities are consistently at low levels in the non-HLA genetic regions such as those between the alpha and beta blocks in the class I region, and in the class III region where the number of alleles for each of the class III genes are often <20, and comparable to the allele numbers detected for non-classical HLA genes, like HLA-F, and HLA pseudogenes (Table 2).
Fewer SNPs are detected between two aligned homologous or highly similar sequences (e.g., Fig. 2B, PGF versus LD2B) than between different haplotypes (e.g., Fig. 2A, PGF v COX) because they are identical by descent with no recombination. However, some nucleotide differences either as de novo mutations and/or sequencing or assembly errors are evident across the alignment between fully matched HLA loci (conserved haplotypes). In contrast, sequence alignments of recombinant haplotypes (e.g., Fig. 2C–E) reveal an extended sequence block that is rich in SNPs adjoining an extended block of homologous sequences with no or few SNPs (labelled as a SNP poor or SP) that are seen to be SNP rich in other haplotype comparisons (Fig. 2A). The junction between the SNP rich and SNP poor blocks are the SNP crossover junctions suggesting that they are in close proximity to chromosomal recombination crossover regions13,14, as outlined in Fig. 1C. With recombinations and crossovers, a considerable amount of opportunistic hitchhiking may occur particularly near the HLA loci53, and with the integration and rearrangement of Alu, LTR and HERV elements54.
Supergene expression, eQTL, epistasis and disease
Since undertaking our earlier analyses of MHC gene variants, epistatic interactions, expression activity and associations with various diseases taken from publications and records in public databases such as the Gene Expression Omnibus (GEO), Online Mendelian Inheritance in Man (OMIM) and the Genetic Association Database (GAD)1,2, these types of genome-wide MHC association studies have progressed much further with the more formidable bioinformatic analyses of phenotype associations, known as MHC PheWAS55. However, regulatory elements can act over long distances and in a cell-type specific manner that hamper the easy identification of the causal genes for a given pathological condition56,57. In this regard, haplotyped homozygous cell-lines also can be used to study gene interactions or epistasis both inside and outside the MHC genomic region16,58,59. Expression quantitative trait locus (eQTL) studies associate genomic and transcriptomic data sets from the same individuals to identify loci that affect mRNA expression by linking SNPs to changes in gene expression58. Thus, eQTL analysis can be an useful procedure for annotating GWAS variants.
A number of recent studies using homozygous cell-lines and/or biological samples have demonstrated that the expression of various clusters of genes inside or outside the MHC genomic region can be affected by the expression of one or more haplotypic genes within the MHC genomic region58,59,60,61. Lam et al. used eight homozygous cell-lines, six with Chinese haplotypes (A*33:03-C*03:02-B*58:01-DRB1*03:01 or A*02:07-C*01:02-B*46:01-DRB1*09:01), and two with European haplotypes (A*01:01-C*07:01-B*08:01-DRB1*03:01)58. They used haplotypic RNA and DNA-sequencing data to show that haplotype sequence variations represented by eQTL SNP alleles can function as cis-acting regulatory variants for multiple MHC genes. The enriched haplotype-specific transcriptional eQTLs were localised especially within four segmental regions containing HLA-A (alpha block), HLA-C (beta block), C4A (gamma block) and HLA-DRB (delta block). Thirty-six MHC genes from extended MHC and classes I, II and III showed significantly differential expression between the three MHC haplotypes.
Lamontagne et al. used hundreds of lung tissue samples collected from patients in Canada and the Netherlands to show that gene expression within the extended MHC region and class I, II and III regions correlated with lung disease/trait specific local- and distant-acting eQTL SNPs60. By using eQTL analysis of a large human cohort with both RNA-sequencing and genotyping data available for HLA alleles in peripheral blood, Sharon et al. found strong trans-regulatory associations between the HLA-DR, HLA-DQ, or HLA-DP β chains and the T cell receptor (TCR) α chains61. Their results suggest that MHC genotypes have a key role in shaping the TCR repertoire by determining the V gene usage profiles of an individual’s TCR repertoire. In a recent in-depth interrogation of associations between genetic variation, gene expression and disease, D’Antonio et al. showed that eQTL analyses of HLA haplotypes provided substantially greater statistical power than only using single variants59. They examined the association between AH8.1 and delayed colonisation in Cystic Fibrosis, and suggested that downregulation of RNF5 expression was the likely causal mechanism. Taken together, these pioneering eQTL studies incorporating HLA haplotypes are a powerful approach to identify causal genetic mechanisms underlying disease associations both inside and outside the MHC region. In this regard, we recently developed a new RNA-sequencing method to capture differential allele-level expression and genotypes of all the classical HLA loci and haplotypes in the Japanese population for further in-depth studies of graft rejection after transplantation and HLA-related diseases28.
Structural variants: indels and transposable elements in MHC genomic evolution and regulation of expression
The human MHC structural variants and indels have received far less attention than SNPs and minor variants with respect to health and disease. In comparative genomic analyses between different MHC haplotypes, the indel diversity is two to seven times greater than SNP diversity53,62. Structural variants and indels have a potential gain and loss of functions that can affect phenotypes, susceptibility and resistance to disease via many different molecular, cellular and pathogenic independent and interrelated mechanisms. Figure 3 shows an ~55-kb deletion within the alpha block of a haplotype with HLA-A*24:0213 that has the highest allele frequency of 35.6% in the Japanese population (http://hla.or.jp/med/frequency_search/en/allele/). HLA-A*24:02:01 apparently has a protective effect against Stevens-Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) that are life-threatening acute inflammatory vesiculobullous reactions of the skin and mucous membranes63.
The locations of HLA-H, HLA-T, HLA-K pseudogenes (labelled green boxes) HLA-A (horizontal arrow) and some TE are indicated on the GR_A03C07 sequence. The yellow box labelled C9 represents Charlie9. The location of the intact telomeric HLA-G gene and the deleted pseudogene HLA-U centromeric of HLA-K are not shown. All interspersed repeats in the upper sequence are indicated with the symbols used by Kulski et al.13.
Transposable elements (TEs) have important, albeit, often poorly defined roles in generating haplotypes via recombination mechanisms such as integration (insertion), duplication, rearrangements, deletions and gene conversion64,65. TEs and other repeat sequences appear to have been integral in the generation of MHC segmental duplications of the class I and class II regions6,66, and of different haplotypes, mainly by acting both as recombination acceptor and suppression sequence regions for DNA binding Rec proteins and enzymes such as PRDM9 depending on their genomic distribution, sequence conservation or diversity, and evolutionary age of integration and transposition13,14. The association of particular TEs and repeats with MHC segmental duplications were reported previously for the genomic structural organisation of MHC duplicated genes in humans6, chimpanzees38,62 and rhesus macaques67. Both old and young Alu insertions generate point mutations, microsatellites and SNPs within the flanking regions of the insertion sites68. TEs such as Alu, SVA, HERVs and LTR have been used as genetic markers to estimate the evolutionary age of MHC gene duplication events and for discerning the evolutionary interrelationships between different human haplotypes54,66,69. For example, ten young AluY indels that are either present or absent in particular human MHC class I and class II haplotypes are useful evolutionary genetic markers of past recombination events, as well as excellent markers for elucidating population phylogenetics and genetic interrelationships70,71,72. In this regard, Cun et al. recently showed that five different MHC class II dimorphic Alu elements either alone or linked together as haplotypes with HLA-DRB1 alleles can differentiate 12 Chinese minority ethnic groups according to their geographic locations, and correlate them with their population characteristics of language family, migration and sociality73.
TE insertions within the MHC genomic region might act like surgical sutures or band-aids that help to repair and rejoin double-strand DNA breaks during recombination events41, such as those involved with the ‘mismatch repair system’ or via various other repair mechanisms of damaged DNA17. In this regard, it seems that TEs like Alu, L1, SVA and LTR are involved intimately with recombination, DNA repair, as well as contributing to nucleotide point mutations between different sequences6,13,41. Moreover, some of these TE indels have been strongly associated with the regulation of gene expression and disease74,75. Much work is needed to characterise which MHC TEs have contributed to past recombination events, affect gene expression, and have a role in MHC related diseases, and various important traits and phenotypes associated with pathogen defence.
Population MHC haplotypes
Although homozygous cell-lines can provide phased genomic sequences for analysis of haplotypic structures, population studies are necessary for information about the frequency and distribution of the MHC haplotypes and their association with disease, and for obtaining cross-matching data for organ and cell transplantations. Most frequency data of population MHC haplotypes are based on genotyping HLA alleles of heterozygotes and applying statistical and computation methods such as the expectation-maximisation algorithm or LD values of non-random, multi-allelic correlations between pairs of loci to estimate the correct phase of the haplotypes76. The LD statistical analysis of heterozygotes might be reasonably accurate for estimating high frequency or common haplotypes, but the reliability decreases for low frequency or minor haplotypes. Confounders to haplotype estimations include typing ambiguity, sample size, incompleteness of HLA data, allele frequency errors, recombination and especially unknown gamete phase.
A number of family-based population studies were published in the 1980s and 1990s on extended MHC haplotype frequencies for Caucasians in Australia77, and the United States78, as well as for American non-dominant European Caucasian and non-Caucasian or admixed Caucasian/non-Caucasians18. Since then, the HLA haplotype frequencies have been determined for many more different worldwide populations79,80, and ethnic groups using pedigrees or statistical inference (http://www.allelefrequencies.net/default.asp). Table 5 lists examples of the six most common HLA haplotype frequencies for Japanese, Chinese, Saudi, British Caucasians, European Americans (Caucasians) and African Americans deduced by LD inference or segregation by pedigree analysis. Although we used the British Caucasian population as an example of the common European haplotypes such as AH7.1, AH8.1 and AH44.1 (Table 5), the European HLA haplotype frequencies vary markedly among European populations across the European continent80. According to Dawkins and Lloyd46, the five most common MHC AH haplotypes (at five HLA loci) in Australian Europeans living in Perth, Western Australia are AH8.1 (13.2%), AH7.1 (12.9%), AH44.1 (5.5%), AH44.2 (2.6%) and AH57.1 (2.6%), frequencies which tend to reveal a large immigratory bias towards their British ancestors (Table 5).
The conserved or fixed haplotypes that have little diversity and no evidence of recombination within their genomic sequences such as AH7.1 or AH8.1 of Caucasian individuals (Table 5) can be studied and described as ‘identity by descent’ (IBD) haplotypes81, which are distinct from ‘identity by state’ (IBS) haplotypes, that is, those that have emerged by convergence. The highly conserved haplotypes that are shared between generations (haplotype sharing) might remain fixed or frozen over long periods of evolutionary time because of founder effects and population bottlenecks82, as well as efficient DNA repair mechanisms, negative population selection, or as yet unknown mutation inhibitory mechanisms. To what degree are conserved haplotypes frozen or fixed? Although this question is not resolved fully, available data suggest that many inherited haplotypes are not completely identical and that de novo mutations, SNPs and/or indels, in MHC genomic sequence comparisons do exist between the same conserved haplotypes83,84,85,86. The identification of variants between the same haplotypes might have importance in assisting with optimal donor-recipient selection for allogeneic stem cell transplantation and with reducing acute and chronic graft-versus-host disease26.
On the other hand, heterozygous haplotypes or those that are very different between individuals (e.g., AH7.1 and AH8.1) are likely to have been inherited by an interplay of various genetic and population evolutionary processes including recombination, positive selection of benign mutations or SNPs, gene flow, genetic drift, frequency-dependent selection, admixture and trans-speciation over long periods of evolution15,16,80. For example, the known MHC class I haplotype sequences of Japanese, Africans, Asians, Arabs and Europeans generally are all different to each other in phylogenetic analyses86,87. Despite haplotype sharing of high frequency conserved polymorphic sequences by IBD such as those for AH8.1 or AH7.110,52, most haplotypes among Europeans and other populations (Table 5) generally are markedly different in structure, organisation and frequency as a consequence of various hypothetical genetic and population evolutionary processes80.
Conclusion: third generation sequencing
The new knowledge gathered during the past decade on the architectural complexity and diversity of MHC haplotype genomic sequences stems largely from DNA and RNA sequencing methods, but remains incomplete because it is difficult to assign SNPs correctly to loci and assemble structural variants of numerous duplicated genes within individuals by using the first generation Sanger sequencing method or the short read NGS technology88,89. Despite the large number of genomes produced by second generation sequencing, their quality is compromised by the relatively short reads (usually <250 bp) used to construct them (typically from Illumina sequencing by synthesis)89. Long-read sequencing by third generation sequencing (TGS) together with the many improved bioinformatic tools allow the longer regions of genomic sequence with repetitive elements to be assembled for more reliable haplotype reconstruction90,91,92,93,94. Pacific Biosystems (PacBio) and Oxford Nanopore can generate reads over 10 kb91, which makes TGS ideal for assembling genomes in areas with gene duplications27,28, repetitive elements90 and for generating long haplotype blocks91,92,93. Thus, TGS along with pan-genome bioinformatic analyses have the potential to better assist with haplotype phasing, and for elucidating haplotype regulatory modules within the HLA super-locus and their association with a wide range of complex diseases, including infectious and autoimmune diseases.
References
Shiina, T., Inoko, H. & Kulski, J. K. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens 64, 631–649 (2004).
Shiina, T., Hosomichi, K., Inoko, H. & Kulski, J. K. The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 54, 15–39 (2009).
Wang M., Claesson M. H. Immunoinformatics (eds. De R. K. & Tomar N.). Immunoinformatics, pp 309–317 (Springer New York, 2014).
Trowsdale, J. & Knight, J. C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genom. Hum. Genet. 14, 301–323 (2013).
Campoy, E., Puig, M., Yakymenko, I., Lerga-Jaso, J. & Cáceres, M. Genomic architecture and functional effects of potential human inversion supergenes. Philos. Trans. R. Soc. B 377, 20210209 (2022).
Kulski, J. K., Gaudieri, S., Martin, A. & Dawkins, R. L. Coevolution of PERB11 (MIC) and HLA class genes with HERV-16 and retroelements by extended genomic duplication. J. Mol. Evol. 49, 84–97 (1999).
Black, D. & Shuker, D. M. Supergenes. Curr. Biol. 29, R615–R617 (2019).
Porubsky, D. et al. Recurrent inversion polymorphisms in humans associate with genetic instability and genomic disorders. Cell 185, 1986–2005.e26 (2022).
Alper, C. A., Raum, D., Karp, S., Awdeh, Z. L. & Yunis, E. J. Serum complement ‘supergenes’ of the major histocompatibility complex in man (complotypes). Vox Sanguinis 45, 62–67 (1983).
Dawkins, R. et al. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol. Rev. 167, 275–304 (1999).
Norman, P. J. et al. Sequences of 95 human MHC haplotypes reveal extreme coding variation in genes other than highly polymorphic HLA class I and II. Genome Res. 27, 813–823 (2017).
Traherne, J. A. Human MHC architecture and evolution: implications for disease association studies. Int. J. Immunogenet. 35, 179–192 (2008).
Kulski, J. K., Suzuki, S. & Shiina, T. SNP-density crossover maps of polymorphic transposable elements and HLA genes within MHC class I haplotype blocks and junction. Front. Genet. 11, 594318 (2021).
Kulski, J. K., Suzuki, S. & Shiina, T. Haplotype shuffling and dimorphic transposable elements in the human extended major histocompatibility complex class II region. Front. Genet. 12, 665899 (2021).
van Oosterhout, C. A new theory of MHC evolution: beyond selection on the immune genes. Proc. R. Soc. B 276, 657–665 (2009).
Meyer, D., C. Aguiar, V. R., Bitarello, B. D., C. Brandt, D. Y. & Nunes, K. A genomic perspective on HLA evolution. Immunogenetics 70, 5–27 (2018).
Radman, M. Speciation of genes and genomes: conservation of DNA polymorphism by barriers to recombination raised by mismatch repair system. Front. Genet. 13, 803690 (2022).
Alper, C. A. The path to conserved extended haplotypes: megabase-length haplotypes at high population frequency. Front. Genet. 12, 716603 (2021).
Sella, G. & Barton, N. H. Thinking about the evolution of complex traits in the era of genome-wide association studies. Annu. Rev. Genom. Hum. Genet. 20, 461–493 (2019).
Crux, N. B. & Elahi, S. Human leukocyte antigen (HLA) and immune regulation: how do classical and non-classical hla alleles modulate immune response to human immunodeficiency virus and hepatitis C virus infections? Front. Immunol. 8, 832 (2017).
Wieczorek, M. et al. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. https://doi.org/10.3389/fimmu.2017.00292 (2017).
Mosaad, Y. M. Clinical role of human leukocyte antigen in health and disease. Scand. J. Immunol. 82, 283–306 (2015).
La Gruta, N. L., Gras, S., Daley, S. R., Thomas, P. G. & Rossjohn, J. Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol. 18, 467–478 (2018).
Sznarkowska, A., Mikac, S. & Pilch, M. MHC class I regulation: the origin perspective. Cancers 12, 1155 (2020).
Matzaraki, V., Kumar, V., Wijmenga, C. & Zhernakova, A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 18, 76 (2017).
Tait, B. D. The importance of establishing genetic phase in clinical medicine. Int .J. Immunogenet. 49, 1–7 (2022).
Suzuki, S. et al. Reference grade characterization of polymorphisms in full-length HLA class I and II genes with short-read sequencing on the ION PGM system and long-reads generated by single molecule, real-time sequencing on the PacBio platform. Front. Immunol. 9, 2294 (2018).
Yamamoto, F. et al. Capturing differential allele-level expression and genotypes of all classical HLA loci and haplotypes by a new capture RNA-seq method. Front. Immunol. 11, 941 (2020).
Jensen, J. M. et al. Assembly and analysis of 100 full MHC haplotypes from the Danish population. Genome Res. 27, 1597–1607 (2017).
Cullen, M., Perfetto, S. P., Klitz, W., Nelson, G. & Carrington, M. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am. J. Hum. Genet. 71, 759–776 (2002).
Murphy, N. M. et al. Haplotyping the human leukocyte antigen system from single chromosomes. Sci. Rep. 6, 30381 (2016).
Lokki, M. & Paakkanen, R. The complexity and diversity of major histocompatibility complex challenge disease association studies. HLA 93, 3–15 (2019).
Kulski, J. K., Shiina, T. & Dijkstra, J. M. Genomic diversity of the major histocompatibility complex in health and disease. Cells 8, 1270 (2019).
The MHC sequencing consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature 401, 921–923 (1999).
Horton, R. et al. Variation analysis and gene annotation of eight MHC haplotypes: The MHC Haplotype Project. Immunogenetics 60, 1–18 (2008).
Xie, T. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 13, 2621–2636 (2003).
Zhou, D., Lai, M., Luo, A. & Yu, C.-Y. An RNA metabolism and surveillance quartet in the major histocompatibility complex. Cells 8, 1008 (2019).
Kulski, J. K., Shiina, T., Anzai, T., Kohara, S. & Inoko, H. Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol. Rev. 190, 95–122 (2002).
Amadou, C. Evolution of the Mhc class I region: the framework hypothesis. Immunogenetics 49, 362–367 (1999).
Thompson, P. J., Macfarlan, T. S. & Lorincz, M. C. Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol. Cell 62, 766–776 (2016).
Kulski J. K., Gaudieri S., Dawkins R. L. Major Histocompatibility Complex. (eds Kasahara M.), p. 158–177 (Springer Japan, 2000).
Kulski, J. K., Gaudieri, S., Inoko, H. & Dawkins, R. L. Comparison between two human endogenous retrovirus (HERV)-rich regions within the major histocompatibility complex. J. Mol. Evol. 48, 675–683 (1999).
Kulski, J. K. et al. Human endogenous retrovirus (HERVK9) structural polymorphism with haplotypic HLA-A allelic associations. Genetics 180, 445–457 (2008).
Kulski, J. K. et al. HLA-A allele associations with viral MER9-LTR nucleotide sequences at two distinct loci within the MHC alpha block. Immunogenetics 61, 257–270 (2009).
Bodmer, W. Ruggero ceppellini: a perspective on his contributions to genetics and immunology. Front. Immunol. 10, 4 (2019).
Dawkins, R. L. & Lloyd, S. S. MHC genomics and disease: looking back to go forward. Cells 8, 944 (2019).
Dorak, M. T. et al. Conserved extended haplotypes of the major histocompatibility complex: further characterization. Genes Immun. 7, 450–467 (2006).
Schneider, V. A. et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 27, 849–864 (2017).
Houwaart, T. et al. Complete sequences of six Major Histocompatibility Complex haplotypes, including all the major MHC class II structures. Cold Spring Harbor Laboratory, bioRxiv. Posted May 06, 2022. Preprint at https://www.biorxiv.org/content/10.1101/2022.04.28.489875v2.
Traherne, J. A. et al. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2, e9 (2006).
Gaudieri, S., Leelayuwat, C., Tay, G. K., Townend, D. C. & Dawkins, R. L. The major histocompatibility complex (MHC) contains conserved polymorphic genomic sequences that are shuffled by recombination to form ethnic-specific haplotypes. J. Mol. Evol. 45, 17–23 (1997).
Smith, W. P. et al. Toward understanding MHC disease associations: Partial resequencing of 46 distinct HLA haplotypes. Genomics 87, 561–571 (2006).
Shiina, T. et al. Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics 173, 1555–1570 (2006).
Kulski, J. K., Shigenari, A. & Inoko, H. Genetic variation and hitchhiking between structurally polymorphic Alu insertions and HLA-A, -B, and -C alleles and other retroelements within the MHC class I region. Tissue Antigens 78, 359–377 (2011).
Hirata, J. et al. Genetic and phenotypic landscape of the major histocompatibilty complex region in the Japanese population. Nat. Genet. 51, 470–480 (2019).
Handunnetthi, L., Ramagopalan, S. V., Ebers, G. C. & Knight, J. C. Regulation of major histocompatibility complex class II gene expression, genetic variation and disease. Genes Immun. 11, 99–112 (2010).
van Heyningen, V. & Bickmore, W. Regulation from a distance: long-range control of gene expression in development and disease. Philos. Trans. R. Soc. B 368, 20120372 (2013).
Lam, T. H., Shen, M., Tay, M. Z. & Ren, E. C. Unique allelic eQTL clusters in human MHC haplotypes. G3 7, 2595–2604 (2017).
D’Antonio, M. et al. Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease. eLife 8, e48476 (2019).
Lamontagne, M. et al. Susceptibility genes for lung diseases in the major histocompatibility complex revealed by lung expression quantitative trait loci analysis. Eur. Respir. J. 48, 573–576 (2016).
Sharon, E. et al. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat. Genet. 48, 995–1002 (2016).
Anzai, T. et al. Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc. Natl Acad. Sci. USA 100, 7708–7713 (2003).
Nakatani, K. et al. Identification of HLA-A*02:06:01 as the primary disease susceptibility HLA allele in cold medicine-related Stevens-Johnson syndrome with severe ocular complications by high-resolution NGS-based HLA typing. Sci. Rep. 9, 16240 (2019).
Kent, T. V., Uzunović, J. & Wright, S. I. Coevolution between transposable elements and recombination. Philos. Trans. R. Soc. B 372, 20160458 (2017).
Chénais, B. Transposable elements and human diseases: mechanisms and implication in the response to environmental pollutants. Int J. Mol. Sci. 23, 2551 (2022).
Andersson, G., Svensson, A.-C., Setterblad, N. & Rask, L. Retroelements in the human MHC class II region. Trends Genet. 14, 109–114 (1998).
Kulski, J. K., Anzai, T., Shiina, T. & Inoko, H. Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol. Biol. Evol. 21, 2079–2091 (2004).
Kulski, J. K. et al. The evolution of MHC diversity by segmental duplication and transposition of retroelements. J. Mol. Evol. 45, 599–609 (1997).
Kulski, J. K., Shigenari, A. & Inoko, H. Polymorphic SVA retrotransposons at four loci and their association with classical HLA class I alleles in Japanese, Caucasians and African Americans. Immunogenetics 62, 211–230 (2010).
Kulski, J. K. & Dunn, D. S. Polymorphic Alu insertions within the Major Histocompatibility Complex class I genomic region: a brief review. Cytogenet. Genome Res. 110, 193–202 (2005).
Kulski, J. K., Mawart, A., Marie, K., Tay, G. K. & AlSafar, H. S. MHC class I polymorphic Alu insertion (POALIN) allele and haplotype frequencies in the Arabs of the United Arab Emirates and other world populations. Int. J. Immunogenet. 46, 247–262 (2019).
Shi, L. et al. Association and differentiation of MHC class I and II polymorphic Alu insertions and HLA-A, -B, -C and -DRB1 alleles in the Chinese Han population. Mol. Genet. Genomics 289, 93–101 (2014).
Cun, Y. et al. Haplotypic associations and differentiation of MHC class II polymorphic alu insertions at five loci with HLA-DRB1 alleles in 12 minority ethnic populations in China. Front. Genet. 12, 636236 (2021).
Wang, L., Norris, E. T. & Jordan, I. K. Human retrotransposon insertion polymorphisms are associated with health and disease via gene regulatory phenotypes. Front. Microbiol. 8, 1418 (2017).
Savage, A. L. et al. Retrotransposons in the development and progression of amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 90, 284–293 (2019).
Mack S. J., Gourraud P-A, Single R. M., Thomson G., Hollenbach J. A. Immunogenetics. (eds Christiansen F. T. & Tait B. D.) p 215–244 (Humana Press, 2012).
Degli-Esposti, M. A. et al. Ancestral haplotypes: conserved population MHC haplotypes. Hum. Immunol. 34, 242–252 (1992).
Awdeh, Z. L., Raum, D., Yunis, E. J. & Alper, C. A. Extended HLA/complement allele haplotypes: evidence for T/t-like complex in man. Proc. Natl Acad. Sci. USA 80, 259–263 (1983).
Mack, S. J. et al. HLA-A, -B, -C, and -DRB1 allele and haplotype frequencies distinguish Eastern European Americans from the general European American population. Tissue Antigens 73, 17–32 (2009).
Sanchez-Mazas, A., Buhler, S. & Nunes, J. M. A new HLA map of Europe: regional genetic variation and its implication for peopling history, disease-association studies and tissue transplantation. Hum. Hered. 76, 162–177 (2013).
Zhou, Y., Browning, B. L. & Browning, S. R. Population-specific recombination maps from segments of identity by descent. Am. J. Hum. Genet. 107, 137–148 (2020).
Martin, A. R. et al. Haplotype sharing provides insights into fine-scale population history and disease in Finland. Am. J. Hum. Genet. 102, 760–775 (2018).
Baschal, E. E. et al. Congruence as a measurement of extended haplotype structure across the genome. J. Transl. Med. 10, 32 (2012).
Sun, Y. et al. Recombination and mutation shape variations in the major histocompatibility complex. J. Gen. Genome (2022). https://doi.org/10.1016/j.jgg.2022.03.006.
Koskela, S. et al. Hidden genomic MHC disparity between HLA-matched sibling pairs in hematopoietic stem cell transplantation. Sci. Rep. 8, 5396 (2018).
Nakaoka, H. & Inoue, I. Distribution of HLA haplotypes across Japanese Archipelago: similarity, difference and admixture. J. Hum. Genet. 60, 683–690 (2015).
Kulski, J. K., AlSafar, H. S., Mawart, A., Henschel, A. & Tay, G. K. HLA class I allele lineages and haplotype frequencies in Arabs of the United Arab Emirates. Int. J. Immunogenet. 46, 152–159 (2019).
Kulski J. K. Next Generation Sequencing - Advances, Applications and Challenges. (ed. Kulski J. K.) (InTech, 2016).
Shiina T., Suzuki S., Kulski J. K. Next Generation Sequencing - Advances, Applications and Challenges. (ed. Kulski J. K.) (InTech, 2016).
van Dijk, E. L., Jaszczyszyn, Y., Naquin, D. & Thermes, C. The third revolution in sequencing technology. Trends Genet. 34, 666–681 (2018).
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018).
Chin, C.-S. et al. A diploid assembly-based benchmark for variants in the major histocompatibility complex. Nat. Commun. 11, 4794 (2020).
Dilthey, A. T. State-of-the-art genome inference in the human MHC. Int. J. Biochem. Cell Biol. 131, 105882 (2021).
Hu, T., Chitnis, N., Monos, D. & Dinh, A. Next-generation sequencing technologies: an overview. Hum. Immunol. 82, 801–811 (2021).
Li, Y. et al. Human leukocyte antigen (HLA) A-C-B-DRB1-DQB1 haplotype segregation analysis among 2152 families in China and the comparison to expectation-maximization algorithm result. Chin. Med. J. 134, 1741–1743 (2021).
Jawdat, D., Uyar, F. A., Alaskar, A., Müller, C. R. & Hajeer, A. HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 allele and haplotype frequencies of 28,927 saudi stem cell donors typed by next-generation sequencing. Front. Immunol. 11, 544768 (2020).
Neville, M. J. et al. High resolution HLA haplotyping by imputation for a British population bioresource. Hum. Immunol. 78, 242–251 (2017).
Maiers, M., Gragert, L. & Klitz, W. High-resolution HLA alleles and haplotypes in the United States population. Hum. Immunol. 68, 779–788 (2007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kulski, J.K., Suzuki, S. & Shiina, T. Human leukocyte antigen super-locus: nexus of genomic supergenes, SNPs, indels, transcripts, and haplotypes. Hum Genome Var 9, 49 (2022). https://doi.org/10.1038/s41439-022-00226-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41439-022-00226-5
This article is cited by
-
Low-frequency and rare genetic variants associated with rheumatoid arthritis risk
Nature Reviews Rheumatology (2024)
-
Sequenced-based GWAS for linear classification traits in Belgian Blue beef cattle reveals new coding variants in genes regulating body size in mammals
Genetics Selection Evolution (2023)
-
Variant calling and benchmarking in an era of complete human genome sequences
Nature Reviews Genetics (2023)