Abstract
Leber congenital amaurosis (LCA) is a severe autosomal recessive retinal degenerative disease. The current study describes exome sequencing results for two unrelated Indian LCA patients carrying novel nonsense p.(Glu636*) and frameshift p.(Pro2281Leufs*63) mutations in the ALMS1 gene. Although ALMS1 gene mutations are associated with Alstrom syndrome (AS), the current patients did not exhibit typical syndromic features of AS. These data suggest that ALMS1 should be included in the candidate gene panel for LCA to improve diagnostic efficiency.
Similar content being viewed by others
Leber congenital amaurosis (LCA) is one of the most severe forms of hereditary retinal dystrophy causing early infantile blindness. Affected infants show signs of night blindness, roving/pendular nystagmus, photophobia, and digito-ocular signs that are characterized by a nonrecordable electroretinogram (ERG)1. Accounting for 10–20% of childhood blindness, LCA is clinically heterogeneous; indeed, it is reported as an isolated clinical entity as well as in certain syndromes, such as Joubert syndrome2, thiamine-responsive megaloblastic anemia3, Senior-Loken syndrome4, and Batten’s disease5. Genetically, LCA is predominantly inherited as an autosomal recessive disease and rarely as an autosomal dominant disease. To date, mutations in at least 25 genes have been identified as causing LCA6. More recently, biallelic ALMS1 gene mutations have been shown to be associated with nonsyndromic LCA7.
The ALMS1 gene (2p13)8 encodes a basal body and centrosome-associated protein found in ciliated cells. The ALMS1 protein is involved in processes such as microtubule organization, cilium formation and maintenance, extracellular matrix production, cell migration, intraciliary transport, and cell cycle regulation9. This ciliary protein is reportedly expressed in multiple tissues, and as such, improper translation of ALMS1 may result in Alstrom syndrome characterized by cone-rod dystrophy (CRD), obesity, type 2 diabetes mellitus, cardiomyopathy, hearing loss, and multiple organ failure10. Thus far, ALMS1 (NM_015120) is the only gene associated with Alstrom syndrome (AS).
A proper clinical evaluation along with relevant genetic molecular testing for pathogenic disease-causing variants are necessary for a conclusive and definitive diagnosis of retinal dystrophies such as LCA, retinitis pigmentosa (RP), and CRD. We report herein two novel homozygous pathogenic genetic variants in the ALMS1 gene in two unrelated Indian families presenting with clinical features similar to LCA, without extraocular phenotypes.
Consultation with the Vitreo Retinal Services of a tertiary eye care center was sought for the patients, who underwent complete ophthalmic and clinical evaluation, including documentation of family history, birth history, refraction, visual acuity testing, retinal examination, nystagmus, photophobia, oculo-digital signs, full-field electroretinogram (ffERG), fundus photography, and optical coherence tomography (OCT). This study was approved by the institutional Ethics Committee, and informed consent was obtained from the patients’ families. Peripheral blood samples were collected from the proband and family members, and genomic DNA was extracted using a NucleoSpin Blood XL kit (Macherey-Nagel, GmbH, Germany). In our previous study, we performed targeted resequencing of 20 candidate genes in 92 Indian LCA patients (AIPL1, CABP4, CEP290, CRB1, CRX, GUCY2D, IQCB1, IMPDH1, KCNJ13, LCA5, LRAT, MERTK, NMNAT1, OTX2, RD3, RDH12, RPE65, RPGRIP1, SPATA7, and TULP1) using an Agilent HaloPlex target enrichment assay11. The probands (case 1 and case 2) were negative for disease-causing pathogenic variants when screened by targeted resequencing. Hence, samples from the proband along with one parent were subjected to whole-exome sequencing (WES) using the Agilent SureSelectXT Human All Exon V5 + UTRs enrichment kit and sequenced using an Illumina HiSeq 2500 at 80–100x depth.
We obtained 8 Gb data per sample. The pipeline for processing the data involved a Burrows-Wheeler Aligner (BWA) for mapping to the GRCh37/hg19 genome build, and GATK-lite was used for realignment and recalibration of the obtained reads after duplicate removal. Variant calling was performed using the GATK-lite Unified Genotype caller and annotated using VariMAT (internal data analysis pipeline curated by MedGenome Labs, Bangalore).
Only Q30 data were considered for further annotation. The annotated variants were cross-checked with open databases such as dbSNP (v.2.0, Build 153), GenomAD, LOVD, 1000 Genomes Project, and Ensemble Variation Table to filter variants with MAF ≤ 0.01. As LCA is predominantly an autosomal recessive disease, data for the proband and the respective parent were compared to analyze variants that were found to be homozygous in the proband and heterozygous in the parent and possible compound heterozygous in the proband(s). Heterozygous variants of known dominant and recessive gene(s) [Supplementary Table 1] were also considered and ruled out after segregation and phenotypic correlation.
For Sanger sequencing of the identified variants in exon 8, primers were designed using the ALMS1 transcript ENST00000264448.6 using Primer 3 (v.0.4.0), as follows: for g.73675557 G>T, forward 1-5′TGACCAGACAACTGGCATGTC3′ and reverse 1-3′GACTGTCTGCTAAGTCCTGTG5′; for g.73680491delT, forward 2-5′CTCAGGCTGATGACAGAGTTG3′ and reverse 2-3′GGTGTAGTGGAACCATTGGG5′. PCR was standardized using a touchdown protocol of 63.5–56.5 (−0.5 °C) for g.73675557 G>T and 59.5 °C annealing temperature for g.73680491delT with a reaction mixture comprising 10 pmol/µl primers (forward and reverse), 1X Taq buffer, 50 ng DNA template, 0.5 mM dNTPs, and 0.5 U Taq polymerase. The PCR products were purified by Exo-SAP and cycle sequenced using a Big dye Terminator v3.1 kit (Applied Biosystems, USA). The purified products were sequenced using an ABI 3500 Avant genetic analyzer.
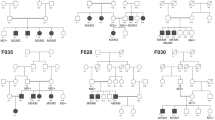
Regarding case 1, the child was first seen at 18 months of age with a history of profound visual loss (counting fingers [CF] at one meter in both eyes) and nystagmus since 6 months of age. The pedigree, as indicated in Fig. 1A shows an isolated family history. The refractive error was +3 D sphere in both eyes. Full-field ERG was performed under general anesthesia and inferred to be unrecordable under both scotopic and photopic conditions (Fig. 1B). Fundus examination was performed under anesthesia and revealed disc pallor with marked arteriolar attenuation, a salt and pepper appearance of the retina, and sparse pigmentation (Fig. 1C). The macula showed a dull reflex, but no macular scarring, pigmentation, or bull’s eye lesion was seen. At a follow-up visit at 12 years of age, we observed that the vision of the child was the same as before, and ERG continued to be unrecordable. The child was cooperative for OCT at this visit, which revealed the presence of a shallow foveal dip with thinning of the outer nuclear and ellipsoid layers (Fig. 1D). This presentation is similar to LCA with extinguished responses on ffERG in the first year of life. Whole-exome sequencing for the proband and unaffected father identified homozygous novel nonsense mutation (g.73675557 G>T; c.1906G>T (NM_015120); p.(Glu636*)), leading to premature truncation of the ALMS1 protein (Fig. 1E). Sanger sequencing identified the parents and unaffected siblings to be heterozygous (Fig. 1F) for the variant.
[A] Family pedigree: affected status is indicated by a shaded circle, and carrier status is indicated by a dot (●). [B] ERG is nonrecordable. [C] Fundus photograph of case 1 (i) showing a near normal looking macula (ii) with RPE granularity in the mid periphery. [D] OCT shows a preserved foveal dip with thinning of the outer nuclear and ellipsoid layer. [E] The analysis, validation, and identification of WES data. [F] Electrophoretogram showing the reverse-strand sequence of the g.73675557 G>T; c.1906G>T; p.(Glu636*) variant, homozygous (II3) in the proband and heterozygous in the (I1) mother, (I2) father, and (II1) unaffected sibling.
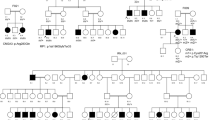
Case 2 was brought to the clinic at 2 and half years of age, and the parents described observing poor vision in the child since the first year of life. The pedigree indicated no family history (Fig. 2A). BCVA was very poor in the proband, with both eyes exhibiting only fixation and light following. The child had nystagmus and photophobia at presentation, with bilateral hyperopia of +8.00 D. Nonrecordable photopic responses for both 3.0 and 30 Hz flickers were obtained by ffERG. Dark-adapted (DA) responses were extinguished for the DA 0.01 flash but were recordable, though with markedly reduced amplitudes for the DA 3.0 flash. These findings are suggestive of severe and widespread effects of the rod and cone system, with the cone system being more severely affected (Fig. 2B). Fundus examination revealed the presence of disc pallor with arteriolar attenuation, widespread peripheral granularity, and early macular involvement in the form of macular pigment mottling with bony spicule pigmentation (Fig. 2C). The child at this age was not cooperative for OCT. Molecular testing by WES identified a novel homozygous deletion variant (Fig. 2D), g.73680491delT; c.6840delT (NM_015120), leading to a frameshift and thereby truncated protein (p.Pro2281Leufs*63). The deletion variant segregated in the parents, with a heterozygous genotype (Fig. 2E).
[A] Family pedigree: affected status is indicated by a shaded square and carrier status by a dot (●). [B] ERG shows severely reduced rod and cone responses, with cone responses almost extinguished. [C] Fundus photograph of case 2 (i & ii): right and left eye of the child showing a large macular scar, arteriolar attenuation, and RPE granularity. [D] The analysis, validation, and identification of WES data. [E] Electrophoretogram showing the g.73680491delT; c.6840delT; (p. Pro2281Leufs*63) variant, homozygous (II1) in the proband and heterozygous in the (I1) father and (I2) mother.
Control samples (N = 100) exhibited the wild-type genotype for both variants by Sanger sequencing. The variants were not found in open databases, and in silico tools such as Mutation Taster12 and CADD13 predicted them to be disease-causing. Both variants might lead to nonsense-mediated decay (NMD) because they are located ~50–55 nucleotides upstream of the last exon–exon junction14; nevertheless, characterization studies are essential to prove this functionality. The variants are classified as strongly pathogenic according to ACMG guidelines (PVS1, PM-1, 2, 4, PP1, 3)15.
In conclusion, we identified two novel pathogenic variants in exon 8 of the ALMS1 gene in patients with LCA but without any symptoms of AS. Similar to the current study, WES has revealed compound heterozygous null mutations or homozygous mutations in ALMS1 in LCA or early-onset severe cone-rod dystrophy cases without extraocular abnormalities16, thus indicating ALMS1 as a candidate gene for isolated LCA/early-onset retinal diseases other than AS. As LCA is congenital, it could be the primary manifestation of AS, and careful clinical examination along with confirmatory genetic testing is necessary to elucidate the underlying gene, especially in the absence or late onset of other syndromic features. The manifestations of AS phenotypes begin as early as a few days after birth in retinal degeneration or cardiomyopathy and later in adulthood/the second decade, such as the indication of type 2 diabetes and renal or hepatic abnormality17.
Analysis of genotype and phenotype correlation in AS cases harboring a mutation in exon 8 of the ALMS1 gene has suggested an association between exon 8 mutations and a normal18 or delayed renal disease phenotype19. This emphasizes the phenotypic heterogeneity for this protein depending on the mutation. In the current study, except for delayed milestone presentation in case 2, the patients did not show any systemic features specific for AS (current ages are 19 yrs for case 1 and 18 yrs for case 2). Nevertheless, biochemical tests for hepatic and renal problems or to diagnose early diabetes were not performed. Annual monitoring of the cardiac, hepatic, and renal phenotypes of such patients will be helpful for differential diagnosis, early treatment, better disease management, and rehabilitation of future cases.
References
Mamatha, G., Srilekha, S., Meenakshi, S. & Kumaramanickavel, G. Screening of the RPE65 gene in the Asian Indian patients with leber congenital amaurosis. Ophthalmic Genet. 29, 73–78 (2008).
King, M. D. & Stephenson, J. B. Association of Joubert’s syndrome with Leber’s congenital amaurosis. Arch. Neurol. 41, 123 (1984).
Srikrupa, N. N., Meenakshi, S., Arokiasamy, T., Murali, K. & Soumittra, N. Leber’s congenital amaurosis as the retinal degenerative phenotype in thiamine responsive megaloblastic anemia: a case report. Ophthalmic Genet. 35, 119–124 (2014).
Stone, E. M. et al. Variations in NPHP5 in patients with nonsyndromic leber congenital amaurosis and Senior-Loken syndrome. Arch. Ophthalmol. 129, 81–87 (2011).
Casteels, I. S. W. et al. Leber congenital amaurosis–differential diagnosis, ophthalmological and neuroradiological report of 18 patients. Neuropediatrics 27, 189–193 (1996).
Daiger, S. P., Greenberg, B. R. J., Christoffels, A. & Hide., W. Data services and software for identifying genes and mutations causing retinal degeneration. Investig. Ophthalmol. Vis. Sci. 39, S295 (1998).
Wang, X. et al. Whole-exome sequencing identifies ALMS1, IQCB1, CNGA3, and MYO7A mutations in patients with Leber congenital amaurosis. Hum. Mutat. 32, 1450–1459 (2011).
Collin, G. B., Marshall, J. D., Cardon, L. R. & Nishina, P. M. Homozygosity mapping at Alstrom syndrome to chromosome 2p. Hum. Mol. Genet. 6, 213–219 (1997).
Hearn, T. ALMS1 and Alström syndrome: a recessive form of metabolic, neurosensory and cardiac deficits. J. Mol. Med. 97, 1–17 (2019).
Hearn, T. et al. Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes 54, 1581–1587 (2005).
Srikrupa, N. N. et al. Genetic profile and mutation spectrum of Leber congenital amaurosis in a larger Indian cohort using high throughput targeted re-sequencing. Clin. Genet. 93, 329–339 (2018).
Schwarz, J. M., Cooper, D. N., Schuelke, M. & Seelow, D. MutationTaster2: mutation prediction for the deep- sequencing age. Nat. Methods 11, 361–362 (2014).
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894 (2019).
Nagy, E. & Maquat, L. E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23, 198–9 (1998).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–24 (2015). ACMG Laboratory Quality Assurance Committee.
Xu, Y. et al. ALMS1 null mutations: a common cause of Leber congenital amaurosis and early-onset severe cone-rod dystrophy. Clin. Genet. 89, 442–447 (2016).
Marshall, J. D., Maffei, P., Collin, G. B. & Naggert, J. K. Alstrom syndrome: genetics and clinical overview. Curr. Genom. 12, 225–235 (2012).
Sathya Priya, C. et al. Mutation spectrum in BBS genes guided by homozygosity mapping in an Indian cohort. Clin. Genet. 87, 161–166 (2015).
Marshall, J. D. et al. Spectrum of ALMS1 variants and evaluation of genotype-phenotype correlations in Alström syndrome. Hum. Mutat. 28, 1114–1123 (2007).
HGV Database
The relevant data from this Data Report are hosted at the Human Genome Variation Database at https://doi.org/10.6084/m9.figshare.hgv.2975; https://doi.org/10.6084/m9.figshare.hgv.2978.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Srikrupa, N.N., Sripriya, S., Pavithra, S. et al. Whole-exome sequencing identifies two novel ALMS1 mutations in Indian patients with Leber congenital amaurosis. Hum Genome Var 8, 12 (2021). https://doi.org/10.1038/s41439-021-00143-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41439-021-00143-z