Abstract
The fruits of Physalis (Solanaceae) have a unique structure, a lantern-like fruiting calyx known as inflated calyx syndrome (ICS) or the Chinese lantern, and are rich in steroid-related compounds. However, the genetic variations underlying the origin of these characteristic traits and diversity in Physalis remain largely unknown. Here, we present a high-quality chromosome-level reference genome assembly of Physalis floridana (~1.40 Gb in size) with a contig N50 of ~4.87 Mb. Through evolutionary genomics and experimental approaches, we found that the loss of the SEP-like MADS-box gene MBP21 subclade is likely a key mutation that, together with the previously revealed mutation affecting floral MPF2 expression, might have contributed to the origination of ICS in Physaleae, suggesting that the origination of a morphological novelty may have resulted from an evolutionary scenario in which one mutation compensated for another deleterious mutation. Moreover, the significant expansion of squalene epoxidase genes is potentially associated with the natural variation of steroid-related compounds in Physalis fruits. The results reveal the importance of gene gains (duplication) and/or subsequent losses as genetic bases of the evolution of distinct fruit traits, and the data serve as a valuable resource for the evolutionary genetics and breeding of solanaceous crops.
Similar content being viewed by others
Introduction
The family Solanaceae is an important source of nutritional and culinary diversity. Along with several well-characterized model crops, such as potato (Solanum tuberosum), tomato (Solanum lycopersicum), and peppers (Capsicum spp.), the nightshade family contains many neglected orphan crops1,2,3. Members of the genus Physalis include well-known species such as Cape gooseberry (Physalis peruviana) and tomatillo or husk tomato (Physalis philadelphica). Physalis is one of the largest genera within Solanaceae, with 70–90 species4. Species in this genus have emerged as a new class of solanaceous horticultural crops as well as new model plants for studies in ecology, evolution, and development3,5, as the genus is characterized by novel morphological traits and biochemical diversity in fruits. Understanding how such complex traits originated during evolutionary history is a fundamental question, since fruit evolution has long been considered a key contributor to the success of angiosperms6. Moreover, the origin of these evolutionary novelties is a fascinating subject in itself, and the processes giving rise to them are largely unresolved evolutionary mysteries.
Most Physalis species have 12 chromosomes, as observed in most solanaceous species7. Members of the genus have a distinct fruit morphology with a papery husk as an accessory trait8,9; this characteristic structure is known as inflated calyx syndrome (ICS) or Chinese lantern5,8. Within Solanaceae, at least five genera (Physalis, Withania, Przewalskia, Margaranthus, and Nicandra) of Physaleae share this morphological novelty10. Physalis floridana is a representative species of Physalis characterized by solitary flowers with dark maculations and berries that are enveloped by an inflated fruiting calyx (Fig. 1A–E). The adaptive advantages of this novelty have been well documented at developmental, physiological, mechanical, and ecological levels, and selection apparently favors the fixation of this trait to improve plant fitness11,12. The lantern trait appears as an inflated fruiting calyx; fertilization/hormonal signals trigger its formation in Physalis and Withania5,11,13. The question of how this novelty arose has attracted the attention of botanists and evolutionary biologists. Previous studies revealed that the origin of the Chinese lantern is associated with the heterotopic expression of Physalis MADS-box gene 2 (MPF2) in floral organs5. Moreover, floral calyx identity and Chinese lantern size are determined by Physalis MADS-box gene 3 (MPF3), which interacts with MPF2 both physically and genetically14. The Darwinian selection of MPF2-like genes involved directional selection for ICS15,16. ICS is considered a plesiomorphic trait based on the evaluation of MPF2-like expression in flowers10; however, the recently elucidated evolutionary history of Physalideae, which contains the vast majority of Solanaceae species with inflated calyces (146 of 222 in total), using four neutral genetic markers (ITS, LEAFY, trnL-F, and waxy) suggested strong directionality in the origin of ICS17. To better understand the origin and evolutionary and developmental mechanisms of the Chinese lantern within Solanaceae, further investigation at the genomic level is needed.
As a result of adaptive evolution, plants produce secondary metabolites as protective agents18 or to attract pollinators19. Some of the secondary metabolites in Physalis fruits and plants render them commercially valuable, and many such compounds have antibacterial, anti-inflammatory, and anticancer activities that are useful in medical applications. The whole herb of Physalis is used in traditional medicine to treat fever, diabetes, pharyngitis, boils, coughs, and mastitis20,21,22. Fruits, including the fruiting calyces of a few Physalis species, often have curative and culinary uses1,23, and berries are used as both medicine and food. Increasing attention has been paid to Physalis in phytochemical and pharmacological studies, as its chemical constituents include withanolides, sucrose esters, flavonoids, labdane diterpenes, ceramides, and chlorogenic acids24, which are crucial for plant development and growth25. To date, a total of 351 natural ergostane steroid compounds with novel and unique structures, including withanolides, have been identified in the genus Physalis26. The two general five-carbon (C5) isoprene units of phytosterols and subsequent steroid biosynthesis are isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) that are produced via the cytoplasmic mevalonate (MVA) pathway and the chloroplastic methylerythritol phosphate (MEP) pathway27. The biosynthetic pathways of these phytosterols and the structure-activity correlations of all isolated biochemical compounds have been proposed, all of which seem to be conserved in Solanum and Physalis28,29. However, the diversity, biosynthesis, and metabolic pathways of steroids and steroid derivatives in Physalis fruits are largely unexplored.
In the present study, we generated a chromosome-level genome assembly of P. floridana through the combined application of Pacific Biosciences (PacBio) sequencing and chromosome conformation capture (Hi-C) technologies. Using this resource combined with evolutionary and functional analyses, we obtained new insights into the evolutionary genetic basis of the iconic fruit morphological novelty (the Chinese lantern) and characteristic chemical composition (physalins) of Physalis fruits, pinpointing the roles of gene gains (duplication) and/or subsequent losses in the origin and evolution of the morphological novelty and biochemical variation. This first high-quality reference genome of P. floridana will serve as a valuable resource for breeding and improving the horticultural, nutritional, and medicinal value of Physalis crops.
Results
High-quality genome assembly of P. floridana
We selected the diploid and self-compatible P. floridana P106 accession for the first trial aimed at decoding the genome of a Physalis species because this accession shares the typical flower and fruit morphology of Physalis species, characterized by a solitary flower and a berry covered with a lantern-like fruiting calyx (Fig. 1A–E; see Supplementary Table 1 for details). Chromosomal karyotype analysis revealed that the P106 genome contained 12 pairs of chromosomes (2n = 24) (Supplementary Fig. 1). First, we generated 153 Gb of whole-genome data from shotgun sequences of P106 via the Illumina sequencing of genomic libraries with an insert size of 500 bp (Supplementary Fig. 2), from which 149 Gb of clean data was yielded after removing low-quality reads. The main Poisson-shaped distribution peak (depth = 91), with a minor peak and long tail (depth >163), represented the unique 21-mers found in the P. floridana genome (Supplementary Fig. 3), suggesting that the genome is nearly homozygous, with substantial repeat sequences. The genome size was estimated to be ~1.40 Gb by K-mer analysis and flow cytometry (Supplementary Figs. 3 and 4; Supplementary Table 2). Then, two PacBio libraries were constructed and sequenced on five cells using the PacBio Sequel platform, yielding 125 Gb of subreads corresponding to ~90-fold coverage of the genome (Supplementary Fig. 5 and Supplementary Fig. 6A, B). The PacBio reads were assembled into 922 contigs with a contig N50 of 4.87 Mb (Supplementary Table 3). Furthermore, ~215 million uniquely mapped paired-end reads (25% of 858 million cleaned Hi-C reads) were generated, among which ~196 million (91.35%) were valid paired-end reads and were used to generate chromosome contact information (Supplementary Table 4). Finally, a total of 1.37 Gb of data were clustered into 12 pseudochromosomes, which contained 98.82% of the total assembly length (Supplementary Figs. 7 and 8).
We then remapped the Illumina reads to the assemblies (mapping rate ~ 99.93%; mean depth 86.75x) (Supplementary Table 5). Single nucleotide polymorphisms were called to further estimate the level of heterozygosity, which was ~0.19% for the P106 genome (Supplementary Table 6). A guanine-cytosine (GC) depth analysis revealed that the Physalis genome had a mean GC content of 42% (Supplementary Fig. 6C, D).
We also generated the full-length transcriptome of P. floridana based on PacBio long reads and Illumina transcriptomes from various biological tissues. By mapping the transcriptome reads to the genome assembly, we found that the mapping rates were ~89.44%–99.61% (Supplementary Table 7), indicating the high completeness of the genome assembly. We further evaluated the completeness of the scaffold assembly using the BUSCO (v.3) plant datasets and identified 97.9% of the 1,375 single-copy core sets of orthologous genes in the Physalis genome assembly (Supplementary Table 8). Taken together, the genome assembly assessments (Fig. 1F) suggest that we obtained a high-quality reference genome for Physalis.
Genome annotation and gene prediction
The prediction and density of genes (Fig. 2A), including protein-coding genes and the tRNA and rRNA genes (Fig. 2B); transposable elements (TEs), i.e., Copia and Gypsy (Fig. 2C); and other information, such as the GC distribution (Fig. 2D) and genome rearrangement events of collinear blocks (Fig. 2E), were evaluated and integrated into the assembled Physalis genome. For this purpose, ab initio and homology-based methods were combined to annotate protein-coding sequences aided by the transcriptome sequences. In total, 32,075 complete protein-coding genes were predicted in the current genome version, with an average gene length, coding sequence length, and exon number of 4,023 bp, 1,134 bp, and 4.63, respectively (Fig. 2A and Supplementary Figs. 9 and 10; Supplementary Table 9). Among these annotated genes, 90.1% of the 1,375 single-copy core sets of orthologous amino acid sequences could be identified (Fig. 1F), and 93.34% (29,938) could be annotated based on publicly available databases (Supplementary Fig. 11). We further predicted 3,655 ribosomal RNA (rRNA), 997 transfer RNA (tRNA), 375 microRNA (miRNA), and 3,047 small nuclear RNA (snRNA) genes in the P. floridana genome (Fig. 2B and Supplementary Table 10).
A Gene density plotted in 1000-kb sliding windows. B tRNA (purple) and rRNA (orange) locations. C TE/Copia (purple) and TE/Gypsy (orange) content per Mb. D GC content per 300-kb block. E Genome segmental duplication and internal rearrangement events. Orange, intercollinear gene pairs; purple, intracollinear gene pairs
The total length of the identified repetitive sequences in the P. floridana genome was 1142.5 Mb, occupying 82.24% of the assembled genome sequences (Supplementary Table 11). This proportion was similar to that observed in S. pennellii (82%)30 and higher than that in the C. annuum genome (76.4%)31, the S. lycopersicum genome (61.3%)32, and the S. tuberosum genome (61.6%)33, while TEs accounted for 81.6% of the assembled sequence (Fig. 1F). Among the identified TEs, LTR retrotransposons (LTR-RT), which usually play a substantial role in genome size variation34,35, represented 65.68% of the P. floridana genome (Fig. 2 and Supplementary Fig. 12A; Supplementary Table 12). This proportion of LTR-RTs in the P. floridana genome was lower than that in C. annuum (70%) and higher than that in S. tuberosum (47%), S. lycopersicum (50%), and S. pennellii (45%)30,31,32,33. We therefore assessed the recent activity of LTR-RTs in five Solanaceae species using a sequence alignment of full-length LTR-RTs and estimated their insertion times. The timing of the main LTR-RT burst was earliest in C. annuum (~2.0 MYA), while S. lycopersicum showed an intermediate insertion time (~0.8 MYA), and S. pennellii, S. tuberosum, and P. floridana exhibited many more recent LTR-RT bursts (~0.4 MYA) (Supplementary Fig. 12B). Similar to the C. annuum genome, the substantial proliferation of the Gypsy family (10-fold more than the Copia family) identified in the Physalis genome (Fig. 2C) might be the main cause of genome expansion, as previously proposed31. Unlike LTR retrotransposons, DNA transposons comprised only 0.71% of the Physalis genome, whereas 15.41% of the genome corresponded to uncharacterized repeats (Supplementary Table 12). Moreover, significant genome segmental duplication and rearrangements occurred during the evolution of the P. floridana genome (Fig. 2E). Thus, the different genome dynamics in these Solanaceae species might be largely due to repetitive sequences, particularly Gypsy TE variations, and to genome segmental duplication and internal rearrangements.
Genome evolution analyses
Phylogenomic analysis using 7,553 single-copy gene families revealed the topology of P. floridana and 12 other representative species (Supplementary Fig. 13). We found that within this subclade, P. floridana and C. annuum diverged from each other ~23.8 (18.0–29.9) million years ago (MYA), while S. lycopersicum, S. pennellii, and S. tuberosum belonged to another subclade that diverged from the P. floridana–C. annuum subclade ~29.9 (23.9–36.6) MYA (Supplementary Fig. 14). These results were in line with recent molecular divergence estimates in which the fossils used were considerably older than the ~30 MYA crown of the entire Solanaceae family36 but were inconsistent with the report that Physalis infinemundi sp. nov. represents a derived lineage of Solanaceae from Gondwanan South America from 52.2 MYA, thereby considerably pushing back the timing of the evolutionary origin of this plant family37.
To understand the whole-genome duplication (WGD) history of P. floridana, we investigated the distribution of the synonymous substitution rate (Ks) between syntenic gene pairs based on comparisons among P. floridana, C. annuum, S. lycopersicum, and Vitis vinifera. The Ks peaks of paralogous syntenic gene pairs in P. floridana, C. annuum, and S. lycopersicum were ~0.65, which was greater than the Ks peak of Solanaceae speciation (~0.21) and less than that of the Vitis-Solanaceae divergence (~1.25) (Fig. 3A). In intragenomic syntenic comparisons of P. floridana and S. lycopersicum, both showed clear 1:2 patterns (Supplementary Fig. 15A, B), suggesting a whole-genome triplication (WGT) event in the evolutionary history. Further comparison of the genomes of P. floridana and V. vinifera showed a clear 3:1 syntenic ratio, indicating that P. floridana WGT occurred after divergence from V. vinifera (Supplementary Fig. 15C). Moreover, syntenic comparisons clearly showed a 1:1:1 relationship among the P. floridana, C. annuum, and S. lycopersicum genomes (Fig. 3B and Supplementary Fig. 15D). All of these observations highlighted the occurrence of a recent WGT event before the divergence of Solanaceae species and after the core eudicot common hexaploidization event31,32,38.
A Synonymous substitution rate (Ks) distributions of syntenic blocks for P. floridana paralogs and orthologs with other eudicots are indicated by colored lines. B Synteny analyses of three closely related genomes of S. lycopersicum, P. floridana and C. annuum. Lines between chromosomes indicate syntenic regions. Gray wedges in the background highlight syntenic blocks between two species. C Identification of genomic regions with Physalis-specific gains or losses compared with Solanum. Most genes within one nonsyntenic region have no orthologs in another species, suggesting that these genes have been lost. The numbers and ratios of these lost genes in each species are indicated in corresponding pie charts with a light color
Given the common WGD history of Solanaceae species, we further investigated genome structural evolution following the speciation of P. floridana, S. lycopersicum, and C. annuum (Fig. 3B). A clear one-to-one syntenic relationship among the three genomes was detected at the whole-chromosome level, and the overall gene collinearity among the three genomes was largely conserved, although their genome size varied (Fig. 3B and Supplementary Fig. 16), implying that these species did not experience large amounts of chromosome fusion or recent WGD events after species divergence. Specifically, we identified 367 and 529 large syntenic gene blocks in the P. floridana genome compared with S. lycopersicum (occupying 78% Physalis genes) and C. annuum (occupying 60% Physalis genes), respectively (Supplementary Table 13). Therefore, the genomes of three Solanaceae species were relatively conserved; only several small-scale chromosomal rearrangements and specific genomic regions were observed (Fig. 3B, C).
We investigated these nonsyntenic genomic regions among the three studied Solanaceae genomes as regions likely to provide insights into their specific genomic evolutionary history. In total, we found that 145 P. floridana genomic regions containing 3,314 genes of P. floridana and 211 S. lycopersicum genomic regions, including 5,891 genes and 226 C. annuum genomic regions comprising 5,149 genes, were nonsyntenic (Fig. 3C). Furthermore, an all-against-all BLAST search revealed that 2,767/3,314 genes in these nonsyntenic regions in P. floridana did not identify orthologous genes in the genomes of S. lycopersicum or C. annuum (Supplementary Table 14), indicating that they were likely P. floridana-specific genes. Notably, we found that several steroid biosynthesis-related squalene epoxidase (SQE) genes were located in these nonsyntenic regions in P. floridana and seemed to be specifically expanded in P. floridana (Supplementary Table 14). More strikingly, a few floral development-associated MADS-box genes were located in the S. lycopersicum-specific nonsyntenic regions (Supplementary Table 15), likely indicating key gene losses through segmental deletion in the P. floridana and C. annuum genomes.
Evolutionary analyses of gene families
Comparisons among multiple genomes can reveal the distribution of orthologous genes and the expansion and contraction of gene families. We first focused on gene family analyses during the evolutionary history of Solanaceae using grape as an outgroup. The genomes of eight solanaceous species, including P. floridana, C. annuum, S. pennellii, S. lycopersicum, S. tuberosum, Nicotiana attenuata, Petunia axillaris, and Lycium barbarum, together with V. vinifera, were employed to construct orthogroups using OrthoMCL. We found that most genes were clustered into orthologs or paralogs among these Solanaceae genomes (Fig. 4A and Supplementary Table 16), and a core set of 9,980 gene families were shared among these genomes (Fig. 4B). In addition, there were 884 families specific to P. floridana, 854 gene families specific to C. annuum, 820 specific to N. attenuata, 582 specific to Pe. axillaris, 1,038 specific to S. pennellii, and 618 specific to L. barbarum (Fig. 4B and Supplementary Table 17). We found 160 gene families specifically shared by P. floridana and C. annuum, which was greater than the number of specifically shared gene families between the P. floridana and any of the other Solanaceae genomes (Fig. 4B), further supporting the notion that C. annuum is the closest relative to P. floridana.
A Distribution of single-copy orthologs (dark blue), multiple-copy orthologs (pale blue), unique genes (red), other paralogs (orange), and other unclassified genes (green) in P. floridana, C. annuum, S. lycopersicum, S. tuberosum, S. pennellii, L. barbarum, N. attenuata, Pe. axillaris, and V. vinifera. B Venn diagram showing unique and shared gene families among P. floridana and the indicated species. The numbers of orthologous gene families shared by all species, gene families specific to each species, and specifically shared gene families between the P. floridana and other Solanaceae genomes are given. C Gene family expansion and contraction across a phylogenetic tree of 9 plant species. The number at the root (11,087) represents the number of gene families in the most recent common ancestor (MRCA). Bootstrap support numbers are given in parenthesis. Green numbers indicate the number of gene families that expanded in a species relative to the closest species during evolution, and red numbers indicate the number of gene families that contracted. The pie charts on the right show the proportions of these categories (green, expanded; red, contracted; blue, unchanged.)
Expansions and contractions of certain gene families have occurred frequently during evolution. A total of 291 gene families expanded before the divergence of Solanaceae, whereas 464 families were found to have contracted when the representative genomes of six genera (Physalis, Capsicum, Solanum, Nicotiana, Lycium, and Petunia) within Solanaceae were included (Fig. 4C). Moreover, 847 gene families were expanded (360 by a significant margin, P ≤ 0.05), while 1,134 gene families were contracted (57 by a significant margin, P ≤ 0.05) in P. floridana (Supplementary Tables 18 and 19). We conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of the 3,797 genes in the 360 significantly expanded gene families in Physalis. GO categories including flower morphogenesis, terpenoid biosynthetic process, and cytokinin metabolic process were significantly enriched in these expanded gene families (Supplementary Fig. 17A and Supplementary Table 20), while pathways including zeatin biosynthesis, steroid biosynthesis (squalene monooxygenase), sesquiterpenoid and triterpenoid biosynthesis, and brassinosteroid biosynthesis (CYP450 85A3-like proteins) were significantly enriched in the KEGG analysis (Supplementary Fig. 17B and Supplementary Table 21). Among the 57 significantly contracted gene families, we found that GO categories including plant−type hypersensitive response, proteolysis, signal transduction, and low−affinity nitrate transport (nitrate/nitrite transporter) were significantly enriched (Supplementary Fig. 18A and Supplementary Table 22), while pathways including plant hormone signal transduction, linoleic acid metabolism, and isoflavonoid biosynthesis were significantly enriched in the KEGG analysis (Supplementary Fig. 18B and Supplementary Table 23).
To further reveal the uniqueness of the Physalis genome, we constructed orthogroups using P. floridana with 12 other sequenced plant species. We found that orthologous genes, paralogous genes, and nonclustered genes were distributed in a similar manner among the 13 compared plant species (Supplementary Fig. 19). In total, 27,115 annotated protein-coding genes were classified into 15,724 gene families, 1,013 of which were P. floridana species specific relative to other investigated species (Supplementary Tables 24 and 25). We next conducted a GO enrichment analysis of the 1,013 Physalis-specific gene families and found that these genes participated in multiple biological and metabolic processes (Supplementary Fig. 20 and Supplementary Table 26), suggesting diverse roles of the Physalis-specific genes. By examining these genes, we found that some gene families of MADS-box genes and steroid biosynthesis-related genes were unique to P. floridana. Additional and unique copies of a few MADS-box genes, including several families from each of the AGL61-like, AGL80-like, Arabidopsis nitrate regulated 1 (ANR1)-like, MADS50-like, SOC1-like, and AGL29-like gene families, were found in Physalis (Supplementary Table 25), perhaps as a result of unequal gene duplications or losses among different species. Four steroid biosynthesis-related gene families were identified: gene family 12781 (12 genes) and gene family 24583 (2 genes), encoding SQE, as well as gene family 20969 (3 genes) and gene family 25195 (2 genes), related to brassinosteroid biosynthesis (Supplementary Table 25). In accordance with this, KEGG enrichment analysis revealed that “steroid biosynthesis” was the most significantly enriched pathway in P. floridana (Supplementary Fig. 21). Eleven of 35 steroid biosynthesis (map00100) pathway-related genes were significantly enriched, and 10 of the 11 genes were SQE genes (Supplementary Table 27).
A higher dN/dS ratio (>1) indicates positive Darwinian selection on proteins, which may thus have undergone more rapid protein evolution. We therefore performed this analysis at the genomic level, revealing 127 positively selected (dN/dS > 1, Branch-site-specific model M2A, p < 0.05) genes in the P106 genome, including genes encoding zinc finger, ribosomal, ABC transporter, cytochrome P450, small GTPase superfamily, and glycoside hydrolase family proteins (Supplementary Table 28), suggesting that positive selection on these genes during evolution might be involved in Physalis plant responses to different stresses. The pf02G034050 (GO: 0006694, p ≤ 0) and pf03G074230 (GO: 0016126, p ≤ 9e-06) genes, related to steroid biosynthetic processes, were found to be positively selected (Supplementary Table 28). Some steroid compounds isolated from Physalis plants contain chloride ions39 that are thought to be formed by reaction with an epoxide group21. Consistent with this, the positively selected gene pf03G066370 (GO: 0006821, p = 0) was found to be a chloride transport-related gene (Supplementary Table 28). These results suggest a role of these positively selected genes related to steroid biosynthesis.
Through the above evolutionary analyses at multiple levels, we found that the solanaceous genomes are conserved overall; however, small-scale mutations were also observed among the genomes of Physalis, Capsicum, and Solanum. The evolutionary consequences of these genetic variations deserve further functional investigation. Here, potential genetic variations of genes related to key steps in the biosynthetic pathways of steroid-related compounds, especially the MADS-box genes involved in Chinese lantern formation, were of primary interest to us and were thus further explored.
Evolution of genes involved in steroid compound biosynthetic pathways
Steroids are essential for all eukaryotes25,40,41. Withanolides are a class of polyoxygenated steroids based on a C28 ergostane skeleton; they are the most abundant steroids found in the genus Physalis24,26. This is a striking characteristic of chemical diversity in Physalis fruits, and the genes in the biosynthetic pathways of steroid-related compounds were repeatedly captured in the above analyses. We therefore checked the key genes in the entire steroid biosynthetic pathways27,28,29,42, which were shown in a simplified framework (Fig. 5A). Thirty-three gene families, including those encoding the rate-limiting enzymes 3-hydroxy-3-methylglutaryl-coenzyme A (HMGR) and SQE, were checked in the related genomes. We found that the copy-number variation (CNV) of most gene families was comparable among the species of Physalis, Capsicum, and Solanum. In contrast, significant changes in the CNV of annotated HMGR and SQE genes (Fig. 5A and Supplementary Table 29) showed direct expansion of these genes in Physalis. In particular, a total of 20 SQE-like genes were found in P. floridana (P106), and the copy number was ~4-fold higher than that in the non-Physalis genomes examined (Supplementary Table 30). These SQEs showed Physalis-specific expansion (Fig. 5B). In addition to the conserved SQE-located regions among Solanaceae, these expanded SQE genes were mainly located in Physalis-specific regions (Supplementary Fig. 22), and most of them shared high homology with pf03G072270, which might be restricted to Physalis and Capsicum (Supplementary Fig. 23), indicating that these expanded SQE genes may have been derived from pf03G072270. Moreover, most Physalis-specific SQEs belonged to the two subfamilies of this P. floridana (PfSQE) gene family (24583 and 12781) identified in the gene expansion analysis (Supplementary Fig. 23 and Supplementary Table 18). SQE genes encode flavoprotein monooxygenases that act as rate-limiting enzymes in the steroid biosynthetic pathway41, and the roles of the CNV of these genes in the natural variation of steroid metabolites among different species related to Physalis need further investigation. However, genetic variations related to MADS-box genes involved in the formation of the Chinese lantern, the most striking novel morphological trait in Physalis, were explored in more detail from functional and molecular perspectives.
A A simplified steroid-related derivative biosynthetic pathway. The solid arrows indicate the confirmed sections, while the dashed arrows represent the proposed sections. The examined enzymes are given, and the gene numbers in different species are indicated by the histogram. MVA, mevalonic acid; MEP, methylerythritol 4-phosphate; IPI, isopentenyl pyrophosphate isomerase; GPPS, geranyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; SQS, squalene synthase; SQE, squalene epoxidase; CAS, cycloartenol synthase; LAS, lanosterol synthase; HSD, 3-beta hydroxysteroid dehydrogenase/isomerase. B A schematic diagram displaying the origin and expansion of SQE genes in P. floridana. The blue square represents 15 Physalis-specific SQE genes that expanded from a conserved Solanaceae SQE (pf03G072270) but were lost in Solanum. The triangle represents the other four Solanaceae-conserved SQE genes. Detailed information is available in Supplementary Figs. 22 and 23
MBP21 loss might have contributed to the origination of the Chinese lantern in Physalis
The role of MADS-box genes in Chinese lantern formation was reported previously, and these genes include MPF2 and MPF3 and possibly their MADS-domain protein-interacting factors5,14,43. Genome sequencing showed that the P. floridana genome contained 136 putative MADS-box genes, including 92 type I and 44 type II genes (Supplementary Table 31). Mγ in the type I class was significantly multiplied in P. floridana, but both MIKC and MIKC* in the type II class appeared to have been phased out in P. floridana relative to Solanum (Supplementary Table 31). The copy or subclade numbers of most MIKC genes were generally invariant among solanaceous species; however, several showed changes, including AGL12, FLOWERING LOCUS C (FLC), and SEPALLATA (SEP)-like genes (Supplementary Fig. 24 and Supplementary Table 31). No evidence supports a putative role of other lost or expanded MADS-box genes, such as AGL12 or FLC, in Chinese lantern formation; these genes play roles in flowering time control and root meristem development in Arabidopsis44,45. However, SEP-like proteins in Physalis or Arabidopsis dimerize with MPF2 or MPF3, and the loss of a certain SEP-like interacting protein is correlated with sepal inflation16,43. In line with these results, a genomic region harboring a SEP-like gene was identified as having been lost in Physalis relative to Solanum (Fig. 3C and Supplementary Table 15). Therefore, the genomic variation related to SEP-like loss might be essential in the origin of ICS. To obtain further evidence supporting this assumption, we investigated SEP-like genes in Solanaceae. Six putative SEP-like MADS-box genes were isolated from P. floridana, S. pimpinellifolium, and C. annuum, and the full length of CaCMB1 was significantly shorter than those of other SEP-like MADS-box genes (Fig. 6A and Supplementary Table 32). The encoded proteins were mainly localized in the nucleus (Supplementary Fig. 25). Moreover, we further demonstrated that nearly all SEP-like MADS-box proteins from the three species and StMBP21 from S. tuberosum interacted with MPF2 and MPF3 in both yeast two-hybrid and bimolecular fluorescence complementation (BiFC) analyses (Fig. 6A and Supplementary Figs. 26 and 27), further hinting at their roles in ICS formation. However, the observed copy number is evidence against SEP-like gene loss in Physalis.
A The phylogeny of SEP-like MADS-box genes from the indicated plant species. Different subclades are highlighted with different colors. The circles represent the interactions between MPF2 (left)/MPF3 (right) and the corresponding SEP-like proteins. Blue represents the interaction signals detected in yeast two-hybrid assays. Yellow represents the fluorescent signals detected in BiFC assays. Gray indicates no detectable interaction signal. White indicates not analyzed. The percentage at the bottom of each subclade is the sequence identity of amino acids among the sequences from the four species. Pf, P. floridana; Ca, C. annuum; Sl, S. lycopersicum; St, S. tuberosum; At, Arabidopsis thaliana. B Southern-blotting analyses of MBP21 genes between Solanum and Physalis. Total DNA was isolated from S. tuberosum and P. floridana and digested with EcoR I and Hind III. C The expression of the indicated genes in Solanum and Physalis revealed by northern-blotting analysis. Total RNA was isolated from the leaves (Le), floral buds (FB), and flowers (Fl) of S. tuberosum and P. floridana. MADS-box gene-specific probes from S. tuberosum or P. floridana, as indicated, were used. D Microsynteny of the MBP21 subclade-containing region among the four indicated species from Solanum, Capsicum, and Physalis. The brown linear region represents the syntenic gene blocks of the MBP21 gene, and the gray linear region represents the syntenic regions of genes near the target gene. E StMBP21 expression in OE-StMBP21 P. floridana plants. Total RNA was isolated from the flowers of two independent transgenic lines and subjected to northern-blotting analyses. The indicated gene-specific probes were used. F Flower morphology. Compared to wild-type (WT) flowers, flowers from StMBP21-overexpressing transgenic plants (OE-StMBP21) were withered, implying that fertilization was inhibited. G Fruit morphology comparison. Wild type, WT; StMBP21-overexpressing transgenic Physalis plants, OE-StMBP21. Self-pollination (SP) occasionally occurred in the OE-StMBP21 flowers, and artificial pollination (AP) produced berries of similar size to the WT, indicating that female fertility was normally functional and that male function was reduced in OE-StMBP21 plants. Part of the Chinese lantern was removed to show the berry. Bar = 1.0 cm in (F, G)
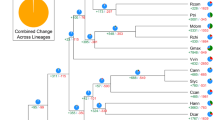
To clarify this issue, we investigated the evolutionary relationships of SEP-like genes. A protein sequence-based phylogenetic tree was constructed using the neighbor-joining (NJ) method (Fig. 6A). The SEP-like genes in S. tuberosum were analyzed in silico, and it was noteworthy that Sotub05g006010 contained two sections, each with a MADS-domain and a K domain (Supplementary Fig. 28A). The phylogenetic analysis revealed that Sotub05g006010-N fell into the SEP-like (RIN) group and that Sotub05g006010-C was an A-class MADS-box protein (ortholog of LeMADS-MC and MPF3) (Supplementary Fig. 28B). These findings are in line with the observations in tomato and Physalis that the LeMADS RIN–LeMADS MC and PfRIN–MPF3 orthologs show good maintenance of microsynteny14,46. The phylogenetic tree suggested that solanaceous SEP-like genes were divided into six subclades, and the sequence identity of each subclade varied in a phylogeny-dependent manner (Fig. 6A). One gene from each of the M29, MADS1, and CMB1 subclades was maintained in all examined solanaceous species. One duplication in the SEP3 subclade of P. floridana and one duplication in the RIN subclade of C. annuum were found. However, no genes in the MBP21 subclade were found in P. floridana or C. annuum (Fig. 6A). Nevertheless, one-to-one orthologs of MPF2-like and MPF3 genes existed in all examined solanaceous species (Supplementary Fig. 29), suggesting the specific loss of the MBP21 subclade from the SEP-like gene family in Physalis and Capsicum.
To verify this hypothesis, we examined the existence of the four closely related SEP-like genes in S. tuberosum and P. floridana via gel-blotting analyses since the variation in sequence identity allowed us to generate subclade-specific probes (Supplementary Table 33). Southern-blotting analyses did not detect the MBP21 subclade in the Physalis genome but did detect it in Solanum, while the other three genes were detected in both species (Fig. 6B). Northern-blotting results suggested that the MBP21 subclade was florally expressed in Solanum, whereas it was not detected in Physalis, and the other three closely related genes were expressed in the floral organs of both species (Fig. 6C). Microsynteny analysis was further performed for each subclade, and the results showed that microsynteny differed among each subclade in the four examined solanaceous species. Most subclades maintained perfect synteny, as observed in the MADS1, RIN, M29, CMB1, and SEP3 subclades (Supplementary Figs. 30–34), but the extent of the conservation of the synteny of each subclade seemed to be independent of phylogenetic relationships (Fig. 6A). In the SEP3 subclade, PfSEP3-1 was located close to PfSEP3-2 (Supplementary Fig. 34), implying that these two Physalis genes might have resulted from a tandem duplication. In the MBP21 subclade, the MBP21-orthologous genes were located in syntenic blocks on S. lycopersicum chromosome 12 (chr12) from 24.22 to 59.29 Mb and S. tuberosum chromosome 12 from 13.40 to 57.54 Mb (Fig. 6D). However, the corresponding orthologous regions in P. floridana and C. annuum were separated and were located on different chromosomes, indicating that significant genomic rearrangements occurred in P. floridana and C. annuum (Fig. 6D). Notably, we found that no orthologous syntenic region in P. floridana matched the region of chr12 from 35.85 to 54.46 Mb in S. lycopersicum, and 81% of the orthologous genes (216/264) in this region of S. lycopersicum were lost in P. floridana and in C. annuum, which included a key floral development gene, MBP21 (Supplementary Table 34). To confirm MPB21 gene loss, we further conducted homology searches against all Illumina sequencing data from P. floridana and against the entire genomic sequences of C. annuum, using the SlMBP21 coding sequence as a query. We found no valid hits in any of the searches, and we therefore excluded the possibility of genomic misassembly (Supplementary Table 35). These results confirm the loss of the MBP21 gene in P. floridana and C. annuum and imply that the loss event might have resulted from chromosomal rearrangements during evolution.
Functional conflict between MPF2 and MBP21 in Physalis
The downregulation of the MBP21 gene in Solanum as well as its homologs contributes to inflated sepal development47,48,49,50, and the failure to detect protein–protein interactions (PPIs) of MPF2-like with certain SEP-like proteins is associated with the loss of the Chinese lantern or enlarged sepals16,43, suggesting a role of the SEP-like gene family in the evolution of the Chinese lantern. Overall, the Physalis SEP-like genes shared similar expression patterns with their counterparts in Solanum and Capsicum during flower and fruit development (Supplementary Fig. 35); however, virus-induced gene silencing (VIGS) analyses revealed a highly redundant role of Physalis SEP-like genes. Organ size (flower, fruit, ICS, and seed) and seed number per berry were decreased in the PfMADS1 knockdown plants (Supplementary Figs. 36 and 37). Furthermore, two transgenic Physalis lines overexpressing StMBP21 were generated and showed poor male fertility (Fig. 6E, F). Self-pollination occasionally occurred, and artificial pollination with wild-type pollen could produce fruits, but the resulting fruits had abnormal lanterns with small berries or small lanterns that tightly enveloped the berry (Fig. 6G), indicating that the coexpression of MPF2 and MBP21 genes affected male sterility and calyx growth in Physalis. Such a functional conflict implies that coordination between the two genes is required for their proper function in ICS development.
MBP21 loss is correlated with ICS occurrence within Solanaceae
To identify the correlation of MBP21 loss and ICS occurrence within Solanaceae, northern and Southern-blotting analyses were conducted in additional species; the results further indicated the possible loss of MBP21-orthologous genes in Physaleae (Fig. 7), in which the ICS trait occurs frequently10,17. The gene existed in most Solanum species lacking ICS but was lost in the species with inflated leaf-like sepals (Fig. 7), reinforcing the essential role of MBP21 loss in calyx accrescence and inflation. The MBP21 gene was also found in other representative solanaceous genomes, such as those of Lycium, Petunia, and Nicotiana (Supplementary Fig. 38), whose species do not exhibit ICS. Thus, MBP21 loss was correlated with ICS-like occurrence overall but with a few exceptions, including the Capsicum, Vassobia, and Tubocapsicum genera (Fig. 7), which are explicable10,15. These observations again support a role of MBP21 loss in the origin of ICS within Solanaceae.
A Northern-blotting analyses. Total RNA was isolated from the leaves (L) and flowers (F) of the 13 indicated species. B Southern-blotting analysis. Genomic DNA of the indicated 24 species was digested by EcoR I. SEP-like MADS-box gene-specific probes of the MADS1, M29, SEP3, and MBP21 subclades from S. tuberosum were used. +, presence of the indicated genes or the calyx accrescence/ICS; -, not detectable, or the gene or ICS trait has been lost; *, accrescent or enlarged leaf-like sepals. Overall, MBP21 loss was correlated with ICS occurrence (marked in both green and blue). The observed exceptions (highlighted in orange) could be explained by secondary mutations in ICS biosynthesis pathways10,15
Taken together, the results indicate that the loss of the MBP21 subclade likely represented a novel mutation involved in the origin of the Chinese lantern.
Discussion
The first high-quality Physalis genome assembly
Physalis is quickly becoming an important genus within Solanaceae in the context of the current focus of the plant sciences, not only because of its iconic fruit morphology but also due to its unique biochemical profile of steroid metabolites3,6. The genus has drawn attention from both evolutionary biologists and berry breeders. No complete genome of Physalis has been made publicly available to date, thus hampering relevant evolutionary genetic research and breeding efforts. Here, we report the genome of P. floridana, providing data on a conserved representative of Physalis species that originated in the New World. The genome size of P. floridana is ~1.40 Gb, and the 12 assembled pseudochromosomes covered ~98.82% of the genome. The number of protein-coding genes was predicted to be 32,075, comparable to the numbers in Solanum and Capsicum30,31,32,33. Nevertheless, the genome size and gene content varied, possibly due to small-scale gene duplications, TE variations, and genome rearrangements after species divergence. Among the reported solanaceous genomes, the current genome of P. floridana was the first to be deciphered by using PacBio sequencing combined with Hi-C technology, where the latter method overcomes the limitations imposed by the lack of genetic maps for genome assembly51. The assembled genome of P. floridana exhibits higher chromosome-level completeness (98.82%) than those of C. annuum (86%), S. lycopersicum (84%), S. pennellii (97%), and S. tuberosum (86%)30,31,32,33 as well as greater contiguity, with an N50 of ~4.87 Mb, and was further anchored onto pseudochromosomes. Considering the novelty, contiguity, accuracy, and completeness of our sequences, our genome constitutes the first available high-quality genome of Physalis. The data should be able to promote further genetic and genomic studies of Physalis species and other solanaceous plants.
Through comparative genomic analyses at different levels, we characterized genomic structural variations, Physalis-specific genes, expanded and contracted gene families, and positively selected genes to understand the novel genetic variation occurring in the Physalis genome relative to the genomes of Solanum and Capsicum, although the gene collinearity of the solanaceous genomes was conserved overall. We found that some gene families were repeatedly detected in these analyses; many varied in either copy number or the coding sequences that could be associated with the development and evolution of Physalis from various aspects. For example, the special expansion of ANR1-like MADS-box genes and the significantly enrichment of low−affinity nitrate transport (nitrate/nitrite transporter) among the contracted gene families may be related to the fact that Physalis plants are adapted to poor soil conditions52,53. The enriched hormone-related genes (i.e., those involved in cytokinin metabolic processes in Physalis-specific gene families) were in line with the observation that Chinese lantern development is triggered by fertilization/hormonal signals5,11. However, these genetic variations observed in Physalis need further extensive investigation through comparative and functional analyses among solanaceous species to understand their evolutionary significance. In this work, we focused on the evolution of SEP-like MADS-box genes and SQE genes and their potential impacts on both the morphological and biochemical evolution of the novel fruit traits of Physalis.
On the origin of the Chinese lantern, a morphological novelty within Solanaceae
Consecutive genomic mutations are believed to drive the process of morphological evolution. Gene copy-number gains and losses and variation in the spatiotemporal expressional pattern or dosage impacts are crucial mutational events in evolution3,8,45,54. The pioneering hypothesis regarding the genetic basis of Chinese lantern formation was inspired by the tunicate mutants of Zea mays8,55,56, in which the ectopic expression of a ZMM19-like MADS-box gene caused an increase in sepal size8. In Physalis, MPF2, a closely related homolog of ZMM19, was characterized as playing a role in the development of a novel trait based on RNA interference (RNAi)-mediated silencing5 and overexpression approaches8,11. The evolutionary genetic mutation related to the heterotopic expression of an MPF2-like gene from vegetative to floral contexts is due to cis-element variation in the promoter of this gene5,14,57. We further demonstrated that MPF2 is involved in calyx organ identity and size control together with MPF3, and the role of MPF2 in increasing calyx size is dependent on the expression of normal MPF314. The knockdown of both MPF2 and MPF3 also affects male fertility5,14. Heterotopically expressed MPF2 was demonstrated to establish PPIs with proteins integral to floral pathways such as PFMAGO, MPF3, AG, and SEP43. The role of MPF2-like variation in ICS formation seems to be conserved in Solanaceae15,16,58. Within Solanaceae, the heterotopic expression of MPF2-like genes is a plesiomorphic trait10, and mutations in MPF2-related PPIs affect the evolution of ICS16,43. Among these mutations, SEP-like variation is likely essential for Chinese lantern formation, since the knockdown of several SEP-like genes in Solanum led to enlarged sepals despite their major roles in floral meristem determination47,48,49,50,59. In this work, through genome comparisons and various experimental approaches, we revealed the evolutionary trajectory of the SEP genes within Solanaceae and confirmed the loss of the MBP21 subclade in Physalis and Capsicum. Moreover, the MBP21 subclade may have been lost in Physaleae, a lineage grouping most species exhibiting ICS within Solanaceae. SEP-like genes are also essential for fertility in various plants47,60,61. We further showed that overexpressing StMBP21 in Physalis led to poor male fertility and small lanterns, resembling the floral phenotypic variation observed in MPF2-RNAi transgenic P. floridana lines5. We therefore inferred that the loss of the MBP21 subclade was a major event giving rise to ICS.
All genes related to lantern development characterized thus far have also been found to be required for male fertility5,14,58 (this work), leading to the “genetic erosion-restoration of male fertility” hypothesis for the origin of the Chinese lantern. The origin of this structural novelty might have resulted from two genetic changes: MPF2 floral expression and MBP21 loss. The overexpression of MPF2 or STMADS16 created enlarged leaf-like sepals but did not produce self-fertilized berries in transgenic S. tuberosum plants11. We are not able to evaluate the effects on male fertility in that case; however, in our StMBP21 transgenic Physalis plants, in which MPF2 is already expressed5, overexpressing StMBP21 in stamens apparently reduces male fertility, indicating that the combination of MPF2 and MBP21 might be deleterious. We further found that MPF2 and MBP21 shared the interaction partner MPF3, an essential gene in calyx development14. Moreover, MPF2 was shown to heterodimerize with MBP21. Thus, floral MPF2 expression could affect the heterodimerization of MPF3 and MBP21, which forms a normally functional dimer in floral development (Fig. 8)43, or the MPF2-MBP21 heterodimer could have a deleterious effect on male fertility (Fig. 8). Furthermore, in line with observations made in tomato48, we found that SlMBP21 and StMBP21 interacted with MPF3, the LeMADS-MC ortholog of Physalis14. The downregulation of MBP21 can significantly increase sepal size in tomato49; however, the loss of MBP21 might have caused poor male fertility in the ancestral plants before the origination of ICS (Fig. 8). We thus hypothesized that the expression of either MPF2 or MBP21 in stamens (one only) may have been sufficient to guarantee normal male fertility, and their coexistence in the stamen was harmful; however, a single mutation affecting either floral MPF2 expression or MBP21 loss could complement the disadvantageous consequence of the other mutation for fertility so that the two mutations together, incidentally and additively, caused the development of an inflated fruiting calyx (Fig. 8). This also suggests an evolutionary disadvantage of having both genes present in floral organs (i.e., stamens) and an advantage of having only one gene present in stamens. In this scenario, the Chinese lantern might have been a byproduct at the timepoint of its origin, but its adaptive role was selectively maintained during flower evolution thereafter12,37,55.
Three genes (MPF3, MPF2, and MBP21) and two mutational events (floral MPF2 expression and MBP21 loss) were characterized. Male fertility was integral, and the establishment and variation of the PPIs among these three MADS-domain proteins seemed to be an important way to exert their effects on improving impaired male fertility, thus incidentally giving rise to the origin of the Chinese lantern. (1) and (2) represent the molecular scenarios of non-ICS (ics) and ICS phenotypic variation, respectively, within Solanaceae; (3) indicates that the coexpression of MPF2 and MBP21 in stamens and calyx led to male sterility and calyx size reduction, while (4) proposes that the loss of MBP21, which could be replaced by other SEP-like proteins (SEPL), might have impaired male fertility and promoted calyx inflation. Situation (3) is deleterious and might have been eliminated during floral evolution, while (2) and (4) are two mutual compensatory mutation events co-contributing to the origination of ICS in Solanaceae. The orthologs of these genes from other solanaceous species were defined as MPF2-like (MPF2L), MPF3-like (MPF3L), and MBP21-like (MBP21L). The expression and PPIs in the non-ICS (ics) and ICS species are indicated. In the current model, compared to the non-ICS plants (ics), the co-occurrence of two mutation events becomes a hallmark of plants with ICS within Solanaceae
An inflated calyx develops in S. tuberosum and Arabidopsis upon the overexpression of an MPF2-like gene8,11,15,16,56. However, the induction of the development of a perfect Chinese lantern, a masterpiece of nature, in a non-ICS plant (i.e., S. tuberosum) represents a formidable challenge. Moreover, completely disrupting the formation of the inflated calyx in plants showing ICS (i.e., P. floridana) has not yet been achieved5,58, even in large-scale mutagenesis studies62. First, male fertility is integral to the ICS origin5,11,43. Moreover, the Chinese lantern is a complex trait with an intricate biosynthetic pathway5,11, and there are multiple genes associated with the trait, whose functional divergence may have differed after species divergence. Unlike the situation in Solanum, the disruption of each SEP gene could affect floral or fruit development46,47,48,49,50,63,64,65,66,67. The knockdown of any single SEP gene in Physalis did not lead to obvious phenotypic variation, hinting at genetic robustness due to redundancy, compensation, or rewiring in the genetic network. Nevertheless, the overexpression of StMBP21 in Physalis abolished male fertility and repressed ICS development. These findings suggest that the orthologous and paralogous genes could have undergone distinct functional divergence fates and patterns after species divergence, highlighting putative specialized roles of MBP21 in male fertility and repressing calyx inflation. This gene is apparently not a component of the ICS development program, but its loss during evolution may have been one major mutational event promoting the origination of ICS.
MPF2-like floral expression was demonstrated to be plesiomorphic10, while MBP21-like loss seemed to be mostly restricted to Physaleae. Once the biosynthetic pathway of ICS was established after the two major mutational events, secondary mutations in the pathway could have led to the disappearance of ICS, as observed in species that have lost, as observed in Capsicum, Vassobia, and Tubocapsicum. The secondary mutations affecting ICS evolution are largely underestimated throughout Solanaceae, although a few, including mutations abolishing MPF2-like expression in floral organs and polyploidization and subsequent gene loss, have been characterized10,11,15. The two mutation events were unlikely to have occurred simultaneously, and we would assume that MBP21-like loss might have been the primary mutation, since it could have resulted from genome rearrangements and was likely disadvantageous, while floral MPF2-like expression is mainly a consequence of promoter alteration5,14. We could not discriminate between a plesiomorphic origin10 and a directed acquisition17 of ICS, as this would depend on the order and importance of the two identified mutation events (floral MPF2 expression and MBP21 loss) and on the exact spatial expression of MBP21- and MPF2-like genes and their functional characteristics in the floral organs of Solanaceae. The proposed hypothesis for the origin of ICS and its evolutionary pattern might be elucidated once these issues are well-characterized in more representatives of Solanaceae and the corresponding ancestral states are more firmly reconstructed.
Distinct evolution of steroid biosynthetic pathways in Physalis fruits
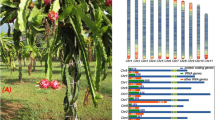
Steroids are essential triterpenoid compounds that are essential for plant growth, development, and differentiation25,29. As a result of adaptive evolution, steroid-derived compounds often play roles in communicating with neighboring plants, attracting pollinators and seed dispersers, and defending against pathogens and herbivores18,19 and occasionally function in host-plant specialization68. Some steroid derivatives are antinutritional factors that disrupt digestion and nutrient absorption in humans and have thus been reduced or eliminated during crop domestication42. However, the steroid biosynthetic pathway is highly conserved and is a key step in eukaryote evolution27,29,69,70. Steroids are present in all eukaryotes, and they modulate the fluidity and flexibility of cell membranes41. Some steroid compounds exhibit potential anticancer, anti-inflammatory, and apoptotic activities20,21. Thus, steroid-based drugs have a broad range of therapeutic applications; consequently, they represent the category of pharmaceuticals with the largest market share71. In most plants, steroid components are low in abundance, while members of the Solanaceae family show relatively high steroid synthesis and accumulation and therefore serve as unique models for studying plant cholesterogenesis29. Physalis species, belonging to Solanaceae, have the ability to oxidize carbons in the steroidal nucleus and the lateral chain, giving rise to a variety of withasteroids, such as withanolides and physalins39,68,72. Withanolides (steroidal lactones) are polyoxygenated ergostane derivatives with a lactone group at C26, while physalins are C28-secosteroid, lactone-type constituents of Physalis species73. Multiple withanolides and physalins have been isolated from the genus Physalis24,74. Based on untargeted metabolome analysis, our preliminary analyses suggested that the accumulation of some steroid components of Physalis fruits (bile acid derivatives, estrane steroids, physalins and derivatives, steroid lactones, and steroid esters) occurs at higher levels than is observed in Capsicum and Solanum fruits (Supplementary Fig. 39 and Supplementary Table 36). The identities of these compounds need further investigation. However, most of these compounds can usually be found in herbs and are produced via multiple biosynthesis pathways and from multiple precursors, including the two general C5 isoprene units for phytosterol biosynthesis via a cytoplasmic pathway (the MVA pathway), a chloroplastic pathway (the MEP pathway)27, and an additional 13 steps of triterpenoid biosynthesis28,29. Nevertheless, the biosynthesis and metabolic pathways of steroids and steroid derivatives in Physalis are largely unknown, and the events downstream of C24 alkyl sterols (phytosterols) are particularly a mystery.
The biosynthesis of steroidal alkaloids and saponins in triterpenoid biosynthetic pathways in solanaceous plants is mainly based on cholesterol29,42. We systematically analyzed the genes involved in steroid biosynthesis and metabolic pathways among various species of Solanum, Capsicum, and Physalis and found that most gene families involved were similar in copy number; however, the copy numbers of the rate-limiting enzyme-encoding genes HMGR and SQE differed among the genera. In particular, SQE genes are dramatically expanded in the P. floridana genome. The SQE enzyme catalyzes the first oxygenation reaction from squalene to 2,3-oxidosqualene in the pathway, leading to the syntheses of steroids and steroid derivatives41,75. Our preliminary experiment involving PfSQE knockdown mediated by group-specific VIGS resulted in a dramatic reduction in steroid constituents, including androstane steroids, cycloartanols and derivatives, estrane steroids, and physalins and their derivatives (Supplementary Figs. 40 and 41; Supplementary Table 37), confirming the essential role of this gene family in the biosynthesis of these steroid-related species in Physalis. Although the functional divergence of the genes related to each steroid biosynthesis, and their duplication history need further investigation, the specific origin and expansion of the PfSQE family is apparently associated with the biochemical diversity of certain steroid-related species in Physalis and might be responsible for the natural variation in these steroid derivatives in different solanaceous species. In accordance with the observed chemical variation among solanaceous species (Supplementary Fig. 39 and Supplementary Table 36), only the levels of physalins and their derivatives were consistently reduced and correlated with the expression of the PfSQE genes in the VIGS analyses, which was independent of the applied statistical approaches (Supplementary Fig. 41 and Supplementary Table 37), suggesting that the PfSQE family may primarily determine the levels of physalins and their derivatives, which are characteristic steroid-related species in Physaleae.
In addition to Physalis, the diversification of ergostane structure has also occurred in Withania, another genus with ICS, and this has mainly produced withanolides15,76. Whether this diversification is directly related to the significant expansion of SQEs remains elusive. Our findings provide a unique example of the natural genetic engineering of SQE genes to give rise to a “high-steroid and steroid derivative” model plant, underscoring the value of our gene toolbox for producing high-value steroidal compounds via synthetic biology. The results also provide a reference genome and a new model for investigating the mysterious process of steroid compound biosynthesis.
Conclusions
We generated the first, high-quality chromosome-level reference genome for P. floridana, a representative species in the genus Physalis of Solanaceae. Based on the novel genomic data, we pinpointed another key genetic mutation involved in the loss of a SEP-like MADS-box gene, in addition to the previously known MPF2 and MPF3 genes5,14, which apparently contributed to the origin of the morphological novelty of ICS, or Chinese lantern. We thus complemented the working model and proposed a new evolutionary scenario in which one mutation compensated for the disadvantageous effect of another mutation during the origin of the Chinese lantern. Morphological innovations such as ICS could be hitchhiking byproducts that significantly improve plant fitness. We also found that the origination and significant expansion of the SQE genes might be responsible for the high accumulation of certain steroid compounds in Physalis fruits. The CNV of this gene family was correlated with the natural variation of these steroids in the fruits of various solanaceous species. We propose that both the complex fruit-related traits of the Chinese lantern and richness in physalins and derivatives are results of adaptive evolution. Although the molecular mechanisms underlying these novel fruit traits resulting from these mutations and the recruited genes need further investigation by using state-of-the-art technologies, including the CRISPR/Cas9 system, the present work highlights the significant roles of both gene gains and gene losses in the evolution and development of novel fruit traits. In addition to advances in functional genomics in Physalis5,14,43,77,78,79,80, the assembled Physalis genome serves as a resource for studies of the 5Gs (genome, germplasm, genes, genomic breeding, and gene editing) in Physalis crops, and the data provide a platform for studying plant evolutionary genetics as well as for the genetic improvement and breeding of other solanaceous crops.
Materials and methods
Plant materials
Diploid, self-compatible Physalis pubescens (syn. P. floridana P106)5 was mainly used for de novo genome sequencing. Other species from Physaleae10 and other plant species were also used for Southern gel-blotting analysis. Before genome sequencing, the karyotype was characterized, and genome size was evaluated using flow cytometric analysis. Genomic DNA from young leaves of a single P. floridana plant was extracted using the QIAamp DNA Mini Kit (QIAGEN, Dusseldorf, Germany) following the manufacturer’s instructions.
DNA library preparation and PacBio sequencing
Eight microgram samples of genomic DNA were sheared using g-Tubes (Covaris, Woburn, MA, USA) and concentrated with AMPure PB magnetic beads. Each SMRT bell library was constructed using the Pacific Biosciences SMRTbell template prep kit (Pacific Biosciences, Menlo Park, USA). Sequencing was carried out on the Pacific Bioscience Sequel platform by Annoroad Gene Technology Co., Ltd. (Beijing, China).
Genome assembly and quality assessment
To obtain a high-accuracy genome for P. floridana, we adopted nearly 100× NGS data and more than 90× PacBio long reads for genome assembly. Falcon81 (falcon-kit = 1.0, https://github.com/PacificBiosciences/FALCON/) was employed to assemble the genome with the parameters [seed_coverage=30, length_cutoff_pr=5000]. To improve the local base accuracy of the contigs, Pilon (v1.22)82 with the default parameters was employed to polish contigs using the Illumina reads. After assembly, three approaches were employed to evaluate the quality of the genome.
High-through chromosome conformation capture (Hi-C)
The cells of P. floridana were retrieved for Hi-C library preparation according to standard procedures. Biotinylated DNA fragments were enriched and sheared to construct a sequencing library. Sequencing was performed on an Illumina HiSeqX-Ten platform with PE150 bp reads. Hi-C data were used to assign the contigs to chromosomes and then to order and orient the contigs within each chromosome. The Hi-C read contact frequency matrix was visualized using Juicebox (version 1.8.8).
Gene family classification
To perform the gene family analysis, the proteins of the selected species were downloaded from the NCBI database. All-versus-all BLASTP (version 2.2.26) was performed with an E-value cutoff of 1e-5 for all proteins. We then used OrthoMCL (Version 2.09)83 to cluster the gene families with an MCL inflation parameter of 1.5.
Phylogenetic analyses of plant species
We constructed a phylogenetic tree for P. floridana and other selected plants based on single-copy orthologous genes. Multiple sequence alignment was performed with MUSCLE (http://www.drive5.com/muscle/)84. Fourfold degenerate sites were extracted from sequences of each single-copy gene family and concatenated into a supergene for each species. PhyML 3.085 was used to construct the phylogenetic tree using fourfold degenerate sites via the maximum likelihood method under the GTR model. MCMCTREE in the PAML package86 (http://abacus.gene.ucl.ac.uk/software/paml.html) was used to estimate divergence times via the BRMC method87 using the soft fossil calibration obtained from the TimeTree website (http://www.timetree.org/).
Gene family expansion and contraction analysis
Gene families were further filtered out if one species had more than 200 genes or fewer than three genes. The remaining gene families were used to run CAFÉ (version 4.1)88 (http://sourceforge.net/projects/cafehahnlab/) with the parameters “-p 0.05 -t 1 -r 10000.” We employed probabilistic graphical models (PGM) to estimate the size of each gene family at each ancestral node of the phylogenetic tree topology using the orthologous genes inferred from OrthoMCL and to obtain a family-wide p-value (p ≤ 0.05; based on a Monte Carlo resampling procedure) to indicate whether there was significant expansion or contraction in each gene family across species.
Whole-genome duplication analysis and genome synteny analyses
A syntenic block was identified by using MCScanX software89. Searches for putative paralogous and orthologous genes were performed for P. floridana and the other species both against themselves and between species by using BLASTP (E-value ≤ 1e-5), and we performed multiple sequence alignment by using MUSCLE84. The synonymous substitution rate (Ks) was calculated using PALM (version 4.9e) with the YN00 model. We plotted the Ks distribution of all gene pairs in syntenic blocks using in-house Perl scripts. Syntenic gene pairs between P. floridana and other species were identified using MCScan software implemented in Python (JCVI v0.84) (https://github.com/tanghaibao/jcvi/wiki/MCscan-Python-version).
Copy-number variation (CNV) survey
To reveal the CNV of MADS-box genes, the complete sequences of Arabidopsis MADS-box genes were collected90, and protein sequences of other solanaceous species were extracted and aligned against Arabidopsis MADS-box genes using BLASTP (v2.2.28+, parameters: 1e-5, -m = 6). To assess the CNV of genes involved in steroid-related compound synthesis, all genes of interest were annotated using hmmer (v3.1b1) with the Pfam database (Pfam-A.hmm, parameter: -noali), and the Pfam id of each gene (E-value ≤ 1e-5) in the Physagulin R pathway in all examined solanaceous species was then extracted for statistical analysis.
Gel-blotting analyses
Total RNA was isolated by using a total RNA reagent kit (Biomol, Hamburg, Germany). The procedures for DNA and RNA gel blots, probe preparation, hybridization, and signal quantification followed previously described methods91. The filters were exposed to a Storage Phosphor Screen (Molecular Dynamics), and signals were quantified with a Typhoon 8600 PhosphorImager (Amersham Pharmacia).
Characterization of SEP-like and SQE genes
Protein–protein interactions were assessed using yeast two-hybrid assays and bimolecular fluorescence complementation (BiFC) assays. VIGS was performed to reveal the developmental roles of the selected genes. Full-length StMBP21 cDNA was overexpressed in Physalis via Agrobacterium-mediated transformation to obtain transgenic P. floridana. Phylogenetic analysis was performed via the neighbor-joining method in MEGA92,93. The cDNA of the reported genes was isolated using PCR amplification, which was carried out using the KOD-Plus-Neo kit (TOYOBO, Japan). All resulting constructs were sequenced by TSINGKE Biological Technology (Beijing, China), and the primers used in this work (Supplementary Table 38) were also synthesized by TSINGKE Biological Technology (Beijing, China).
Phenotypic quantification and statistical analysis
Multiple traits, including the flower radius, berry weight (without ICS), and 100-seed weight, were quantified in the VIGS and transgenic plants. Fruits from the indicated plants were harvested for untargeted metabolome analyses. Without special note, statistical analysis was performed by using IBM SPSS Statistics for Windows, Version 24.0 (IBM Corp, NY, USA).
The full description of the “Materials and methods” is available in the Supplementary Methods.
Data availability
All data are available in the manuscript or the supplementary materials. The P. floridana P106 genome sequence data and annotation reported in the article have been deposited in the Genome Warehouse at the National Genomics Data Center (https://ngdc.cncb.ac.cn/) under accession number GWHANUX00000000. The raw sequencing data and RNA-seq data have been deposited in the NCBI at the Sequence Read Archive (SRA) database under accession numbers PRJNA627991 and PRJNA552437.
References
Hernández, S. M. & Rivera, J. R. A. In Plant Production and Protection (eds Bermejo, J. E. H., León, J.). Series No. 26. 117–122 (FAO, 1994).
Paran, I. & van der Knaap, E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Bot. 58, 3841–3852 (2007).
Wang, L., Li, J., Zhao, J. & He, C. Y. Evolutionary developmental genetics of fruit morphological variation within the Solanaceae. Front. Plant Sci. 6, 248 (2015).
Pretz, C. & Deanna, R. Typifications and nomenclatural notes in Physalis (Solanaceae) from the United States. Taxon 69, 170–192 (2020).
He, C. Y. & Saedler, H. Heterotopic expression of MPF2 is the key to the evolution of the Chinese lantern of Physalis, a morphological novelty in Solanaceae. Proc. Natl Acad. Sci. USA 102, 5779–5784 (2005).
Knapp, S. Tobacco to tomatoes: a phylogenetic perspective on fruit diversity in the Solanaceae. J. Exp. Bot. 53, 2001–2022 (2002).
Menzel, M. Y. The cytotaxonomy and genetics of Physalis. Proc. Am. Philos. Soc. 95, 132–183 (1951).
He, C. Y., Münster, T. & Saedler, H. On the origin of floral morphological novelties. FEBS Lett. 567, 147–151 (2004).
Whitson, M. & Manos, P. S. Untangling Physalis (Solanaceae) from the Physaloids: a two-gene phylogeny of the Physalinae. Syst. Bot. 30, 216–230 (2005).
Hu, J. Y. & Saedler, H. Evolution of the inflated calyx syndrome in Solanaceae. Mol. Biol. Evol. 24, 2443–2453 (2007).
He, C. Y. & Saedler, H. Hormonal control of the inflated calyx syndrome, a morphological novelty, in Physalis. Plant J. 49, 935–946 (2007).
Li, J., Song, C. J. & He, C. Y. Chinese lantern in Physalis is an advantageous morphological novelty and improves plant fitness. Sci. Rep. 9, 596 (2019).
Khan, M. R., Hu, J. Y. & He, C. Y. Plant hormones including ethylene are recruited in calyx inflation in Solanaceous plants. J. Plant Physiol. 169, 940–948 (2012).
Zhao, J. et al. The euAP1 protein MPF3 represses MPF2 to specify floral calyx identity and displays crucial roles in Chinese lantern development in Physalis. Plant Cell 25, 2002–2021 (2013).
Khan, M. R., Hu, J. Y., Riss, S., He, C. Y. & Saedler, H. MPF2-like-A MADS-box genes control the inflated calyx syndrome in Withania (Solanaceae): roles of Darwinian’s selection. Mol. Biol. Evol. 26, 2463–2473 (2009).
Zhang, J. S. et al. Divergences of MPF2-like MADS-domain proteins have an association with the evolution of the inflated calyx syndrome within Solanaceae. Planta 236, 1247–1260 (2012).
Deanna, R., Larter, M. D., Barboza, G. E. & Smith, S. D. Repeated evolution of a morphological novelty: a phylogenetic analysis of the inflated fruiting calyx in the Physalideae tribe (Solanaceae). Am. J. Bot. 106, 270–279 (2019).
Swain, T. Secondary compounds as protective agents. Annu. Rev. Plant Physiol. 28, 479–501 (1977).
Pichersky, E. & Gershenzon, J. The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 5, 237–243 (2002).
Guimarães, E. T. et al. Activity of physalins purified from Physalis angulata in in vitro and in vivo models of cutaneous leishmaniasis. J. Antimicrob. Chemother. 64, 84–87 (2009).
Chen, L. X., He, H. & Qiu, F. Natural withanolides: an overview. Nat. Prod. Rep. 28, 705–740 (2011).
Olivares-Tenorio, M. L., Dekker, M., Verkerk, R. & van Boekel, M. A. J. S. Health-promoting compounds in cape gooseberry (Physalis peruviana L.): review from a supply chain perspective. Trends Food Sci. Technol. 57, 83–92 (2016).
Puente, L. A., Pinto-Munoz, C. A., Castro, E. S. & Cortes, M. Physalis peruviana Linnaeus, the multiple properties of a highly functional fruit: A review. Food Res. Int. 44, 1733–1740 (2011).
Zhang, W. N. & Tong, W. Y. Chemical constituents and biological activities of plants from the genus. Physalis. Chem. Biodivers. 13, 48–65 (2016).
Benveniste, P. Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 55, 429–457 (2004).
Huang, M. et al. Withanolides from the genus Physalis: a review on their phytochemical and pharmacological aspects. J. Pharm. Pharm. 72, 649–669 (2020).
Schaller, H. New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiol. Biochem. 42, 465–476 (2004).
Fukushima, A. et al. Comparative characterization of the leaf tissue of Physalis alkekengi and Physalis peruviana using RNA-seq and metabolite profiling. Front. Plant Sci. 7, 1883 (2016).
Sonawane, P. D. et al. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants 3, 16205 (2016).
Bolger, A. et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 46, 1034–1038 (2014).
Kim, S. et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278 (2014).
The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 (2011).
Feschotte, C., Jiang, N. & Wessler, S. R. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3, 329–341 (2002).
Du, J. C. et al. Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J. 63, 584–598 (2010).
Särkinen, T., Bohs, L., Olmstead, R. G. & Knapp, S. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol. Biol. 13, 214 (2013).
Wilf, P., Carvalho, M. R., Gandolfo, M. A. & Cúneo, N. R. Eocene lantern fruits from Gondwanan Patagonia and the early origins of Solanaceae. Science 355, 71–75 (2017).
Paterson, A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427 (2012).
Pomilio, A. B., Falzoni, E. M. & Vitale, A. A. Toxic chemical compounds of the Solanaceae. Nat. Prod. Commun. 3, 593–628 (2008).
Payne, A. H. & Hales, D. B. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr. Rev. 25, 947–970 (2004).
Pollier, J. et al. A widespread alternative squalene epoxidase participates in eukaryote steroid biosynthesis. Nat. Microbiol. 4, 226–233 (2019).
Itkin, M. et al. Biosynthesis of antinutritional alkaloids in Solanaceous crops is mediated by clustered genes. Science 341, 175–179 (2013).
He, C. Y., Sommer, H., Grosardt, B., Huijser, P. & Saedler, H. PFMAGO, a MAGO NASHI-like factor, interacts with the MADS-domain protein MPF2 from Physalis floridana. Mol. Biol. Evol. 24, 1229–1241 (2007).
Tapia-López, R. et al. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 146, 1182–1192 (2008).
Gu, X. et al. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4, 1947 (2013).
Vrebalov, J. et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346 (2002).
Ampomah-Dwamena, C., Morris, B. A., Sutherland, P., Veit, B. & Yao, J. L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 130, 605–617 (2002).
Liu, D. M. et al. The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J. 77, 284–296 (2014).
Li, N. et al. The MADS-box gene SlMBP21 regulates sepal size mediated by ethylene and auxin in tomato. Plant Cell Physiol. 58, 2241–2256 (2017).
Zhang, J. L. et al. Suppression of a tomato SEPALLATA MADS-box gene, SlCMB1, generates altered inflorescence architecture and enlarged sepals. Plant Sci. 272, 75–87 (2018).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017).
Zhang, H. & Forde, B. G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409 (1998).
El-Tohamy, W. A., El-Abagy, H. M., Abou-Hussein, S. D. & Gruda, N. Response of cape gooseberry (Physalis peruviana L.) to nitrogen application under sandy soil conditions. Gesund. Pflanz. 61, 123 (2009).
Gu, Y. Z., Xing, S. L. & He, C. Y. Genome-wide analysis indicates lineage-specific gene loss during Papilionoideae evolution. Genome Biol. Evol. 8, 635–648 (2016).
Zhang, J. S., Tian, Y., Wang, L. & He, C. Y. Functional evolutionary developmental biology (evo-devo) of morphological novelties in plants. J. Syst. Evol. 48, 94–101 (2010).
Wingen, L. U. et al. The molecular basis of pod corn (Tunicate maize). Proc. Natl Acad. Sci. USA 109, 7115–7712 (2012).
Khan, M. R., Hu, J. Y. & Ali, G. M. Reciprocal loss of CArG-boxes and auxin response elements drives expression divergence of MPF2-like MADS-box genes controlling calyx inflation. PLoS ONE 7, e42781 (2012).
Khan, M. R., Ihsan, H. & Ali, G. M. WSA206, a paralog of duplicated MPF2-like MADS-box family is recruited in fertility function in Withania. Plant Sci. 253, 215–228 (2016).
Soyk, S. et al. Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 169, 1142–1155 (2017).
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S. & Yanofsky, M. F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940 (2004).
Morel, P. et al. Divergent functional diversification patterns in the SEP/AGL6/AP1 MADS-box transcription factor superclade. Plant Cell 31, 3033–3056 (2019).
Lönnig, W. E. Mutagenesis in Physalis pubescens L. ssp. floridana: some further research on Dollo’s law and the law of recurrent variation. Floriculture Ornam. Biotech. 4, 1–21 (2010).
Pnueli, L., Hareven, D., Broday, L., Hurwitz, C. & Lifschitz, E. The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6, 175–186 (1994).
Dong, T. T. et al. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 163, 1026–1036 (2013).
Ito, Y. et al. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat. Plants 3, 866–874 (2017).
Roldan, M. V. G. et al. Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Sci. Rep. 7, 4402 (2017).
Zhang, J. L. et al. A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 8, 3413 (2018).
Barthel, A. et al. Immune modulation enables a specialist insect to benefit from antibacterial withanolides in its host plant. Nat. Commun. 7, 12530 (2016).
Desmond, E. & Gribaldo, S. Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol. Evol. 1, 364–381 (2009).
Gold, D. A., Caron, A., Fournier, G. P. & Summons, R. E. Paleoproterozoic sterol biosynthesis and the rise of oxygen. Nature 543, 420–423 (2017).
Fernández-Cabezón, L., Galán, B. & García, J. L. New insights on steroid biotechnology. Front. Microbiol. 9, 958 (2018).
Damu, A. G. et al. Isolation, structures, and structure−cytotoxic activity relationships of withanolides and physalins from Physalis angulata. J. Nat. Prod. 70, 1146–1152 (2007).
Misico, R. I. et al. In Progress in the Chemistry of Organic Natural Products (eds Kinghorn, A. D., Falk, H., Kobayashi, J.) Vol. 94, pp. 127–229 (Springer, 2011).
Maldonado, E., Alvarado, V. E., Torres, F. R., Martínez, M. & Pérez-Castorena, A. L. Androstane and withanolides from Physalis cinerascens. Planta Med. 71, 548–553 (2005).
Phillips, D. R., Rasbery, J. M., Bartel, B. & Matsuda, S. P. T. Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9, 305–314 (2006).
Lavie, D., Glotter, E. & Shvo, Y. Constituents of Withania somnifera Dun. III. the side chain of withaferin A. J. Org. Chem. 30, 1774–1778 (1965).
Wang, L. et al. Regulatory change at Physalis Organ Size 1 locus correlates to natural variation in tomatillo reproductive organ size. Nat. Commun. 5, 4271 (2014).
Li, Z. C. & He, C. Y. Physalis floridana Cell Number Regulator 1 encodes a cell membrane-anchored modulator of cell cycle and negatively controls fruit size. J. Exp. Bot. 66, 257–270 (2015).
Lemmon, Z. H. et al. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770 (2018).
Gong, P. C. et al. Physalis floridana CRABS CLAW mediates neofunctionalization of GLOBOSA genes in carpel development. J. Exp. Bot. https://doi.org/10.1093/jxb/erab309 (2021).
Chin, C. S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Li, L., Stoeckert, C. J. & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Yang, Z. H. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Sanderson, M. J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19, 301–302 (2003).
De Bie, T., Cristianini, N., Demuth, J. P. & Hahn, M. W. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22, 1269–1271 (2006).
Wang, Y. P. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012).
Paenicová, L. et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551 (2013).
He, C. Y., Zhang, J. S. & Chen, S. Y. A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor. Appl. Genet. 104, 1125–1131 (2002).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Acknowledgements
We sincerely thank Dr. Heinz Saedler (Max-Planck Institute for Plant Breeding Research, Cologne, Germany) for his strong support for initiating the Chinese lantern evo-devo project and for his inspiring ideas and discussions, which contributed significantly to the current model proposed herein. The authors gratefully acknowledge Profs Tingyun Kuang (Institute of Botany, Chinese Academy of Sciences, Beijing, China) and Shouyi Chen (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for bringing about cooperation between CYH and HZW. This work was initiated in 2006, and some experiments were performed by CYH while working as a co-group leader with Heinz Saedler at the Max-Planck Institute for Plant Breeding Research, Cologne, Germany, and the Max-Planck Institute is acknowledged. The technical assistance of Britta Grosardt (Max-Planck Institute for Plant Breeding Research, Cologne, Germany) and Dr. Xiuping Xu (Institute of Botany, Chinese Academy of Sciences, Beijing, China) is appreciated. We also thank Drs. Jing Liu (Institute of Botany, Chinese Academy of Sciences, Beijing, China) and Cheng Qin (Zunyi Academy of Agricultural Sciences, Zunyi, China) for generous gifts of the seeds of the indicated solanaceous species. This work was supported by grants from the National Natural Science Foundation of China (31525003, 31930007) to C.Y.H., grants (31970346) to H.Z.W., the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27010106) to C.Y.H., and grants from the National Natural Science Foundation of China (31470407) to H.Z.W.
Author information
Authors and Affiliations
Contributions
C.Y.H. conceived, designed, and initiated the project. J.J.L., M.F.L., K.P.L., Y.Y.Y., W.F.Y., X.M.Z., Z.M.L., Y.N.J., and C.Y.H. were involved in genome sequencing analyses. H.H.G., Y.Y.Y., W.F.Y., and C.Y.H. were involved in RNA-seq analyses. L.W. prepared plant materials for genome sequencing and performed karyotype and flow cytometric analyses. C.Y.H., M.F.L., and J.Z. performed molecular and functional characterization of MADS-box genes. J.J.L., C.Y.H., X.Z., J.Y.K., T.Y.Y., P.C.G., and H.Z.W. performed PfSQE-VIGS, gene expression, and steroid component analyses. C.Y.H., J.J.L., P.C.G., L.F.W., Q.R.L., M.S.Z., X.Y.L., K.P.L., and Y.N.J. were involved in analyzing the data. J.J.L., M.F.L., L.W., K.P.L., Y.N.J. and H.Z.W. helped with writing the manuscript. C.Y.H. wrote the manuscript. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lu, J., Luo, M., Wang, L. et al. The Physalis floridana genome provides insights into the biochemical and morphological evolution of Physalis fruits. Hortic Res 8, 244 (2021). https://doi.org/10.1038/s41438-021-00705-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-021-00705-w
This article is cited by
-
Genetics and evolutionary insights from Solanaceae genome sequences
Plant Systematics and Evolution (2024)
-
Chromosome-level genome assembly and annotation of the prickly nightshade Solanum rostratum Dunal
Scientific Data (2023)
-
Revealing evolution of tropane alkaloid biosynthesis by analyzing two genomes in the Solanaceae family
Nature Communications (2023)