Abstract
Starch branching enzyme (SBE) has rarely been studied in common starchy banana fruits. For the first time, we report here the molecular characterization of seven SBE (MaSBE) and six SBE (MbSBE) genes in the banana A- and B-genomes, respectively, which could be classified into three distinct subfamilies according to genome-wide identification. Systematic transcriptomic analysis revealed that six MaSBEs and six MbSBEs were expressed in the developing banana fruits of two different genotypes, BaXi Jiao (BX, AAA) and Fen Jiao (FJ, AAB), among which MaSBE2.3 and MbSBE2.3 were highly expressed. Transient silencing of MaSBE2.3 expression in banana fruit discs led to a significant decrease in its transcription, which coincides with significant reductions in total starch and amylopectin contents compared to those of empty vector controls. The suggested functional role of MaSBE2.3 in banana fruit development was corroborated by its transient overexpression in banana fruit discs, which led to significant enhancements in total starch and amylopectin contents. A number of transcription factors, including three auxin response factors (ARF2/12/24) and two MYBs (MYB3/308), that interact with the MaSBE2.3 promoter were identified by yeast one-hybrid library assays. Among these ARFs and MYBs, MaARF2/MaMYB308 and MaARF12/MaARF24/MaMYB3 were demonstrated via a luciferase reporter system to upregulate and downregulate the expression of MaSBE2.3, respectively.
Similar content being viewed by others
Introduction
Plant starch, which consists of two glucan polymers, linear amylose and branched amylopectin, is a major supplier of human nutrition and calories. Starch is commonly found in cereal grains and potato tubers1,2 as well as in some fresh fruits, such as mango (Mangifera indica)3, kiwifruit (Actinidia deliciosa)4, and banana (Musa acuminata)5,6,7. As the main component of the total starch in the storage organs of the abovementioned plant species, amylopectin is primarily catalyzed by soluble starch synthase, debranching enzyme, and starch branching enzyme (SBE)8. Of these three classes of major enzymes, SBE mainly controls the branching pattern of amylopectin and influences the amylopectin yield and, ultimately, the nutritional value of those plants9,10,11.
Variable numbers of SBEs have been isolated in several higher plant species based on genomic data: three have been identified in maize (Zea mays), rice (Oryza sativa), sorghum (Sorghum bicolor), barley (Hordeum vulgare), and potato (Solanum tuberosum); two have been identified in Arabidopsis (Arabidopsis thaliana); six in cassava (Manihot esculenta); and seven in wheat (Triticum aestivum)12,13,14,15. Furthermore, SBEs can be generally classified into no less than three groups—SBEI, II, and III, each of which typically contains a C-terminal all-beta domain and an alpha-amylase domain16. SBEI condenses amylose to long glucan chains13, while SBEII is mainly associated with the biosynthesis of amylopectin using short glucan chains as a substrate13,17. However, the functional role of SBEIII remains ambiguous despite reports of its increased activity in the process of wheat grain development18.
A number of expression and functional studies have further revealed the multifunctional aspect of SBEs in both amylopectin biosynthesis and plant development. For instance, the temporal expression of SBE1 was found to be responsible for amylopectin accumulation in apple fruits and in maize seedling development12,19. SBEII expression is involved in nut development in chestnut (Castanea mollissima)20 and seed development in rice21. A study on the kidney bean (Phaseolus vulgaris) mutant pvsbe2 suggested that SBEII activity is important for both amylopectin biosynthesis and maturation during the middle and late stages of seed development22. Further, the expression of OsSBEIIa was found to regulate the sugary endosperm trait during development and maturity in the Sugary Endosperm rice mutant23. In maize, SBEIIa-defective mutants exhibit a leaf-senescence phenotype as the result of the reduction in the proportion of branched starch24. Taken together, all of these studies imply that the expression of SBE is a critical factor in amylopectin biosynthesis and plant growth and development.
A number of transcription factors (TFs) regulating SBE expression have been identified. In maize, the expression of numerous SBE genes was upregulated when ZmNAC36 was transiently overexpressed25, while the protein levels of maize SBEs were significantly reduced in the endosperm-specific TF opaque2 mutant and prolamin-box-binding factor (PBF) mutant26. In rice, OsbZIP58 was found to upregulate SBE1 expression through its interaction with the SBE1 promoter27. Taken together, all of these studies indicated that SBE expression is regulated by the coordination of multiple TFs.
Banana is a popular fresh starchy fruit worldwide and constitutes the principal staple food in several African countries28. Amylopectin accounts for approximately 70–80% of the total starch content on a dry weight basis in unripe banana fruits6,7,29. The biosynthesis and accumulation of amylopectin in banana fruit is of crucial importance in terms of fruit quality, yield, and nutritional value. Genome-wide identification of key candidate genes associated with amylopectin biosynthesis is therefore necessary to enhance the yield and improve the quality of banana fruits6,30,31. In the current study, we report the identification of seven and six distinct SBE members from the banana A- and B-genomes, respectively, which are henceforth referred to as MaSBE and MbSBE. The molecular characteristics, phylogenetic relationships, conserved domains, gene structures, and spatiotemporal expression patterns were investigated in the two different banana genotypes. The potential functionality of MaSBE2.3 was studied through both transient virus-induced gene silencing (VIGS) and overexpression in banana fruit discs. Three auxin response factors (ARF2/12/24) and two MYBs (MYB3/308) that regulate MaSBE2.3 expression were identified using the yeast one-hybrid (Y1H) method and luciferase (LUC) activity analysis. Together, these studies may enhance our understanding of the members of the SBE gene family involved in amylopectin biosynthesis and fruit development and pave the way for the genetic improvement of the quality, yield, and nutritional traits of banana.
Materials and methods
Plant material
The two banana genotypes used in this experiment were Fen Jiao (FJ, M. acuminata, AAB, cultivar Fenjiao) and BaXi Jiao (BX, M. acuminata, AAA, cultivar Cavendish), which were planted at the Danzhou Banana Germplasm Centre (Hainan, China, 19 N, 109 E). BX fruits are characterized by their high yield, superior quality, and long shelf-life, whereas FJ fruits are characterized by their good flavor, rapid ripening, and strong disease resistance. According to the published classification of banana fruit developmental stages by Ram et al.32, we selected three representative stages corresponding to pulp initiation at 0 day after inflorescence emergence from the pseudostem (DAF), pulp growth (20 DAF), and maturation (80 DAF) for SBE temporal expression analysis. Following harvest at 80 DAF, BX fruits were stored for 0, 8, and 14 days post-harvest (DPH), while FJ fruits were stored for 0, 3, and 6 DPH prior to expression analysis of SBE during fruit ripening. These fruits can also be distinguished by their color: peel green, peel yellowish green, and peel yellow. A spatial gene expression analysis of SBEs was also performed on select banana tissues, including root, leaf, and fruit tissues, at 80 DAF. These expression studies were conducted for three replicates.
Genome-wide investigation and phylogenetic tree construction of SBE family proteins
SBE amino acid sequences were obtained from the M. acuminata (DH-Pahang, AA genotype, 2n = 22) database5 and Musa balbisiana (DH-PKW, BB genotype, 2n = 22) database7. The putative amino acid sequences of SBE from A. thaliana, T. aestivum, Z. mays, O. sativa, Solanum lycopersicum, S. tuberosum, and Vitis vinifera were retrieved from The Arabidopsis Information Resource, the wheat genome, the Maize Genetics and Genomics database, the Rice Genome Annotation Project, the Sol Genomics Network, the Potato Genome Sequence Consortium, and the grapevine genome, respectively. All bioinformatic analytical databases and websites used are listed in Table S1. The banana SBEs, together with SBEs from the A. thaliana, T. aestivum, Z. mays, O. sativa, S. tuberosum, S. lycopersicum, and V. vinifera searches, were analyzed using the BLAST software. The accession numbers of the SBEs from the abovementioned plant species are presented in Table S2. The chromosome distribution of MaSBEs and MbSBEs was analyzed by the Circos software. The SBE protein sequences derived from these diverse plant species were used to construct a phylogenetic tree using the ClustalX 2.0 tool (http://downloads.fyxm.net/Clustal-x-58923.html) and MEGA 5.0 software based on 1000 bootstrap replicates31.
Characterization of conserved domains and exon–intron structure
The isoelectric point and molecular mass of the SBEs were calculated using ExPASy, and the data are presented in Table S3. The conserved domains and SBE proteins were predicted and annotated by using the NCBI database and MEME software. The polypeptides were deduced and analyzed by PeptideMass and BLASTP software (Table S4). The genomic structure of the SBE genes was deduced by using the Gene Structure Display Server. The MaSBE promoter was obtained from the banana A-genome database within the Banana Genome Hub. The predicted transcription start sites and cis-acting elements were identified by searching the Berkeley Drosophila Genome Project database and the PlantCARE website, respectively.
RNA deep sequencing and transcriptomic analysis
Various banana plant organs, including the roots, leaves, and fruits, at 80 DAF in both BX and FJ genotypes were collected for spatial transcriptomic analysis. The pulp of banana fruits at various developmental and ripening stages, including that of BX and FJ at 0, 20, and 80 DAF, that of BX at 8 and 14 DPH, and that of FJ at 3 and 6 DPH, were collected for transcriptome sequencing at different time points.
RNA-sequencing (RNA-seq) libraries were constructed using total RNA samples prepared by the use of a plant RNAout Kit (TIANGEN Biotech, Beijing, China). Deep sequencing was performed by using the GAII platform (Illumina, Inc., San Diego, CA) according to the protocol provided by the manufacturer. The adapter sequences and low-quality sequences were removed by using the FASTX toolkit and FastQC, respectively. After mapping the clean reads to both the A- and B-genome banana databases, transcriptome assemblies were constructed by using Cufflinks. The gene expression level was assessed as reads per kilobase pair of transcripts per million reads (RPKM). The DESeq package was applied to screen differentially expressed genes. There were two technical replicates and three biological replicates.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
The spatiotemporal expression patterns of banana SBEs revealed by transcriptome sequencing were further identified through qRT-PCR using an Mx3000P system (Stratagene, San Diego, CA). The primer pair quality was verified according to melting curve analysis, with amplification efficiencies ranging from 0.9 to 1.1, and agarose gel electrophoresis (Table S5). Actin (GenBank accession No. EF672732) and UBQ2 (GenBank accession No. HQ853254) were used as the internal reference genes. The relative gene expression level was calculated according to the 2−ΔΔCT method33. There were three replicates included.
Vector construction and VIGS in banana fruit discs
For VIGS vector construction, the entire coding region of MaSBE2.3 (2502 bp in length) was amplified by PCR and verified by DNA sequencing (the primers used are listed in Table S6; the sequence is listed in Fig. S1) prior to insertion between the EcoR I and Xho I restriction enzyme sites in pTRV234, yielding pTRV2-MaSBE2.3, which was subsequently introduced into Agrobacterium (Agrobacterium tumefaciens strain GV3101) together with pRTV1. Surface-sterilized BX banana fruit (80 DAF) discs that were 1–2 mm thick were then vacuum infiltrated with the Agrobacterium solution (OD600 = 0.6) prior to cocultivation on Murashige and Skoog (MS) media at 30 °C for 0, 1, 2, or 3 days35, followed by iodine–potassium iodide (I2-KI) staining36. In addition to I2-KI staining, assessment of the transcript levels of the downregulated genes and the measurements of total starch and amylopectin contents in these agroinfiltrated banana fruit discs were also carried out. Biologically independent replicates were assessed in triplicate.
Vector construction and transient overexpression in banana fruit discs
For the transient overexpression vector, the entire coding region of MaSBE2.3 was inserted into a pCAMBIA1304 vector (Cambia, Canberra, Australia) behind the CaMV 35S promoter in the sense orientation using the restriction enzymes Bgl I and Spe I. Following confirmation by DNA sequencing, the recombinant pCAMBIA1304-MaSBE2.3 plasmid was transformed into A. tumefaciens GV3101 cells. The cells of the A. tumefaciens strain were diluted to an OD600 of 0.60 in infiltration buffer (200 mM acetosyringone, 10 mM 2-(N-Morpholino)ethanesulfonic acid, 10 mM MgCl2), in which the BX banana fruit discs (80 DAF) were soaked34. Following a period of culture on MS media at 30 °C for 3 days, the agroinfiltrated banana fruit discs were subjected to I2-KI staining, transcription analysis, and total starch and amylopectin content evaluations. The biologically independent transformations were performed in triplicate.
Measurement of total starch content
The banana pulp was soaked for 10 min in 1.5% sodium bisulfite solution to prevent browning prior to drying for 24 h at 40 °C37, after which the material was ground into powder. One hundred milligrams of powder was washed in 5 mL of 80% ethanol and centrifuged for 5 min at 4000 rpm. Following the removal of the supernatant, the pellet was washed in 5 mL of 80% Ca(NO3)2 twice and then centrifuged for 5 min at 4000 rpm. The total starch content was calculated as the percentage per gram of dry sample, which was calculated according to a 100 μg/mL starch standard solution (Sigma, S-2630) with an absorbance of 620 nm38. Three replicates were measured.
Measurement of amylopectin content
In total, 9 mL of 1 M sodium hydroxide and 1 mL of 95% ethanol were added to a sample of 100 mg of banana powder, after which the mixture was incubated at 40 °C for a period of 24 h. Afterward, 5 mL of the mixture, 2 mL of 0.2 N I2-KI solution, and 1 mL of 1 M acetic acid were mixed together prior to further dilution to 100 mL with distilled water in a new volumetric flask. The amylopectin content was calculated as the percentage per gram of dry sample based on a 1 mg/mL amylopectin standard solution (Sigma-Aldrich, catalog number 10120) measured at 533 nm. The measurement was carried out for three replicates.
cDNA library construction and Y1H library screening
Total RNA was extracted from the BX banana pulp samples at 0, 20, and 80 DAF by a plant RNAout Kit (TIANGEN Biotech) and then applied to cDNA library construction by using a Mathchmaker Gold Yeast One-Hybrid Library Screening System Kit (Clontech, Mountain View, CA). The putative MaSBE2.3 promoter, which consists of a genomic DNA fragment 1824 bp in length, immediately upstream of the MaSBE2.3 open reading frame (ORF; the primers used are listed in Table S6; the sequence is listed in Fig. S2) was inserted into a pAbAi vector (Clontech) using the restriction enzymes Sac I and Xho I. The bait and pGADT7-Rec prey vectors together with the cDNA library were cotransformed into the Y1HGold (Clontech) cells, which were subsequently cultured on SD/-Leu+AbA200 media for 3 days at 30 °C. Selected positive colonies were further verified via PCR and sequence analyses. The identified TF genes were annotated in the banana A-genome database5.
Y1H assays
To identify the individual interaction between the MaSBE2.3 promoter and a single TF, the MaSBE2.3 promoter was inserted into a pAbAi vector to form a bait construct (Clontech). A prey construct was built by inserting the ORFs of candidate TF genes (the primer sequences used are listed in Table S6; the sequences are listed in Fig. S3) into a pGADT7 AD vector. Both the prey and bait gene constructs were cotransformed into cells of the Y1HGold yeast strain (Clontech), which were subsequently grown on SD/-Leu+AbA200 selective media at 30 °C for 3 days.
Analysis of dual LUC activity
The MaSBE2.3 promoter was cloned into a pGreenII 0800-REN-LUC vector as described by Hellens et al.39. The ORFs of MaARF2/12/24 and MaMYB3/308 were each inserted into a pGreenII 62Sk vector separately, which were subsequently transformed into the A. tumefaciens GV3101 strain. A. tumefaciens cultures harboring the pGreen-MaSBE2.3 promoter vector and A. tumefaciens cultures containing pGreenII62Sk-MaARFs or pGreenII62Sk-MaMYBs were mixed together at a ratio of 1:7 (v/v) prior to cocultivation with banana fruit discs by soaking and shaking. The luciferase and REN-LUC activities following cocultivation for a period of 3 days were analyzed by using a dual-LUC reporter assay system (Promega, Madison, WI). Three replicates were measured.
Statistical analysis methods
SPSS 10.0 software (SPSS Inc., Chicago, IL) was used for the statistical analyses. The least significant difference test was used for one-way analysis of variance (ANOVA). Student’s t tests were applied to test the differences between the means revealed by ANOVA. Each sample consisted of three replicates, and p < 0.05 and p < 0.01 were recognized as statistically significant and highly significant, respectively.
Results
Genome-wide characterization and phylogenetic analysis of banana SBE genes
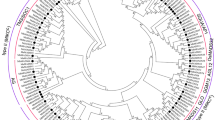
Seven M. acuminata MaSBE (MaSBE-1, -2.1, -2.2, -2.3, -2.4, -2.5, and -3) and six M. balbisiana MbSBE (MbSBE-1, -2.1, -2.2, -2.3, -2.4, and -3) amino acid sequences were obtained from the banana A- and B-genome databases5,7, respectively. Among the 11 chromosomes, MaSBE1/MbSBE1, MaSBE2.1/2.2/MbSBE2.1/2.2, and MaSBE2.3/2.4/2.5/3/MbSBE2.3/2.4/3 were distributed on chromosomes 4, 5, and 6, respectively (Fig. 1a). The number of amino acids in the putative MaSBE and MbSBE proteins varied from 162 (MbSBE2.2) to 924 (MbSBE3), with relative molecular masses ranging from 17.314 (MbSBE2.2) to 106.701 (MbSBE3) kDa and isoelectric points ranging from 4.87 (MbSBE2.1) to 7.74 (MaSBE2.2) (Table S3). The high-level variation of MaSBEs and MbSBEs in the numbers of amino acids, molecular mass, and isoelectric points may possibly reflect their functional divergence in various biological processes.
a Circos image generated by the Circos software. b Phylogenetic tree constructed by MEGA 5.0. The SBEs were divided into three subfamilies: groups I, II, and III. ACHR04/05/06 represent chromosomes 4, 5, and 6 from the banana A-genome, respectively, and BCHR04/05/06 represent chromosomes 4, 5, and 6 from the banana B-genome, respectively. The yellow, green, blue, purple, red, black, orange, light blue, and dark purple circles represent SBEs from Musa acuminata, Musa balbisiana, Arabidopsis thaliana, Oryza sativa, Vitis vinifera, Solanum lycopersicum, Zea mays, Triticum aestivum, and Solanum tuberosum, respectively
To understand the evolution of the SBE proteins in banana, a phylogenetic tree was constructed by comparing all seven MaSEB and six MbSBE amino acid sequences containing GlgB domains (Tables 1 and 2) together with the amino acid sequences of each of the following: three O. sativa OsSBEs, Z. mays ZmSBEs, S. tuberosum StSBEs, V. vinifera VvSBEs, and S. lycopersicum SlSBEs; two A. thaliana AtSBEs; and seven T. aestivum TaSBEs (Fig. 1b). MaSBEs and MbSBEs clustered into three groups (I–III). MaSBE1/MbSBE1 and MaSBE3/MbSBE3 were found in group I and group III, respectively. The majority of banana SBEs, including five MaSBE (MaSBE-2.1, -2.2, -2.3, -2.4, and -2.5) and four MbSBE (MbSBE-2.1, -2.2, -2.3, and -2.4) members, were present in group II. It is likely that the banana SBEs in group II have undergone significant gene duplication and possible functional diversification throughout the evolutionary process.
Exon–intron structural characteristics and conserved domain annotations of the banana SBE family members
The evolutionary characteristics of the MaSBE and MbSBE family members were further verified by exon–intron structural analysis (Fig. 2a, c). The exon–intron organization of the MaSBE and MbSBE genes clearly differed in groups I (5 exons), II (10–24 exons), and III (20–22 exons) (Fig. 2b, d), implying that evolutionary and functional divergence occurred among these three groups of SBE genes in banana.
a Phylogenetic tree of MaSBE genes from the banana A-genome. b Exon–intron structure analysis of MaSBE genes. c Phylogenetic tree of MbSBE genes from the banana B-genome. d Exon–intron structure analysis of MbSBE genes. Three subgroups of MaSBEs and MbSBEs (groups I, II, and III) were formed on the basis of phylogenetic relationships. The black lines indicate introns; the yellow boxes indicate exons
To further assess the protein structural divergence, the transit peptides, polypeptides, and conserved domains of MaSBEs and MbSBEs were further analyzed in silico. Transit peptides were identified in all putative MaSBE and MbSBE proteins (Fig. S4). It was predicted that there are numerous polypeptides with numbers ranging from 13 (MbSBE2.2) to 67 (MbSBE2.3) and that they also have 1,4-alpha-glucan-branching enzyme activity or alpha-amylase activity (Table S4). All three domains, including the alpha-amylase domain, C-terminal all-beta domain, and carbohydrate-binding module 48 (CBM_48), were found in four SBEs, SBE-1, -2.1, -2.3, and -2.4, which is consistent between the A- and B- genomes. However, 0–2 domains were found in the remaining SBE members (Tables 1 and 2).
Spatial expression features of banana SBE genes
SBE expression in different banana plant tissues was investigated by comparative transcriptome analyses using RNAs derived from leaves, roots, and fruits of BX (AAA) and FJ (AAB). In BX, six MaSBE genes, MaSBE-1, -2.1, -2.2, -2.3, -2.4, and -3, were expressed in all the tested tissues of BX, whereas the expression of MaSBE-2.5 was not discernible (Fig. 3a, Table S7). In FJ, seven MaSBEs (MaSBE-1, -2.1, -2.2, -2.3, -2.4, -2.5, and -3) and six MbSBEs (MbSBE-1, -2.1, -2.2, -2.3, -2.4, and -3) were expressed in all the tested tissues (Fig. 3b, c, Table S7); the expression of MaSBE-2.3 and MbSBE-1, -2.3, and -2.4 was significantly greater than that of other members. Among the highly expressed genes, MaSBE2.3 and MbSBE2.3 presented not only the highest expression level (RPKM > 33) but also a high level of tissue specificity in the fruits of both genotypes. The consistently high expression in the fruits of both genotypes may imply that SBE2.3 is a key player in banana fruit development and starch accumulation.
a Expression patterns of MaSBEs in the roots, leaves, and fruits of the BX (AAA genotype) banana variety. b Expression patterns of MaSBEs in the roots, leaves, and fruits of the FJ (AAB genotype) banana variety. c Expression patterns of MbSBEs in the roots, leaves, and fruits of the FJ (AAB genotype) banana variety. d Expression patterns of MaSBEs throughout the processes of fruit development and ripening in the BX (AAA genotype) variety. e Expression patterns of MaSBEs throughout the processes of fruit development and ripening in the FJ (AAB genotype) banana variety. f Expression patterns of MbSBEs throughout the processes of fruit development and ripening in the FJ (AAB genotype) variety. BX BaXi Jiao (M. acuminata, AAA, cultivar Cavendish), FJ Fen Jiao (M. acuminata, AAB, cultivar Fenjiao), DAF days after inflorescence emergence from the pseudostem, DPH days post-harvest. The heat map was generated using the RPKM values of the MaSBEs and MbSBEs. The color of the red–white scale shows the differences in gene expression changes. The asterisks indicate significant differences compared to BX-roots, FJ-roots, BX-0DAF, or FJ-0DAF (*p < 0.05; **p < 0.01)
Temporal expression pattern of banana SBE genes
The temporal expression pattern of banana SBEs was analyzed by using developing banana fruits and harvested banana fruits under two different storage durations (Fig. 3d–f, Table S8). In BX, six MaSBE genes, MaSBE-1, -2.1, -2.2, -2.3, -2.4, and -3, were expressed during the processes of fruit development and ripening (Fig. 3d). In FJ, six MaSBEs (MaSBE-1, -2.2, -2.3, -2.4, -2.5, and -3) and six MbSBEs (MbSBE-1, -2.1, -2.2, -2.3, -2.4, and -3) exhibited various expression changes during fruit development and ripening, among which MaSBE-1 and -2.3 or MbSBE-1 and -2.3 showed significant upregulation during fruit development from 0 to 80 DAF and downregulation during the ripening process. In both genotypes, MaSBE2.3 and MbSBE2.3 maintained the highest expression levels throughout fruit development (RPKM > 11). This genotype-based temporal expression pattern warrants further functional characterization of SBE2.3 in banana.
Differential expression analysis of banana SBE genes via qRT-PCR
The RNA-seq results showed that six MaSBEs (MaSBE-1, -2.1, -2.2, -2.3, -2.4, and -3) and six MbSBEs (MbSBE-1, -2.1, -2.2, -2.3, -2.4, and -3) exhibited variable expression in the different tissues or at various fruit developmental stages. This transcriptomic expression profile was verified by qRT-PCR (Figs. 4 and 5). Following normalization, the expression level of each gene of BX-root, FJ-root, BX-0DAF or FJ-0DAF was considered “1,” and the relative expression level of each gene in other tissues or at other development and ripening stages was calculated by comparing the expression to that of BX-root, FJ-root, BX-0DAF, or FJ-0DAF. All of the tested banana SBEs, except MaSBE3 and MbSBE3 in the FJ fruit (Fig. 4) and MaSBE2.1 in the BX fruit at 8 DPH (Fig. 5), exhibited consistent expression patterns according to the RNA-seq expression analysis. Moreover, the correlation coefficients for both RNA-seq and qRT-PCR in the different tissues and at different developmental stages were greater than 0.8783 and 0.8386, respectively (Table S9), implying that the RNA-seq analysis in this experiment could provide a suitable expression assessment for both banana genotypes.
BX BaXi Jiao (M. acuminata, AAA, cultivar Cavendish), FJ Fen Jiao (M. acuminata, AAB, cultivar Fenjiao), DAF days after inflorescence emergence from the pseudostem, DPH days post-harvest. Biological replicates were tested in triplicate, and the asterisks indicate significant differences compared to BX-0DAF or FJ-0DAF (*p < 0.05; **p < 0.01)
MaSBE2.3 is essential for fruit amylopectin biosynthesis in banana fruit discs based on transient VIGS and overexpression analyses
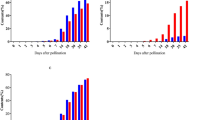
Based on the temporal and spatial expression patterns, MaSBE2.3 showed not only significantly higher expression in the developing banana fruits than in the somatic tissues (Fig. 3a) but also a trend of increasing expression during fruit development (Fig. 3d). To further investigate the function of MaSBE2.3 in amylopectin biosynthesis, the expression of MaSBE2.3 was modulated through VIGS in BX fruit discs (80 DAF). As illustrated in Fig. S5, following 3 days of incubation on MS media, relative to the fruit discs transformed with empty vector controls, MaSBE2.3-VIGS banana fruit discs showed lighter colors of I2-KI staining (Fig. 6a). Similarly, MaSBE2.3 expression, total starch, and amylopectin contents decreased in the VIGS fruit discs, as evidenced by the respective qRT-PCR analysis and total starch and amylopectin content determination compared with those of the control fruit discs (Fig. 6b–d). On the other hand, the fruit discs transiently overexpressing MaSBE2.3 appeared to be darker following I2-KI staining compared with those expressing the empty vector control (Fig. 6e). Further quantitative analysis revealed that the expression level of MaSBE2.3 and the total starch and amylopectin contents increased by approximately 80-, 1.3- and 1.4-fold, respectively, in the MaSBE2.3-infiltrated BX fruit discs (80 DAF) compared with the control fruit discs (Fig. 6f–h).
a I2-KI staining of MaSBE2.3-downregulated banana fruit discs. b Expression level of the MaSBE2.3 gene in MaSBE2.3-downregulated banana fruit discs. c Change in total starch content in MaSBE2.3-downregulated banana fruit discs. d Change in amylopectin content in MaSBE2.3-downregulated banana fruit discs. e I2-KI staining of MaSBE2.3-overexpressing banana fruit discs. f Expression level of the MaSBE2.3 gene in MaSBE2.3-overexpressing banana fruit discs. g Change in total starch content in MaSBE2.3-overexpressing banana fruit discs. h Change in amylopectin content in MaSBE2.3-overexpressing banana fruit discs. Biological replicates were tested in triplicate, and the asterisks indicate significant differences compared to pTRV1+pTRV2 (empty vector control) or pCAMBIA1304 (empty vector control) (*p < 0.05; **p < 0.01)
Activation of the MaSBE2.3 promoter by MaARF2/12/24 or MaMYB3/308 in yeast
In the Y1H-based library screening when the MaSBE2.3 promoter was used as bait, 30 colonies were obtained and characterized by plasmid DNA sequencing (Table S10). Among them, there were three MaARFs and two MaMYBs. In subsequent verification using individual Y1H assays, the yeast clones harboring pAbAi-MaSBE2.3+pGADT7-MaARF2, pAbAi-MaSBE2.3+pGADT7-MaARF12, pAbAi-MaSBE2.3+pGADT7-MaARF24, pAbAi-MaSBE2.3+pGADT7-MaMYB3, and pAbAi-MaSBE2.3+pGADT7-MaMYB308 grew normally on SD/-Leu+AbA200 selective media, which is consistent with the library screening results (Fig. 7a). In the MaSBE2.3 promoter region, a TGA element (AACGAC) compatible with ARFs and a MYB-binding site (TAACCA) compatible with MYBs were identified (Fig. S2), providing evidence for the potential molecular interactions between the MaSBE2.3 promoter and either MaARF2/12/24 or MaMYB3/308.
a Physical interactions between MaARFs, MaMYBs, and the MaSBE2.3 promoter according to yeast one-hybrid analysis. The autoactivation of the promoters was determined on SD/-Ura+AbA media, while the interaction between TFs and the promoter was tested on SD/-Leu+AbA media. b Schematics of transient expression vectors. c, d LUC activity resulting from the transient coexpression of the MaSBE2.3 promoter and MaARFs or MaMYBs in Agrobacterium tumefaciens strains. There were three replicates. The data are presented as the means ± SDs. The statistical significance of the differences was assessed by ANOVA (*p < 0.05; **p < 0.01)
Activation of the MaSBE2.3 promoter by MaARF2/12/24 or MaMYB3/308 in banana fruit discs
Given the interaction between the MaSBE2.3 promoter and either MaARF2/12/24 or MaMYB3/308, as revealed by the Y1H approach, we further investigated whether MaARF2/12/24 or MaMYB3/308 directly participate in the regulation of MaSBE2.3 expression in BX banana fruits. For this purpose, MaARF2, MaARF12, MaARF24, MaMYB3, and MaMYB308 were transiently overexpressed in banana fruit discs by Agrobacterium-mediated transformation using the LUC reporter system (Fig. 7b). The LUC activity was elevated by the binding of the MaSBE2.3 promoters by MaARF2 (Fig. 7c) or MaMYB308 (Fig. 7d). In contrast, the LUC activity was significantly reduced by the interaction of the MaSBE2.3 promoters with either MaARF12/24 or MaMYB3 (Fig. 7c, d).
Discussion
Despite the social and economic importance of banana for its production of fresh starchy fruit, there have been relatively few studies on banana than on other high-starch crop species, such as cereals, particularly in terms of amylopectin biosynthesis and fruit development processes6. As a vital enzyme catalyzing amylopectin biosynthesis, SBE has also been found to play a role in seed germination, seedling growth, grain/fruit development, and maturation in many crops species14,19,20,21,22,40,41. In this study, through genome-wide identification, we obtained seven and six SBEs from M. acuminata and M. balbisiana, respectively. The putative MaSBEs and MbSBEs could be divided into three distinct subgroups according to phylogenetic relationships. The banana SBE classification is consistent with that of A. thaliana, Populus trichocarpa, and O. sativa12. Moreover, these banana SBEs in groups I and III were closely evolutionarily related to their orthologs in A. thaliana (AtSBEs) and O. sativa (OsSBEs), respectively. In group II, the closest orthologs of the banana SBEs were those from T. aestivum, O. sativa, and Z. mays (Fig. 1b). It appears that SBEs in group II had undergone numerous duplications in both the A- and B-genomes in banana, as there were 4–5 members, which is in sharp contrast to other plant species that have only one or two SBEs.
The phylogenetic and protein structural features of MaSBEs and MbSBEs were further characterized by their gene exon–intron organization and conserved domain structure and via polypeptide analysis. The banana SBEs could be classified into three distinct subfamilies according to the exon–intron organization of their structural genes (Fig. 2a, c), which is consistent with a previous report in Malus × domestica12. Alpha-amylase domains, C-terminal all-beta domains, and CBMs were also identified in four MaSBEs (MaSBE-1, -2.1, -2.3, and -2.4) and four MbSBEs (MbSBE-1, -2.1, -2.3, and -2.4) (Tables 1 and 2), which are the hallmarks of SBE proteins15,16. This finding is in agreement with findings in previous reports in other plant species, such as millet15 and pea16. However, some other members, including MaSBE-2.2 and -2.5 and MbSBE-2.2, lack one or more such core domains; consequently, the molecular mass of these members is <58.0 kDa (Table S3). In the literature, the size of active SBE enzymes was reported to be as small as 51/50 kDa in barley (Hordeum vulgare) caryopses42. Whether low-molecular-mass MaSBEs and MbSBEs are encoded by pseudogenes or have diverse functionalities distinct from classic SBEs remains unknown.
The findings that SBEs may play a crucial role in the fruit development process have been reported in several plant species, such as M. × domestica, P. vulgaris, and C. mollissima12,20,22. The M. × domestica SBE1 was reported to be expressed at a high level in developing fruits, suggesting that it plays regulatory role in fruit development and amylopectin accumulation12. It has also been found that high levels of SBEI and SBEII expression are important to seed development in C. mollissima20 and P. vulgaris22. In the current study, among the seven MaSBEs and six MbSBEs, SBE2.3 was identified as the only gene whose expression was upregulated during fruit development and downregulated during fruit ripening in the BX and FJ genotypes (Fig. 3d–f), implying that SBE2.3 may be a pivotal player in banana fruit development. Moreover, the expression of other SBE genes varied greatly between these two genotypes. For example, the expression level of MaSBE1 at 0, 20, and 80 DAF was 7-, 9-, and 3-fold higher in BX than in FJ. Similarly, MaSBE2.4 showed a significantly higher level of expression in BX than in FJ throughout the entire fruit development process. Compared with FJ (AAB genotype), BX (AAA genotype) is a banana variety known to produce higher yields, and the quality of its fruits is better, with a longer shelf-life31. This finding is in agreement with findings of previous reports suggesting that, in banana, the A-genome contains relatively more functional genes that are associated with fruit quality and yield traits than the B-genome contains; thus the A-genome could be used as a valuable source for potential target genes in banana breeding programs6,31.
It has been established that SBEs are key players in starch biosynthesis13. In pea mutants, the loss of SBEII resulted in a wrinkled phenotype and 50% reduction in starch biosynthesis in the seed compared with those of wild type43. In maize, the loss of SBEIIb caused a 20% reduction in starch content and alterations to the morphology of the starch granules44. The production of high-amylose wheat starch, which comprises >70% amylose, was associated with SBEII suppression45. The simultaneous loss of both SBEII genes resulted in the failure of starch synthesis in A. thaliana17. In the present study, we demonstrated that the genetic downregulation of MaSBE2.3 expression resulted either directly or indirectly in the increased turnover of total starch and amylopectin, as revealed by the reduced I2-KI staining (Fig. 6a–d). Such results are generally in agreement with those of reports in pea43, maize44, and A. thaliana46. In addition, consistent with previous studies in potato1 and chestnut20, the transient overexpression of MaSBE2.3 in BX banana fruit discs at 80 DAF resulted in significant increases in the contents of total starch and amylopectin (Fig. 6g, h). However, it has been previously suggested that SBE might not be a rate-limiting enzyme in amylopectin biosynthesis, as observed in some stable transformation experiments13. Further, the unusually high level of MaSBE2.3 observed in the transient expression study may not be able to reflect in planta scenarios, which warrants further exploration in future studies.
As an auxin signaling regulator, ARF affects almost every aspect of plant development, including lateral root development, embryogenesis, flower development, and seed setting46,47. However, it has never been implicated in starch biosynthesis. In the present study, three members of the banana ARF family, MaARF2/12/24, were found to bind the promoter of MaSBE2.3 in Y1H experiments (Fig. 7a). For the first time, we revealed that MaARF2 activates and upregulates MaSBE2.3 expression in banana fruit, while MaARF12 and MaARF24 suppress the activity of the MaSBE2.3 promoter in banana fruits (Fig. 7c). MYBs compose a large family of transcriptional regulators that have been shown to be involved in starch biosynthesis in hull-less barley48 and maize49. In hull-less barley, the coexpression of MYB and SBE2b was found to significantly contribute to starch biosynthesis during grain development48. Furthermore, maize ZmMYB14 could bind directly to the ZmSBE1 promoter region in yeast and promote the expression of ZmSBE1 in maize endosperm49. Here we identified two MaMYB members, MaMYB3 and MaMYB308, both of which were found to bind directly to the MaSBE2.3 promoter in yeast. We demonstrated that MaMYB308 and MaMYB3 could promote and suppress MaSBE2.3 expression, respectively, in banana fruit discs (Fig. 7d). There is no doubt that the identification of these TFs would shed additional light on the molecular regulatory mechanism of MaSBE2.3 expression in the amylopectin biosynthesis process in banana fruits.
Conclusions
This report represents the first study on the SBE gene family in banana through a genome-wide approach based on A- and B-genome data. The spatial and temporal expression profiles of individual SBE members were investigated in two distinct banana genotypes, and SBE2.3 was shown to play a vital role in amylopectin biosynthesis. Using both transient downregulation and overexpression techniques, we further demonstrated that MaSBE2.3 is essential for amylopectin biosynthesis in banana fruit. Furthermore, the identification of two classes of TFs, MaARFs and MaMYBs, which interact with the MaSBE2.3 promoter provides further evidence of the regulatory mechanism of MaSBE2.3 expression in amylopectin biosynthesis in banana. This study clearly improves our understanding of the molecular mechanism underlying the functionality of SBE in banana and also provides a set of useful tools for quality improvement of banana fruit through molecular breeding and genetic engineering.
References
Jobling, S. A. et al. A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: cloning and characterization of multiple forms of SBE A. Plant J.18, 163–171 (1999).
Wang, J. et al. Gradually decreasing starch branching enzyme expression is responsible for the formation of heterogeneous starch granules. Plant Physiol.176, 582–595 (2018).
Millan-Testa, C. E. et al. Determination of the molecular and structural characteristics of okenia, mango, and banana starches. J. Agric. Food Chem.53, 495–501 (2005).
Nardozza, S. et al. Metabolic analysis of kiwifruit (Actinidia deliciosa) berries from extreme genotypes reveals hallmarks for fruit starch metabolism. J. Exp. Bot.64, 5049–5063 (2013).
D’Hont, A. et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature488, 213–217 (2012).
Miao, H. et al. Soluble starch synthase III-1 in amylopectin metabolism of banana fruit: characterization, expression, enzyme activity, and functional analyses. Front. Plant Sci.8, 454 (2017).
Wang, Z. et al. Musa balbisiana genome reveals subgenome evolution and functional divergence. Nat. Plants5, 810–821 (2019).
Myers, A. M. et al. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol.122, 989–997 (2000).
Kim, K. N. & Guiltinan, M. J. Identification of cis-acting elements important for expression of the starch-branching enzyme I gene in maize endosperm. Plant Physiol.121, 225–236 (1999).
Li, C. & Gilbert, R. G. Progress in controlling starch structure by modifying starch-branching enzymes. Planta243, 13–22 (2016).
Pan, T. et al. Long branch-chains of amylopectin with B-type crystallinity in rice seed with inhibition of starch branching enzyme I and IIb resist in situ degradation and inhibit plant growth during seedling development: Degradation of rice starch with inhibition of SBEI/IIb during seedling development. BMC Plant Biol.18, 9 (2018).
Han, Y. et al. A gene encoding starch branching enzyme I (SBEI) in apple (Malus × domestica, Rosaceae) and its phylogenetic relationship to Sbe genes from other angiosperms. Mol. Phylogenet. Evol.43, 852–863 (2007).
Tetlow, I. J. & Emes, M. J. A review of starch-branching enzymes and their role in amylopectin biosynthesis. IUBMB Life66, 546–558 (2014).
Pei, J. et al. Phylogeny and expression pattern of starch branching enzyme family genes in cassava (Manihot esculenta Crantz) under diverse environments. Mol. Cell. Biochem.406, 273–284 (2015).
Tyagi, R. et al. Transcriptome wide identification and characterization of starch branching enzyme in finger millet. Bioinformation13, 179–184 (2017).
Burton, R. A. et al. Starch branching enzymes belonging to distinct enzyme families are differentially expressed during pea embryo development. Plant J.7, 3–15 (1995).
Dumez, S. et al. Mutants of Arabidopsis lacking starch branching enzyme II substitute plastidial starch synthesis by cytoplasmic maltose accumulation. Plant Cell18, 2694–2709 (2006).
Kang, G. et al. Molecular cloning and expression analysis of the starch-branching enzyme III gene from common wheat (Triticum aestivum). Biochem. Genet.51, 377–386 (2013).
Xia, H. et al. Deficiency of maize starch-branching enzyme I results in altered starch fine structure, decreased digestibility and reduced coleoptile growth during germination. BMC Plant Biol.11, 95 (2011).
Chen, L. et al. Identification and expression analysis of starch branching enzymes involved in starch synthesis during the development of chestnut (Castanea mollissima Blume) cotyledons. PLoS ONE12, e0177792 (2017).
Mizuno, K. et al. Characterization of an isoform of rice starch branching enzyme, RBE4, in developing seeds. Plant Cell Physiol.42, 349–357 (2001).
Hamada, S. et al. Two starch-branching-enzyme isoforms occur in different fractions of developing seeds of kidney bean. Biochem. J.359, 23–34 (2001).
Lee, Y. et al. Sugary endosperm is modulated by starch branching enzyme IIa in rice (Oryza sativa L.). Rice10, 33 (2017).
Yandeau-Nelson, M. D. et al. Starch-branching enzyme IIa is required for proper diurnal cycling of starch in leaves of maize. Plant Physiol.156, 479–490 (2011).
Zhang, J. et al. Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Plant Mol. Biol.84, 359–369 (2014).
Zhang, Z. et al. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl Acad. Sci. USA113, 10842–10847 (2016).
Wang, J. C. et al. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot.64, 3453–3466 (2013).
Paul, J. Y. et al. Golden bananas in the field: elevated fruit pro-vitamin A from the expression of a single banana transgene. Plant Biotechnol. J.15, 520–532 (2017).
Miao, H. et al. Identification of genes encoding granule-bound starch synthase involved in amylose metabolism in banana fruit. PLoS ONE9, e88077 (2014).
Asif, M. H. et al. Genome-wide identification and expression analysis of the mitogen-activated protein kinase gene family from banana suggest involvement of specific members in different stages of fruit ripening. Func. Integr. Genomics14, 161–175 (2014).
Hu, W. et al. Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci. Rep.6, 30203 (2016).
Ram, H. Y. M., Ram, M. & Steward, F. C. Growth and development of the banana plant: 3. A. The origin of the inflorescence and the development of the flowers: B. The structure and development of the fruit. Ann. Bot.26, 657–673 (1962).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods25, 402–408 (2001).
Liu, Y., Schiff, M. & Dinesh-Kumar, S. P. Virus-induced gene silencing in tomato. Plant J.31, 777–786 (2002).
Wang, F. et al. Use of TRV-mediated VIGS for functional genomics research in citrus. Plant Cell Tiss. Org.139, 609–613 (2019).
Muchui, M. N. et al. Effect of perforated blue polyethylene bunch covers on selected postharvest quality parameters in tissue-cultured bananas (Musa spp.) cv. Williams in Central Kenya. J. Stored Prod. Postharvest Res.1, 29–41 (2010).
Chiang, B. H., Chu, W. C. & Chu, C. L. A pilot scale study for banana starch production. Starch/Stärke39, 5–8 (1987).
Xu, C. et al. A simple method for determining the content of starch-iodine colorimety. Biotechnology8, 41–43 (1998).
Hellens, R. G. et al. Transient expression vectors for functional genomics, qualification of promoter activity and RNA silencing in plants. Plant Methods1, 1–14 (2005).
Wang, Z. et al. Starch accumulation, activities of key enzyme and gene expression in starch synthesis of wheat endosperm with different starch contents. J. Food Sci. Technol.51, 419–429 (2014).
Khoshnoodi, J. et al. The multiple forms of starch-branching enzyme I in Solanum tuberosum. Eur. J. Biochem.242, 148–155 (1996).
Sun, C. et al. Demonstration of in vitro starch branching enzyme activity for a 51/50-kDa polypeptide isolated from developing barley (Hordeum vulgare) caryopses. Physiol. Plant96, 474–483 (1996).
Bhattacharyya, M. K. et al. The wrinkled-seed character of peas described by Mendel is caused by a transposon-like insertion in a gene encoding starch branching enzyme. Cell60, 115–122 (1990).
Boyer, C. D., Daniels, R. R. & Shannon, J. C. Abnormal starch granule formation in Zea mays L. endosperms possessing the amylose-extender mutant. Crop Sci.16, 298–301 (1976).
Regina, A. et al. High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc. Natl Acad. Sci. USA103, 3546–3551 (2006).
Okushima, Y. et al. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell19, 118–130 (2007).
Ori, N. Dissecting the biological functions of ARF and Aux/IAA genes. Plant Cell31, 1210–1211 (2019).
Tang, Y. et al. Transcriptomics analysis of hulless barley during grain development with a focus on starch biosynthesis. Funct. Integr. Genomics17, 107–117 (2017).
Xiao, Q. et al. ZmMYB14 is an important transcription factor involved in the regulation of the activity of the ZmBT1 promoter in starch biosynthesis in maize. FEBS J.284, 3079–3099 (2017).
Acknowledgements
The work was sponsored by the National Key R&D Program of China (No. 2018YFD1000200 and 2019YFD1000200), the Modern Agro-industry Technology Research System of China (No. CARS-31), the Central Public-interest Scientific Institution Basal Research Fund (Nos. 1630052020002, 1630052017010, and 1630052016006), and the National Natural Science Foundation of China (NSFC, No. 31401843).
Author information
Authors and Affiliations
Contributions
H.M., B.X., and Z.J. conceived and designed the experiments. H.M., P.S., Q.L., and D.Z. performed the experiments, analyzed the data, wrote the paper, and prepared figures and/or tables. J.L., C.J., D.Z., H.M., B.X., and Z.J. contributed reagents/materials/analysis tools and reviewed drafts of the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miao, H., Sun, P., Liu, Q. et al. Molecular identification of the key starch branching enzyme-encoding gene SBE2.3 and its interacting transcription factors in banana fruits. Hortic Res 7, 101 (2020). https://doi.org/10.1038/s41438-020-0325-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-0325-1
This article is cited by
-
Starch branching enzymes as putative determinants of postharvest quality in horticultural crops
BMC Plant Biology (2021)
-
Transient virus-induced gene silencing of MaBAM9b efficiently suppressed starch degradation during postharvest banana fruit ripening
Plant Biotechnology Reports (2021)