Abstract
Anthocyanins are important secondary metabolites in plants, but information on anthocyanin biosynthesis mechanisms in Chinese cabbage is limited. The new purple head Chinese cabbage cultivar 11S91 was analyzed, and an R2R3-MYB regulatory gene BrMYB2, located on chromosome A07, controlling the dominant purple-head trait was isolated. High expression of BrMYB2 generated a large accumulation of anthocyanins in 11S91, accompanied by highly upregulated BrTT8, BrF3′H, BrDFR1, BrANS1, BrUGTs, BrATs, and BrGSTs. 11S91 inherited the purple locus from purple trait donor 95T2-5, and they shared consensus CDSs and gDNAs with those of BrMYB2 (cBrMYB2 and gBrMYB2). Two SNPs in cBrMYB2 in 11S91 did not cause loss of function; in addition to several SNPs at both ends of intron 1, a large deletion had occurred in intron 1 of gBrMYB2 in 11S91. Genetic transformation of Arabidopsis showed that gBrMYB2 overexpression lines presented deeper purple color and higher expression than did the cBrMYB2 and cBrmyb2 lines, whereas gBrmyb2 with a long intron 1 did not cause the purple phenotype. We first show that BrMYB2 promotes anthocyanin biosynthesis under the control of the short intron 1 of gBrMYB2 in purple head Chinese cabbage, and gBrmyb2 with a long intron 1 represses anthocyanin production in white head Chinese cabbage. This evidence provides a new understanding of anthocyanin biosynthesis and purple germplasm generation in Brassica vegetables.
Similar content being viewed by others
Introduction
Anthocyanins are water-soluble pigments synthesized based on the phenylpropanoid pathway involved in secondary metabolism in plants; anthocyanins not only provide vibrant colors to plants for attracting animal pollinators and for seed dispersal but also provide strong radical-scavenging abilities to protect against biotic and abiotic stresses1,2. Anthocyanin-rich vegetables and fruits are also gaining in popularity because of their health attributes in humans3.
The mechanism of anthocyanin biosynthesis and regulation has been well studied in model plant species such as Arabidopsis, maize, petunia, and snapdragon4,5 and in economically important plant species such as apple, pear, strawberry, and peach6. Investigations of the genetic and molecular mechanisms of anthocyanin biosynthesis in plants have led to substantial scientific breakthroughs: it has been shown that three main steps are involved in these processes6. First, the primary phenylpropanoid metabolic pathway supplies precursor substrates for subsequent flavonoid synthesis, including phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate: CoA ligase (4CL) involved in it1. Second, the early biosynthesis pathway provides precursor substrates for flavonol and anthocyanin synthesis, including chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavanone 3′-hydroxylase (F3′H), and flavonol synthase (FLS) involved in this step7. Third, the late biosynthesis pathway gives rise to the biosynthesis and modification of anthocyanins via processing of dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), UDP-glucosyltransferase (UGT), and acyltransferase (AT)4. Competition between FLS and DFR produces either flavonols or anthocyanins in the following processes, respectively1.
Functional R2R3-MYB transcription factors (such as MYB11, MYB12, and MYB111) usually participate in the direct activation of early biosynthesis genes (EBGs) such as CHS, CHI, F3H, F3′H, and FLS, whereas F3′H and late biosynthesis genes (LBGs) such as DFR, ANS, UGTs, and ATs are activated by a MYB-bHLH-WD40 ternary complex (MBW), which is formed by an R2R3-MYB factor, a bHLH factor, and a WD40-repeat factor8. These regulators or complexes are able to bind to the promoters of anthocyanin biosynthesis genes (ABGs) and activate their expression; the regulatory network of flavonoid biosynthesis usually functions as a positive feedback mechanism with the participation and interaction of both positive regulators and negative regulators, governing the accumulation of and reduction in anthocyanin or proanthocyanidin5. In Arabidopsis, the following positive regulators mainly participate in the formation of the MBW complex: the R2R3-MYBs AtPAP1, AtPAP2, AtMYB113, AtMYB114, and AtTT2; the bHLHs AtTT8, AtEGL3, and AtGL3; and the WD40 factor AtTTG14. Overexpression of AtPAP1, AtPAP2, AtMYB113, and AtMYB114 leads to a large accumulation of anthocyanins in a TTG1- and bHLH- cooperative way8; the MBW complex formed by AtTT2, AtTT8, and AtTTG1 stimulates proanthocyanidin production in Arabidopsis seeds9. Moreover, AtTT8, AtGL3, and AtEGL3 show redundant functions during regulation5. Negative regulatory factors contain R3-MYBs AtMYBL2 and AtCPC and lateral organ boundary domain (LBD) factors AtLBD37, AtLBD38, and AtLBD39; repression processes of AtMYBL2 and AtCPC in anthocyanin biosynthesis include inhibition of ABGs activation and suppression of MBW complex activity, and similar regulatory mechanisms associated with AtLBD37, AtLBD38, and AtLBD39 have been reported10. Usually, activation of positive R2R3-MYB factors results in anthocyanin accumulation in plants, and their functional loss will lead to color loss in plants6. For example, loss of function of the MYB promoter occurs by a retrotransposon insert in grape11 and by methylation in pear12. Similarly, purple cabbage is created by either substitution of the promoter or deletion of BoMYBL2-113. In addition, insertion of DNA transposons into bHLH2 leads to a dramatic decrease in red pigments and pale flowers in morning glory14. These findings suggest that the diversity of mutations in anthocyanin biosynthesis is different and nondirective in plants.
Research on anthocyanins has mainly focused on mapping, component identification, and identifying expression patterns of ABGs in natural mutant Brassica plants. Mapping has shown that the purple trait is a dominant trait; however, the loci differ across various Brassica species. The purple trait in purple cauliflower (Brassica oleracea L. var. botrytis) is controlled by BoMYB215,16; red cabbage (B. oleracea L. var. capitata), BoMYB2 or BoMYBL2.113,17,18; zicaitai (B. rapa L. ssp. chinensis var. purpurea), BrbHLH49, BrEGL3.2 and BrMYBL2.119,20,21; purple bok choy (B. rapa L. ssp. chinensis), BrMYB7322; purple tumorous stem mustard (Brassica juncea var. tumida Tsen et Lee), BjTT823; purple Chinese cabbage (created by interspecific hybridization), the R2R3-MYB gene c3563g1i2 from the B genome of Brassica7; purple Kohlrabi (B. oleracea var. gongylodes L.), BoPAP2 and BoTT824,25; and purple B. juncea, BjP1126. The main anthocyanins in Brassica plants are highly glycosylated and acylated cyanidins with high stability under acidic and low-temperature conditions27. The main mechanism of ABGs activated by the MBW complex during anthocyanin biosynthesis in Brassica crops is believed to be similar across the aforementioned plant species6. Thus, information on new locus mining, component identification, and regulatory mechanism prediction will facilitate the discovery of new genes and the understanding of anthocyanin biosynthesis in Brassica species. Chinese cabbage (B. rapa L. ssp. pekinensis) is an important vegetable with white, yellow, orange or green heads and is widely cultivated in Asian countries; however, anthocyanin-rich Chinese cabbage is important due to the absence of novel natural mutants. Hence, purple head Chinese cabbage is usually bred from other purple varieties and species. For example, a purple head Chinese cabbage generated from interspecific hybridization between Chinese cabbage (2n = AA = 20) and red leaf mustard (B. juncea, 2n = AABB = 36) had the purple gene on chromosome A0228, which was different from our previous predictions27,29. Therefore, exploration of different purple head Chinese cabbage lines with various anthocyanin accumulation mechanisms is important.
In this study, the new purple head Chinese cabbage bred by the intersubspecific hybridization between white head Chinese cabbage (2n = AA = 20) and purple flowering Chinese cabbage (2n = AA = 20) provides an excellent opportunity to explore the molecular mechanism of anthocyanin biosynthesis. We first performed fine mapping and homology gene screening to determine the candidate gene of purple head Chinese cabbage and verified it using genetic transformation and complementary hybridization approaches. The goal was to gain new insight into the anthocyanin biosynthesis mechanism and provide a theoretical basis for the future breeding of purple Brassica plants.
Results
Genetic and physical mapping of BrPur
Head color investigations showed that the segregation ratio of purple individuals to nonpurple individuals in the F2 population was consistent with the ratio of 3:129, and two other F2 populations also yielded the same results, which together indicated that the purple-head trait was controlled by a single dominant gene. Two markers, CL-12 and B214-87, were closely linked to BrPur, with genetic distances of 3.1 cM and 3.5 cM (physical distance of 577.235 kb), respectively29. Additional molecular markers were designed and tested, and fine genetic mapping showed that two markers, QJ-46 and SSR14-36, were closely linked to BrPur, with genetic distances of 0.1 cM and 0.6 cM (physical distance of 47.91 kb), respectively (Fig. 1f). Four predicted genes existed within a 10.93 kb region between markers QJ-46 and LY-2 on chromosome A07 in B. rapa genome, namely, Bra004161, Bra004162, Bra004163, and Bra004164 (Fig. 1g). Among them, Bra004162 encoded an R2R3-MYB transcription factor that is highly homologous to AtPAP1 and AtPAP2, which are involved in the formation of the MBW complex in Arabidopsis6. In combination with recent reports that expression of BrMYB2 (Bra004162) was significantly positively correlated with total anthocyanin content in purple head Chinese cabbage10,27, our results suggest that BrMYB2 is the candidate gene (BrPur) of the purple-head trait.
a 94S17 was the female parent of 11S91, with a white head. b 95T2-5, a purple flowering Chinese cabbage, was the male parent. c 11S91, with stable inheritance of purple head leaves, was bred from the hybridization of 94S17 and 95T2-5 and continuous self-breeding. d 14S162 was the female parent used for creating the mapping population. e Selected individuals of the F2 population formed by crossing 14S162 and 11S91. f Genetic map of BrPur based on fine mapping with markers QJ-46 (0.1 cM) and SSR14-36 (0.6 cM). g Physical map showing the candidate region of the BrPur locus, which was delimited to a 10.93 kb region on chromosome A07, with four predicted genes between markers QJ-46 and LY-2. The scale bar is 10 cm in (a–d)
Sequence analysis of the candidate gene and functional markers of head color
Sequence analysis showed that BrMYB2 of 95T2-5 and 11S91 had the same coding DNA sequences (CDSs) and genomic DNA (gDNA) sequences, and their CDSs were similar to those of Bra004162 of Chifu (Figs. S1–2). The BrPur CDS was designated cBrMYB2 for 11S91 and 95T2-5 and was designated cBrmyb2 (the Brpur CDS) for 94S17. Two SNPs existed at the 225th bp and 485th bp sites and caused two amino acid substitutions, methionine to isoleucine and aspartic acid to glycine from 94S17 to 11S91 at the 74th and 162nd sites, respectively (Figs. 2b, S2 and S3). Both BrMYB2 and Brmyb2 contained an intact open reading frame and coded a deduced protein comprising 247 amino acids (Figs. 2b, S2 and S3). The MYB protein contained two conserved SANT domains (the important functional areas), namely, R2- and R3-MYB repeats (Fig. 2b). Both gBrMYB2 and gBrmyb2 consisted of three exons and two introns (Fig. 2a), but the gDNA length was distinctly different; the length of gBrMYB2 in 11S91 and 95T2-5 was only 1665 bp but increased to 5441 bp in 94S17. Furthermore, their gDNAs were extremely different from that of Chifu (Figs. 2a and S2d, e). In addition to several SNPs at both ends of the first intron (intron 1) of gBrmyb2, a large insertion fragment of 3772 bp was found in the middle of the intron 1 in 94S17; however, the GT-AG splicing sites at both ends of the intron 1 were invariant (Figs. 2a and S2d, e). In addition, other phenotypes of Chinese cabbage, including three white-head lines (15S1080, 15S1084, and 13S1147; Fig. S1b–d), an orange-head line 13S93 (Fig. S1e), and four purple-head lines (14S838, Z33, Z190, and Z240; Fig. S1f–i), were analyzed for variation within BrMYB2. These lines showed sequence results identical to those of the abovementioned three lines; for example, the CDSs and gDNAs of BrMYB2 of the purple-head lines 14S838, Z33, Z190, and Z240 were the same as those of 11S91, whereas the CDSs and gDNAs of Brmyb2 of the nonpurple-head lines 15S1080, 15S1084, 13S1147, and 13S93 were the same as those of 94S17 (Fig. S2c–e). These results showed that the sequences of BrMYB2 and Brmyb2 in purple and nonpurple Chinese cabbage, respectively, were extremely conserved but different from those of Chifu.
a Comparison of exons and introns of BrMYB2. b Display of predicted BrMYB2 domains. c Phylogenetic tree of MYBs. The numbers next to the nodes indicate bootstrap values from 1000 trials, and the branches with the same color indicate that those MYBs are classified into groups. The GenBank number or BRAD ID of these proteins is supplied in Table S2
Furthermore, based on sequence differences between gBrMYB2 and gBrmyb2, two codominant markers, BrP1 and BrP2, were designed and used for PCR-based testing of the head phenotype of F2 individuals. Both BrP1 and BrP2 amplified large products (4080 bp, and 4122 bp, respectively) in the nonpurple plants, small products (306 bp and 348 bp, respectively) in the purple-head plants, and two products (4080 bp/306 bp and 4122 bp/348 bp, respectively) in the heterozygous plants (Fig. S4c, d). The polymorphic bands amplified with BrP1 and BrP2 were completely consistent with the head color (Fig. S4c, d), which further indicated that BrMYB2 was the candidate gene controlling purple head color in 11S91. Moreover, the codominant markers BrP1 and BrP2 were useful in molecular marker-assisted breeding of purple head Chinese cabbage. Fifty-one MYBs from different plant species were used to construct a phylogenetic tree (Fig. 2c), which could help understand the relationship of BrMYB2 to reported anthocyanin MYBs. The amino acids of the MYBs were highly conserved and contained a common [D/E]Lx2[R/K]x3Lx6Lx3R motif in the R3 domain for interactions with R/B-like bHLH proteins30, but the region downstream from this motif was divergent in both sequence and length. The MYBs were classified into seven different groups (Fig. 2c), and important signature motifs of each group might affect related anthocyanin biosynthesis. MYBs in Group A mainly contained Cruciferae species such as cabbage, Chinese cabbage, radish, and Arabidopsis, and they had a common KPRPRSFT motif (but it was not present within AtMYB114) (Fig. S5); in addition, these BrMYBs also showed high DNA sequence similarity (Fig. S2b). The MYBs of Solanaceae species in Group B were adjacent to the BrMYBs, and the common motif was [K/R]PRPRTFS, with several amino acid changes (Fig. S6). Group C and Group D mainly contained MYBs from Convolvulaceae and Vitaceae plants, respectively. Notably, the MYBs in these groups mainly belonged to a family whose members might have higher sequence identity in the downstream areas (Fig. 2c). Only MYBs in Group G included different categories of plants, such as MYBs of maize and rice, which are monocots, and AtTT2, AtMYB12, AtMYB111, and AtMYB11 of Arabidopsis, a dicot (Fig. 2c). AtMYB111, AtMYB12, and AtMYB11 participate in the activation of EBGs in Arabidopsis5. These results indicated that MYBs related to anthocyanin production in monocots showed higher sequence similarity to MYBs of Arabidopsis controlling EBGs, whereas MYBs involved in anthocyanin synthesis in dicots from one family had a closer genetic relationship. Thus, BrMYB2 might affect anthocyanin biosynthesis, as it is highly similar to previously reported AtMYBs.
Expression patterns of BrMYB2 and related ABGs in different tissues
Generally, the expression patterns of BrMYB2 had organ and genetic specificity and were tightly correlated with the degree of purple color in different tissues. In 94S17, BrMYB2 was significantly expressed at low levels in all nonpurple tissues, such as the roots, stems, rosette leaves, stem leaves, siliques, and flower buds (Fig. 3v). BrMYB2 was highly expressed in the stems, rosette leaves, stem leaves, siliques, and flower buds in 95T2-5 and 11S91; there was greater BrMYB2 expression and deeper purple color in these tissues in 95T2-5 than in 11S91 (except in the siliques, in which the expression was higher in 11S91) (Fig. 3a–u). In regard to color appearance, all tissues of 94S17; the roots of 95T2-5; and the roots, stem leaves, and rosette leaves of 11S91 showed no purple color (Fig. 3a–u). Moreover, BrMYB2 was expressed little in these tissues (Fig. 3v). The bHLH regulatory gene BrTT8 together with three structural ABGs, BrF3′H, BrDFR, and BrANS, was also investigated. Compared with that in 11S91, the degree of upregulation of BrTT8 in 95T2-5 was not high, but the expression levels of the LBGs BrDFR and BrANS in 95T2-5 in all tissues were much higher than those in 11S91; the EBG BrF3′H showed significantly higher expression only in the siliques in 95T2-5 compared with 11S91 (Fig. 3w–z). Similar to BrMYB2, these genes showed significantly low expression in the roots, stems, rosette leaves, stem leaves, and flower buds of 94S17; however, they were highly expressed in the siliques. Previous studies indicated that TT8, F3′H, DFR, and ANS redundantly participated in proanthocyanidin biosynthesis and brown testa formation31,32. In this study, 94S17, 11S91 and 95T2-5 have brown testa. Taken together, the results show that BrMYB2 controls anthocyanin biosynthesis in different tissues and is tightly correlated with purple color emergence in purple Chinese cabbage. In addition, related ABGs, such as BrTT8, BrF3′H, BrDFR, and BrANS, respond to BrMYB2 and participate in anthocyanin biosynthesis, and they might be redundantly involved in proanthocyanidin biosynthesis during brown testa formation in three Chinese cabbage lines.
a–u Phenotypes of different tissues of three Chinese cabbage lines; the images in each vertical sample column (left to right) were collected from 94S17, 11S91, and 95T2-5. a–c Stalks and roots. d–f Transection of stems. g–i Rosette leaves. j–l Stem leaves. m–o Flowers. p–r Siliques. s–u Transection of siliques. v–z Gene expression patterns. The rosette leaves of 94S17 were treated as controls in the data analysis; the values are presented as the means ± SDs (n = 3). The different letters above each column are significantly different at p < 0.05 according to Duncan’s test. The scale bar is 1.5 cm in (a–c) and (g–t) and 200 μm in (d–f) and (s–u).
Similar to the determination of different tissues, 95T2-5 had the highest extent of total anthocyanin content, ranging from 42.675 mg kg−1 in S4 external leaves to 399.454 mg kg−1 in S1 inner leaves during the mature period, followed by 11S91, whose content ranged from 10.080 mg kg−1 in S4 external leaves to 225.544 mg kg−1 in S1 inner leaves; however, 94S17 accumulated a very low amount of anthocyanins in the head (Fig. 4a–d). To better understand the anthocyanin biosynthesis mechanism and the role of BrMYB2, a total of 84 ABGs, including 21 phenylpropanoid metabolic pathway genes (PMPGs) (BrPALs, BrC4Hs, and Br4CLs), 18 EBGs (BrCHSs, BrCHIs, BrF3Hs, and BrF3′H), 19 LBGs (BrDFRs, BrANSs, BrUGTs, and BrATs), and two transport genes (BrGSTs), as well as 13 positive regulatory genes and 11 negative regulatory genes identified recently in Chinese cabbage10, were tested to explore their expression patterns in mature heads of 11S91 (Fig. 4e and Table S3). In the figure, the columns reflect different samples, the rows display various genes, and the samples with similar gene expression patterns are aggregated on a branch. Group A consists of only heading leaves S1–S4 from 94S17, Group B includes leaves S1–S4 from 95T2-5 and S1 leaves from 11S91, and Group C contains only heading leaves S2–S4 from 11S91 (Fig. 4e). These results further showed that a significant change in the anthocyanin regulatory mechanism occurred between 94S17 and purple-head lines during head maturity, and the purple S1 leaves of 11S91 inherited the characteristic of anthocyanin biosynthesis from the purple trait donor 95T2-5.
a Total anthocyanin content of samples; the values are presented as the means ± SDs (n = 3), and the different letters above each column are significantly different at p < 0.05 according to Duncan’s test. b–d The head of 11S91 was divided into four parts: S1, the interior heading leaves with deep purple color; S2, the interior leaves with light purple color; S3, the exterior heading leaves; S4, the outer functional leaves. The leaf size and positions of the 94S17 and 95T2-5 samples were the same as those of 11S91. e Hierarchical clustering analysis of gene expression patterns. The expression data were log2-normalized, clustered using PermutMatrix software61 and analyzed with the Pearson distance and Ward’s method. The magenta boxes indicate upregulation, and the yellow boxes indicate downregulation; the color brightness is directly proportional to the expression ratio. The first capital letters ‘G’, ‘P,’ and ‘S’ are different leaf tissues of 94S17, 95T2-5, and 11S91, respectively. The relative coefficient ‘r’ with ‘*’ and ‘**’ indicates that gene expression is significantly and highly significantly correlated with total anthocyanin content at the levels of 0.05 and 0.01, respectively
In the rows, the genes were classified into two main groups. Group I contains genes with similar expression levels in both white and purple head Chinese cabbage (Fig. 4e). For example, genes in Group I.1 showed relatively high or similar expression in 94S17 and 11S91 and low expression in 95T2-5. These genes include the PMPGs BrPAL1.1, BrPAL1.2, BrC4H2, Br4CL1.0, and Br4CL2.4; the EBGs BrFLS3.2, BrFLS3.3, and BrFLS4.0; the LBGs BrUGT84A3.1 and BrUGT84A3.2; the positive regulatory genes BrMYB1, BrPAP1, BrTTG1, and BrEGL3.2; and the negative regulatory genes BrMYBL2.2, BrLBD37.1, BrLBD37.2 BrLBD37.3, BrLBD38.1, and BrLBD39.1. Notably, the gene expression in this group was upregulated in most samples but negatively correlated with total anthocyanin content (Fig. 4e). The genes in Group I.2 were downregulated in nearly all samples and negatively correlated with total anthocyanin content. These genes included the PMPGs BrC4H1, BrC4H3, BrC4H4, BrC4H5, Br4CL2.2, and Br4CL4.1; the EBGs BrFLS1.0, BrFLS2.0, and BrFLS3.1; the positive regulator genes BrMYB12.2 and BrTT2; and the negative regulator genes BrMYBL2.1, BrCPC1, and BrCPC2. On the contrary, the genes in Group II mainly showed higher expression in the purple-head lines than in the white-head lines. Note that genes in Group II.1 were extremely highly expressed in purple-head lines and were significantly correlated with total anthocyanin content (except for BrPAL2.1 and BrCHS3). These genes included the PMPGs BrPAL2.3 and BrPAL3.1; the EBGs BrCHS1, BrCHS2, BrCHS4, BrF3H3, and BrF3′H; the LBGs BrDFR1, BrANS1, BrUF3GT2, BrUF5GT, Br5MAT, and Brp-COUT; the transport genes BrGST1 and BrGST2; and the positive regulatory genes BrMYB2 and BrTT8 (Fig. 4e and Table S3). However, the genes in Group II.2 had lower expression in the outer leaves than in the inner leaves or were not significantly correlated with total anthocyanin content. These genes included the PMPGs BrPAL2.2, BrPAL4.0, Br4CL2.1, Br4CL2.3, and Br4CL3.0; the EBGs BrCHS5, BrCHI1, BrCHI2, BrCHI3, and BrF3H1; and the LBGs BrDFR3, BrANS2, BrANS3, BrANS4, BrUF3GT1, BrUGT84A1.2, and BrUGT84A2.2. The genes in Group II.3 displayed low correlations with total anthocyanin content or were expressed at low levels in purple head Chinese cabbage. These genes included the PMPGs BrPAL3.2 and Br4CL4.2; the EBG BrF3H2, the LBGs BrDFR2, BrSAT, BrUGT84A1.1, and BrUGT84A2.1; the positive regulatory genes BrMYB12.1, BrMYB111.1, BrMYB111.2, BrEGL3.1, and BrGL3; and the negative regulatory genes BrLBD38.2 and BrLBD39.3. These results showed that the increased upregulation of BrMYB2 promoted a large accumulation of anthocyanins in purple head Chinese cabbage and in its purple trait donor at the head-formation stage. This was accompanied by high upregulated expression of the bHLH regulatory gene BrTT8; the PMPGs BrPAL2.1, BrPAL2.3, and BrPAL3.1; the EBGs BrCHS1, BrCHS2, BrCHS3, BrCHS4, BrF3H3, and BrF3′H; the LBGs BrDFR1, BrANS1, BrUF3GT2, BrUF5GT, Br5MAT, and Brp-COUT; and the transport genes BrGST1 and BrGST2.
Variations upstream of BrMYB2 have no effects on the function of its promoter
First, we speculated that differences existed in the BrMYB2 promoter and questioned whether these variations might lead to different degrees of expression of BrMYB2. A CT repeat deletion was found upstream from the ATG translation start site of BrMYB2 in 11S91, and four CT repeat deletions were found in the same site in 95T2-5 (Fig. S8). The CT repeat area was a 5′-UTR Py-rich stretch that served as a cis-acting element providing high transcription levels, according to the PlantCARE database (Fig. S7). Although different numbers of CT repeats were constructed and transiently transformed into tobacco leaves, samples treated with different constructs from three Chinese cabbage lines showed identical degrees of blue color (Fig. 5b–l). Moreover, activity analyses of the BrMYB2 promoter, including a series of cis-acting element deletions (such as CAAT-boxes, TATA-boxes, and AS-2-boxes) from −791 to −2270 bp, revealed the same degree of color (Fig. 5b–l). In addition, BrMYB2 promoter activity was not changed when the 5′-UTR Py-rich stretch region was deleted (Fig. 5o); the BrMYB2 promoter activity was lower than the cauliflower mosaic virus 35S (CaMV35S) promoter activity, as the positive control had a deeper blue color (Fig. 5). These results indicated that variations upstream of BrMYB2 may not cause loss of promoter activity and do not affect the expression of BrMYB2.
aGUS constructs containing different lengths of BrMYB2 promoter sequences (based on 11S91). Schematic diagram of constructs utilized for transient transformation of tobacco ‘NC89’ leaves that were 6 cm in size (~28 DAS [days after sowing]). Tobacco leaves were infiltrated with different plasmids from bacteria at a concentration of OD600 = 0.530 and cultured for 72 h under a 12-h light/12-h dark photoperiod at 23 °C (125 mmol m–2 s–1). The leaves were imaged after GUS solution staining and alcohol decolonization. b, f, j Constructs with a 2270 bp long BrMYB2 promoter (based on that of 11S91). c, g, k Constructs with a 2008 bp long BrMYB2 promoter (based on that of 11S91). d, h, l Constructs with a 791 bp long BrMYB2 promoter (based on that of 11S91). n Construct with 642 bp long BrMYB2 promoter. o Construct with a 498 bp long BrMYB2 promoter. e Negative control: noninfiltrated leaves. i Positive control: leaves infiltrated with the CaMV35S:GUS construct. m Negative control: leaves infiltrated with the GUS construct without CaMV35S promoter. The data as shown are representative of three independent repeats (n = 3). The scale bar is 2.5 cm
Two SNPs do not lead to loss of function of BrMYB2
In addition to promoter variations, two SNPs in cBrMYB2 were present and conserved between white-head lines and purple-head lines (Fig. S2c). To test whether these SNPs were responsible for the purple-head phenotype, different introductions of cBrMYB and cBrmyb2 into Arabidopsis were performed using genetic transformation. A total of 62 T1 CaMV35S:cBrMYB2 lines and 56 T1 CaMV35S:cBrmyb2 lines were obtained after PCR detection (Fig. S9). Seven cBrMYB2 lines (Line 02, Line 06, Line 14, Line 27, Line 32, Line 46, and Line 49) and four cBrmyb2 lines (Line 01, Line 02, Line 05, and Line 06) with the best purple-color phenotype were identified and selected for generating T3 homozygous plants (Fig. S10). Compared with wild type (WT) Arabidopsis, both transgenic cBrMYB2 and cBrmyb2 lines showed distinct purple premature seeds and purple vascular bundles of the leaves, roots, and stalks (Fig. 6).
a–d, m–p WT Arabidopsis; e–h, q–t T3 CaMV35S:cBrMYB2 lines; i–l, u–x T3 CaMV35S:cBrmyb2 lines. a, e, i Twelve-day-old seedlings. b, f, j Inner leaves close to the growing point. c, g, k Rosette leaf petioles. d, h, l Rosette leaf stalk bases. m, q, u Roots. n, r, v Cross-sections of stalks. o, s, w Seeds at approximately 7 days after pollination. p, t, x Siliques at approximately 7 days after pollination. The scale bars are 200 μm
The expression of BrMYB2 in Line 02, Line 14, Line 27, Line 32, Line 46, and Line 49 was 1.5-4 times higher than that in Line 06 (Fig. S11i); however, no BrMYB2 expression was detected in WT Arabidopsis in either quantitative real-time PCR (qRT-PCR) or semi-qRT-PCR analyses. Accompanied by the high upregulation of AtTT8, EBGs such as AtCHI, AtCHS, AtF3H, and AtF3′H and LBGs such as AtDFR, AtANS, AtUF3GT1, AtUF3GT2, AtUF5GT, At5MAT, and AtGST showed higher expression in the transgenic lines than in the WT (Fig. S11j–x). AtTT2, AtEGL3, and AtTTG1 presented relatively high expression in the transgenic lines (Fig. S11). These results indicated that both cBrmyb2 and cBrMYB2 can activate the anthocyanin biosynthesis pathway and that silent mutations caused by two SNPs in cBrMYB2 cannot cause loss of function.
The short intron 1 with a large deletion in BrMYB2 promotes high transcription, which is responsible for the purple-head phenotype
We questioned whether the different intron 1 of BrMYB2 was responsible for the different phenotypes of anthocyanin accumulation. A total of 46 T1 CaMV35S:gBrMYB2 lines and 59 T1 CaMV35S:gBrmyb2 lines were obtained after PCR detection after Arabidopsis genetic transformation (Fig. S12). Ten gBrMYB2 lines with the best purple coloration were selected for generating T3 homozygous plants; however, the phenotype of all the gBrmyb2 lines was the same as that of WT Arabidopsis (Fig. S13). On the contrary, only the gBrMYB2 lines showed a deeper purple color in the roots, flowers, siliques, rosette leaves, stalks, inflorescences, premature seeds, and seedlings older than 12 days (Fig. 7). Interestingly, the degrees of purple color of these tissues were much deeper than that of the tissues of the cBrMYB2 and cBrmyb2 lines (Figs. 6 and 7). Thus, only gBrMYB2 with a short intron 1 promotes purple color formation and produces a purple phenotype, but gBrmyb2 with a long intron 1 causes purple color deficiency and no purple phenotype.
a–d, m–p WT Arabidopsis; e–h, q–t T3 CaMV35S:gBrmyb2 lines; i–l, u–x T3 CaMV35S:gBrMYB2 lines. a, e, i Twelve-day-old seedlings. b, f, j Inner leaves close to the growing point. c, g, k Inflorescences. d, h, l Flowers. m, q, u Roots. n, r, v Rosette leaf petioles. o, s, w Seeds at approximately 7 days after pollination. p, t, x Siliques at approximately 7 days after pollination. The scale bars are 200 μm
Moreover, we measured the total anthocyanin content and related gene expression in all the transgenic plants. Both the gBrmyb2 lines and WT Arabidopsis showed a nonpurple phenotype in their aboveground parts, which presented extremely low anthocyanin contents (Fig. 8a–g); the phenotypes of the aboveground parts of the cBrMYB2 and cBrmyb2 plants were identical and presented relatively high contents of anthocyanins (Fig. 8a–g). Notably, the whole leaves of gBrMYB2 plants containing intron 1 had a deep purple color and the highest anthocyanin content, followed by those of the cBrMYB2 and cBrmyb2 lines, the gBrmyb2 lines and WT Arabidopsis (Fig. 8a–g).
a–e Thirty-day-old Arabidopsis seedlings. a WT Arabidopsis; b T3 CaMV35S:cBrMYB2 lines; c T3 CaMV35S:gBrMYB2 lines; d T3 CaMV35S:cBrmyb2 lines; e T3 CaMV35S:gBrmyb2 lines. f Semi-qRT-PCR analysis of full-length BrMYB2. g Total anthocyanin contents of samples. h qRT-PCR analysis of BrMYB2, and Line 01 of the T3gBrmyb2 lines served as the control. i–r Expression patterns of ABGs in Arabidopsis, and the WT line served as the control. The values are presented as the means ± SDs (n = 3). The different letters above each column are significantly different at p < 0.05 according to Duncan’s test. The first ‘c’ and ‘g’ letters of the abscissa labels represent cDNA and gDNA, respectively; the second letters ‘M’ and ‘m’ represent BrMYB2 and Brmyb2, respectively
Furthermore, the expression patterns of BrMYB2 and other ABGs were measured in transgenic lines. BrMYB2 was expressed little in gBrmyb2 plants but highly expressed in cBrMYB2, cBrmyb2, and gBrMYB2 plants (Fig. 8f–h); the expression of BrMYB2 was consistent with the total anthocyanin content, and the gBrMYB2 lines had the highest expression of BrMYB2, followed by the cBrMYB2 and cBrmyb2 lines, the gBrmyb2 lines and WT Arabidopsis (Fig. 8f–h). In addition, the reverse transcription results showed that gBrMYB2 with short intron 1 had undergone correct intron splicing and produced the same transcript as that of the cBrMYB2 lines; however, Brmyb2 in the gBrmyb2 lines with long intron 1 was expressed at a low level (Figs. 8f and S14). Moreover, ABGs in gBrmyb2 lines were not activated, which indicated that the function of gBrmyb2 was totally lost (Fig. 8i–r); the expression of ABGs in cBrMYB2, cBrmyb2, and gBrMYB2 plants was also different. For example, the expression levels of the several EBGs (i.e., AtCHI, AtCHS, and AtF3H), the LBG AtUG3GT1, and the regulatory gene AtTTG1 were similar in cBrMYB2 and cBrmyb2 plants, but their expression levels were lower in gBrMYB2 plants with intron 1 (Fig. 8h–r). In contrast, the expression patterns of ABGs directly responsible for downstream anthocyanin biosynthesis were similar to those of BrMYB2, with higher expression in gBrMYB2 plants with intron 1 than in cBrMYB2 and cBrmyb2 plants (Fig. 8h–q). These ABGs included the EBG AtF3′H; LBGs AtDFR and AtANS; transport gene AtGST; and regulatory gene AtTT8 (Fig. 8h–q). Generally, cBrMYB2 and cBrmyb2 transgenic plants with moderate expression levels of BrMYB2 presented higher upregulation of EBGs than did the gBrMYB2 plants; gBrMYB2 plants with the highest expression of BrMYB2 mainly exhibited higher upregulation of the LBGs, AtTT8, and AtGST. Similar to the results of the mature Chinese cabbage heads, these ABGs may be directly controlled by BrMYB2. Hence, these results further illustrated that intron 1 of BrMYB2 plays an important role in affecting anthocyanin biosynthesis and ABG regulation. Though gBrmyb2 plants were under the control of the CaMV35S promoter, the long intron 1 with a large insertion in gBrmyb2 still completely inhibited anthocyanin biosynthesis; the short intron 1 with a large deletion of gBrMYB2 highly promoted anthocyanin production. As a result, gBrmyb2 fully lost its function to activate anthocyanin biosynthesis.
Discussion
The new BrPur locus in Brassica
The purple trait of Brassica vegetables has become a popular research topic in recent years. Purple head Chinese cabbage not only has health benefits but also provides an important agronomic trait for germplasm resources. Interestingly, there are different loci that reportedly control the purple trait of Brassica species19,20,21,22,28,33,34,35. Recent investigations of an F2 population derived from the hybridization of zicaitai and common caixin (B. rapa L. ssp. parachinensis) demonstrated that two candidate genes were speculated to control anthocyanin accumulation in zicaitai, including the bHLH gene BrEGL3.2 (positive) and the R3-MYB gene BrMYBL2.1 (negative) located on chromosomes A09 and A07, respectively19,20. Similar to a rapid-cycling B. rapa, a recessive locus affording no anthocyanins was mapped to chromosome A09 without candidate gene prediction36. However, the mapping of an F2 population generated by green Chinese cabbage and purple bok choy showed that the dominant gene Pur was located at the end of chromosome A0333, and similar results were found in purple nonheading Chinese cabbage and in another B. rapa line22,35. In purple turnip (B. rapa cv. Iyo-hikabu), anthocyanin coloration was shown to be controlled by a single dominant gene on chromosome A07 based on a doubled-haploid population generated by crossing purple turnip and Chinese cabbage, although the responsible gene was not verified37. Another report speculated that BrbHLH49 on chromosome A07 might positively regulate anthocyanin accumulation in zicaitai21. A purple Chinese cabbage created from the hybridization between Chinese cabbage and red leaf mustard revealed that the purple gene was translocated onto chromosome A0228. Though these purple Brassica plants might have similar phenotypes or anthocyanin-rich tissues, their different dominant BrPur loci mainly depend on the genetic background of the purple trait donors, at least in Brassica species. In our work, the first reported R2R3-MYB gene, BrMYB2 (BrPur), located on chromosome A07 of B. rapa, was the key gene responsible for anthocyanin accumulation in heading leaves, stalks, and siliques of purple head Chinese cabbage; moreover, it is a novel, unpublished locus that differs from all reported loci in purple Brassica crops.
Upregulation of BrMYB2 promotes high expression of ABGs to generate purple traits
AtMYB113, AtMYB114, AtPAP1, and AtPAP2 activate anthocyanin biosynthesis in Arabidopsis, which have three orthologs in B. rapa, namely, Bra004162, Bra039763, and Bra001917, located on chromosomes A07, A02, and A03, respectively8. Recent studies of purple head Chinese cabbage revealed that total anthocyanin content is associated with high expression of only BrMYB2 (Bra004162) among these three BrMYBs, accompanied by upregulation of BrTT8 and a series of ABGs, including BrF3’H, BrDFR, BrANS, BrUGTs, and BrGSTs10,27. Recent advances have shown that MYBs activate bHLHs and ABGs through strong interactions with bHLHs to promote anthocyanin biosynthesis in crop plants38,39,40. In purple cauliflower and red cabbage, BoMYB2 is responsible for the purple trait by interacting with BoHLHs and related ABGs15,16,17, and homologous comparisons showed that BrMYB2 is highly homologous with BoMYB2 (Fig. 2c). High upregulation of TT8, PALs, F3′H, DFR, ANS, UGTs, and GSTs in both Arabidopsis transformants and Chinese cabbage strongly indicates that BrMYB2 may cooperate with AtTT8 or BrTT8 to regulate anthocyanin biosynthesis in Arabidopsis and Chinese cabbage, respectively. In addition, BrMYB2 also showed higher expression in young, tender purple tissues such as inner leaves or tissues close to the growing point (flowering buds and siliques); in addition, BrTT8, BrF3′H, BrDFR, and BrANS might function redundantly in proanthocyanidin biosynthesis and brown testa formation in Chinese cabbage, as shown in a previous study31,32.
Expression of BrMYB2 was not affected by mutations in its upstream promoter
Upregulation of MYBs has been considered to be the key factor in anthocyanin production in natural variants or artificially bred plants4, and the different transcription levels of these MYBs are usually caused by their promoters. For example, in apple, a rearrangement in the upstream regulatory region of MdMYB10 is responsible for increased anthocyanin accumulation and for producing a striking phenotype involving red foliage and fruit flesh41. When the PcMYB10 promoters were compared between red-skinned pear and green-skinned pear, a higher methylation level of the PcMYB10 promoter caused lower expression of PcMYB10, and anthocyanin accumulation in the green-skinned pear was tightly associated with the methylation level of the PcMYB10 promoter12. In addition, genetic variation in a harbinger DNA transposon insertion upstream of BoMYB2 crucially affects the upregulation of BoMYB2 to induce a purple phenotype of cauliflower15, whereas a retrotransposon-induced mutation in the 5′-flanking region of VvmybA1 is associated with the loss of anthocyanins in white grape42. Recently, deletion in the upstream promoter of BoMYBL2-1 at the 347 bp site resulted in purple cabbage13. The extremely close relationship among species of Brassica vegetables may provide compatibility between the MBW regulatory machinery and their binding sites in promoters7. Hence, the promoter activity of candidate genes is critical to the novel purple trait, and potential discrepancies may be distributed in cis-regulatory regions or even epigenetic marks12,15. However, unlike the above reports on purple color formation, our results appear to present a different mechanism responsible for the increased expression of BrMYB2 during anthocyanin biosynthesis. Although variations in the 5′-UTR Py-rich stretch existed in the BrMYB2 promoter, its activity remained unchanged. Hence, the differential expression of BrMYB2 is not caused by promoter differences.
Mutations in cBrMYB2 cannot lose gene function
Natural gene mutations are not targeted, and sometimes these nonorientation mutations may produce different gene functions; the final results of mutations include missense mutations, synonymous mutations, silent mutations, frameshift mutations, and nonsense mutations43. For example, mutation of a single proline to leucine substitution in BnRGA led to a semidwarf mutant phenotype of B. napus44. A nonsynonymous SNP in the third exon of Brnym1 produced an amino acid substitution in the highly conserved domain of magnesium dechelatase, which generated a stay-green phenotype of Chinese cabbage45. Similarly, variations in amino acids caused by SNPs in Bror did not affect its gene function, whereas an insertion in Bror caused early termination of translation and loss of gene function in orange head Chinese cabbage46. Coding regions of BrMYB2 in 11S91 and 94S17 shared 99.19% sequence identity; two SNPs were found and were conserved. However, the gene functions of cBrMYB2 and cBrmyb2 were nearly identical because their transformants both produced a purple phenotype. Thus, silent mutations of base substitutions in BrPur are not the key factor in purple head formation.
Intron 1 of gBrMYB2 controls the transcription of BrMYB2 and anthocyanin biosynthesis
Introns regulate gene expression in eukaryotes in various ways, such as positive, negative, and intron-mediated manners47. The regulatory sites of most important genes are not situated in the promoter but rather are located within introns, and genes regulated by introns are often highly expressed in most tissues48. Even when the promoters have been deleted, some introns located several hundred nucleotides downstream of the transcription start site still strongly stimulate mRNA accumulation48. For instance, a 90 bp sequence in intron 1 of AtPAP1 is essential for sucrose responses49; intron 1 of Ostua is the key regulatory element of high accumulation of rice tubulin50; similarly, high expression of OsTubA1 requires intron 1, and OsTubA1 cannot be highly expressed when intron 1 exists in the untranslated region51. The BnFAD2-C5 intron provides an enhancement function as a promoter, and the intron-mediated enhancement regions are mainly concentrated in the +631 bp to +1033 bp site52. Similarly, the 5′-UTR intron of AtVTC1 acts as an enhancer for Vc biosynthesis in Arabidopsis53. A recent study further supported that introns promoted expression from all transcribed positions but had no effect when they were located upstream of the most 5′ transcription start site48. In our research, introduction of gBrMYB2 with the short intron 1 and normal intron 2 showed a deep purple phenotype for all tissues of transgenic Arabidopsis, and their degrees of coloration were much deeper than those of cBrMYB2 and cBrmyb2 transgenic plants without related introns. In contrast, introduction of gBrmyb2 with the long intron 1 and normal intron 2 resulted in phenotypes identical to those of WT Arabidopsis. Moreover, the short intron 1 enhanced the high transcription of BrMYB2 in transgenic gBrMYB2 lines.
In addition, introns have a repressor function for gene self-regulation. For example, a reverse repetitive sequence in intron 1 of the human collagen α1(I) gene inhibits its transcription only when the intron 3′ region is located downstream of α1(I)54. Mutant rice plants harboring a T-DNA insertion in intron 1 of OsCAO1 were deficient in chlorophyll b and produced pale-green leaves55. Without changes in the GT-AG splice site at the both ends of intron 1, Brmyb2 with a long fragment insertion in the middle of intron 1 was completely repressed, which led to failure to produce a purple phenotype. In addition, positive genetic methods and reverse genetic methods are usually used for gene functional verification; in addition to transgenic techniques, mutant complementary hybridization methods are usually and traditionally used in positive genetic research of crop species. In our subsequent research, a series of purple head Chinese cabbage lines were acquired by crossing purple head Chinese cabbage with other nonpurple head Chinese cabbage lines, followed by subsequent selection. The BrPur/Brpur loci that gave rise to purple color were conserved and tightly cosegragated with head color in these Chinese cabbage lines, and both Chinese cabbage lines produced functional transcripts of BrMYB2/Brmyb2 (Figs. S1 and S2).
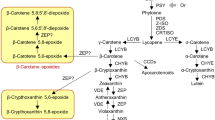
In conclusion, BrMYB2 is the key regulatory gene governing the purple-head trait in Chinese cabbage, and a short intron 1 of this novel gene promotes high expression of BrMYB2 in purple head Chinese cabbage, which activates anthocyanin biosynthesis through substantial upregulation of BrTT8. This process is accompanied by upregulation of the PMPGs BrPAL2.1, BrPAL2.3, and BrPAL3.1; the EBGs BrCHS1, BrCHS2, BrCHS3, BrCHS4, BrF3H3, and BrF3’H; the LBGs BrDFR1, BrANS1, BrUF3GT2, BrUF5GT, Br5MAT, and Brp-COUT; and the transport genes BrGST1 and BrGST2 (Fig. 9). In contrast, the large intron 1 of Brmyb2 completely inhibits its transcription in white head Chinese cabbage, resulting in no activation of the anthocyanin biosynthesis pathway. These findings provide new evidence to understand the mechanism of anthocyanin biosynthesis and lay a good foundation for purple germplasm use in Brassica crops. In the future, we will focus on the regulation of BrMYB2 together with interactions of other anthocyanin biosynthesis factors in purple head Chinese cabbage.
gBrMYB2 with short intron 1 controls anthocyanin biosynthesis and accumulation through coexpression of BrTT8 and activation of a series of structural ABGs, such as BrF3’H, BrDFR1, BrANS1, BrUGTs, BrATs, and BrGSTs, resulting in a purple-head phenotype. Similarly, gBrmyb2 of common white head Chinese cabbage with long intron 1 completely represses anthocyanin biosynthesis by restricting the expression of Brmyb2 and ABGs, resulting in a nonpurple-head trait
Materials and methods
Plant materials and growth conditions
The inbred line 94S17 with white heads and brown testa served as the female parent (Fig. 1a), and the flowering Chinese cabbage 95T2-5 inbred line served as the male parent (purple trait donor, Fig. 1b), which has deep purple phenotype and brown testa. Chinese cabbage 11S91 with a purple head (Fig. 1c) was bred from F1 plants of 94S17 and 95T2-5 with continuous self-crossing for ten generations from select individual plants. An F2 population of head Chinese cabbage generated by crossing 11S91 and 14S162 (Fig. 1d, a yellow head Chinese cabbage served as the female parent) was used for fine mapping (Fig. 1e). In addition, different phenotypes of Chinese cabbage containing three white-head lines (15S1080, 15S1084, and 13S1147; Fig. S1b–d), an orange-head line (13S93, Fig. S1e), and four purple-head lines (14S838, Z33, Z190, and Z240; Fig. S1f–i) were used for the variation analysis of candidate genes. Z33, Z190, and Z240 were F2 purple head Chinese cabbage lines bred from a 14S838 female parent and 15S1080 male parent. All the materials were grown outdoors in autumn in Yangling, Shaanxi Province, China. The head color of individual F2 plants was visually determined at the mature-head stage. In addition, samples were harvested at the mature-head stage (about 4 months after sowing), frozen in liquid nitrogen, and immediately stored in a −80 °C freezer (Sanyo, Japan). Each sample was analyzed in triplicate, and three biological replicates were tested.
The WT Arabidopsis Columbia and tobacco line NC89 were used for genetic transformation. Arabidopsis cultivation was performed as previously described, with modifications56. Briefly, seeds were surface sterilized using 75% alcohol and disinfection solution (consisting of aqueous sodium hypochlorite solution and sterile distilled water at a final volumetric ratio of 1:10), washed using sterile distilled water and sown on Murashige and Skoog (MS) media (2% sucrose, 0.8% agar, and a pH of ~5.5). To unify germination and break dormancy, the seed plates were incubated for 2.5 days in the dark at 4 °C and then transferred into a culture room under a 12-h light/12-h dark photoperiod (125 mmol m–2 s–1) at 23 °C for 12 days. Similarly, tobacco and Chinese cabbage seeds were planted directly after soaking for 24 h. Twelve-day-old Arabidopsis seedlings, tobacco seedlings, and Chinese cabbage seedlings (cultivated for only 10 DAS, after which they were transplanted to the farm) were grown in a culture room in black plastic bowls (10 × 10 cm) containing peat, vermiculite, and perlite at a volumetric ratio of 3:1:1. The photoperiod was changed to 16-h light/8-h dark for Arabidopsis seed propagation during the seed-formation stage.
DNA and RNA extraction, gene mapping, and gene expression
DNA was extracted by the CTAB assay as described previously57. Markers were developed based on the B. rapa genome (BRAD Database) around the BrPur locus according to previous research29. PCR amplification, gel electrophoresis, and genetic map construction were performed as previously reported29,57. Total RNA extraction, cDNA synthesis, and qRT-PCR via an IQ5 optical system (Bio-Rad, USA) were performed following a previously reported protocol27. All the qRT-PCR data were normalized using the cycle threshold value corresponding to BrEF-1α in B. rapa and AtActin2 in Arabidopsis. The gene-specific primers used are listed in Table S1 and were used in a recent report10; the primers were confirmed by melting curve analysis for specific amplification. The relative expression of target genes was calculated using the 2−ΔΔCT method by IQ5 software (Bio-Rad, USA). All reactions were conducted for three replicates.
Sequence isolation, plasmid construction, and plant genetic transformation
The CDS and gDNA of the candidate gene were cloned using gene-specific primers (listed in Table S1) and then transferred into a pVBG2307 binary vector. The constructed vectors were transferred into Agrobacterium tumefaciens strain GV3101 using the freeze-thaw method and then transformed into WT Arabidopsis using the floral-dip method56. T2 progeny with the best purple coloration were selected from independent lines, and T3 homozygous lines were generated for subsequent research.
To construct promoter:GUS vectors, several fragments containing the promoter region of the candidate gene with HindIII and BamI restriction enzyme cutting sites were cloned into a pBI121 vector. The original pBI121 vector with the CaMV35S promoter was treated as the positive control, and the vector without the CaMV35S promoter was treated as the negative control. The primers used in these experiments are listed in Table S1. In transient genetic transformation, different GV3101 cell lines harboring unique vectors were transferred into tobacco line NC89 using the injection infiltration method, with slight modifications58. The injection concentration of GV3101 cells (OD600) was set to 0.530 using an UV-vis spectrophotometer (Thermo Fisher Scientific, USA).
PrimeSTAR Max mix (Takara, Japan) was used in the cloning process to reduce the mismatch rate during the PCR amplification process. The reaction volume and the three-step PCR amplification procedure were performed following the instructions of the mixture. All constructs were verified by sequencing (Auguct, China) for at least three technical repeats.
Sequence analysis of the candidate gene and its promoter
Gene structure analysis of exons and introns was performed using Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/). The conserved domains were predicted by the SMART tool (http://smart.embl-heidelberg.de/). DNA and protein sequence alignment analyses were conducted by the multiple sequence alignment module of DNAMAN software (Lynnon BioSoft, Canada). A phylogenetic tree was constructed with MEGA 5.1 by the neighbor-joining method with 1000 bootstrap values. Promoter functional prediction was performed by tools associated with the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Total anthocyanin analysis
Determination of total anthocyanin content was performed using an UV–vis spectroscopy method as previously described10, and data were calculated using a previous equation59. Approximately 1.0 g of crushed fresh samples were used in each extraction, and the results were expressed as the average of three biological replicates.
Microscopy analysis
Different tissues were prepared for histological observations by manual sectioning as previously described60 and investigated using a fluorescence microscope (Olympus, Japan) at an appropriate magnification under the bright field.
Statistical analysis
Cluster analysis of gene expression patterns in head tissues was performed using a two-way hierarchical clustering methodology via PermutMatrix software61, and Pearson distance and Ward’s method were applied for aggregation. One-way analysis of variance was conducted using SPSS 13.0 (Chicago, USA), and Duncan’s multiple-range test was performed on data at the 0.05 confidence level. Pearson correlation coefficients were calculated via two-tailed tests using bivariate analysis.
References
Li, H. et al. MdMYB8 is associated with flavonol biosynthesis via the activation of the MdFLS promoter in the fruits of Malus crabapple. Hortic. Res.7, 19 (2020).
Zhai, R. et al. PbGA2ox8 induces vascular-related anthocyanin accumulation and contributes to red stripe formation on pear fruit. Hortic. Res.6, 137 (2019).
Mazewski, C., Liang, K. & Gonzalez, dM. E. Comparison of the effect of chemical composition of anthocyanin-rich plant extracts on colon cancer cell proliferation and their potential mechanism of action using in vitro, in silico, and biochemical assays. Food Chem.242, 367–378 (2018).
Albert, N. W. et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell26, 962–980 (2014).
Petroni, K. & Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci.181, 219–229 (2011).
Naing, A. H. & Kim, C. K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol.98, 1–18 (2018).
Xie, L. L. et al. Mining for candidate genes in an introgression line by using RNA Sequencing: the anthocyanin overaccumulation phenotype in Brassica. Front. Plant Sci.7, 1245 (2016).
Guo, N. et al. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genomics15, 71–81 (2014).
Baudry, A. et al. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J.39, 366–380 (2004).
He, Q., Ren, Y. J., Zhao, W. B., Li, R. & Zhang, L. G. Low temperature promotes anthocyanin biosynthesis and related gene expression in the seedlings of purple head Chinese cabbage (Brassica rapa L.). Genes11, 81 (2020).
Walker, A. R. et al. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. Cell Mol. Biol.49, 772–785 (2007).
Wang, Z. et al. The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol.162, 885–896 (2013).
Song, H. et al. Purple Brassica oleracea var. capitataF.rubra is due to the loss of BoMYBL2–1 expression. Bmc Plant Biol.18, 82–97 (2018).
Park, K. I. et al. A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J. Cell Mol. Biol.49, 641–654 (2007).
Chiu, L. W. et al. The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol.154, 1470–1480 (2010).
Chiu, L. W. & Li, L. Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta236, 1153–1164 (2012).
Yuan, Y. X., Chiu, L. W. & Li, L. Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta230, 1141–1153 (2009).
Jin, S. W. et al. Molecular analysis of anthocyanin-related genes in ornamental cabbage. Genome61, 111–120 (2018).
Zhang, X. et al. QTL-Seq and sequence assembly rapidly mapped the gene BrMYBL2.1 for the purple trait in Brassica rapa. Sci. Rep.10, 2328 (2020).
Guo, N. et al. Anthocyanin profile characterization and quantitative trait locus mapping in zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). Mol. Breed.35, 1–11 (2015).
Li, G. H. et al. A high-density genetic map developed by specific-locus amplified fragment (SLAF) sequencing and identification of a locus controlling anthocyanin pigmentation in stalk of Zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). BMC Genomics20, 343 (2019).
Wang, J., Su, T. B. & Yu, Y. J. Molecular characterization of BrMYB73: a candidate gene for the purple-leaf trait in Brassica rapa. Int. J. Agric. Biol.22, 122–130 (2019).
Xie, Q. et al. Accumulation and molecular regulation of anthocyanin in purple tumorous stem mustard (Brassica juncea var. tumida Tsen et Lee). J. Agric. Food Chem.62, 7813–7821 (2014).
Zhang, Y. J. et al. Anthocyanin accumulation and molecular analysis of correlated genes in purple kohlrabi (Brassica oleracea var. gongylodes L.). J. Agric. Food Chem.63, 4160–4169 (2015).
Rahim, M. A. et al. Identification and characterization of anthocyanin biosynthesis-related genes in Kohlrabi. Appl. Biochem. Biotechnol.184, 1120–1141 (2018).
Zhao, Z. et al. Fine mapping the BjPl1 gene for purple leaf color in B2 of Brassica juncea L. through comparative mapping and whole-genome re-sequencing. Euphytica213, 80–90 (2017).
He, Q., Zhang, Z. F. & Zhang, L. G. Anthocyanin accumulation, antioxidant ability and stability, and a transcriptional analysis of anthocyanin biosynthesis in purple heading Chinese cabbage (Brassica rapa L. ssp. pekinensis). J. Agric. Food Chem.64, 132–145 (2016).
Zhang, S. J. et al. Mapping and expression profiling reveal an inserted fragment from purple mustard involved anthocyanin accumulation in Chinese cabbage. Euphytica212, 1–13 (2016).
Wu, J. et al. Genetic analysis and mapping of the purple gene in purple heading Chinese cabbage. Hortic. Plant J.2, 351–356 (2017).
Zimmermann, I. M., Heim, M. A., Weisshaar, B. & Uhrig, J. F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like bHLH proteins. Plant J.40, 22–34 (2004).
Jiang, J. J. et al. Transcriptomic comparison between developing seeds of yellow- and black-seeded Brassica napus reveals that genes influence seed quality. BMC Plant Biol.19, 1–14 (2019).
Zhang, J. F. et al. Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Mol. Biol.69, 553–563 (2009).
Liu, J. et al. Primary mapping of pur, a gene controlling purple leaf color in Brassica rapa. Acta Agric. Boreali-Sin.28, 49–53 (2013).
Rahman, M., Mamidi, S. & Mcclean, P. Quantitative trait loci mapping of seed colour, hairy leaf, seedling anthocyanin, leaf chlorosis and days to flowering in F2 population of Brassica rapa L. Plant Breed.133, 381–389 (2014).
Wang, W. et al. Mapping the BrPur gene for purple leaf color on linkage group A03 of Brassica rapa. Euphytica199, 293–302 (2014).
Burdzinski, C. & Wendell, D. L. Mapping the anthocyaninless (anl) locus in rapid-cycling Brassica rapa (RBr) to Linkage Group R9. Bmc Genet.8, 6456–6468 (2007).
Hayashi, K. et al. Mapping of a novel locus regulating anthocyanin pigmentation in Brassica rapa. Breed. Sci.60, 76–80 (2010).
Xu, Z. S., Yang, Q. Q., Feng, K., Yu, X. & Xiong, A. S. DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol. J.1, 1–13 (2020).
Zhang, X. P. et al. Identification of two novel R2R3-MYB transcription factors, PsMYB114L and PsMYB12L, related to anthocyanin biosynthesis in Paeonia suffruticosa. Int. J. Mol. Sci.20, 1055–1064 (2019).
Peng, Y. Y. et al. Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Hortic. Res.6, 3 (2019).
Espley, R. V. et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell Online21, 168–183 (2009).
Shozo, K., Nami, G. Y. & Hirohiko, H. Retrotransposon-induced mutations in grape skin color. Science304, 982–982 (2004).
Liu, L. X. et al. Current status and outlook perspective of induced mutations for plant improvement. J. Nucl. Agric. Sci.23, 1001–1007 (2009).
Liu, C. et al. A missense mutation in the VHYNP motif of a DELLA protein causes a semi-dwarf mutant phenotype in Brassica napus. Theor. Appl. Genet.121, 249–258 (2010).
Wang, N. et al. Defect in Brnym1, a magnesium-dechelatase protein, causes a stay-green phenotype in an EMS-mutagenized Chinese cabbage (Brassica campestris L. ssp. pekinensis) line. Hortic. Res.7, 8 (2020).
Zhang, J. X. et al. Molecular characterization and transcriptome analysis of orange head Chinese cabbage (Brassica rapa L. ssp. pekinensis). Planta241, 1381–1394 (2015).
Laxa, M. Intron-mediated enhancement: a tool for heterologous gene expression in plants? Front. Plant Sci.7, 1977–1977 (2017).
Gallegos, J. E., Rose, A. B. & Intron, D. N. A. sequences can be more important than the proximal promoter in determining the site of transcript initiation. Plant Cell29, 843–853 (2017).
Broeckling, B. E., Watson, R. A., Blaire, S. & Bush, D. R. Intronic sequence regulates sugar-dependent expression of Arabidopsis thaliana production of anthocyanin pigment-1/MYB75. PLoS ONE11, e0156673 (2016).
Fiume, E., Christou, P., Gianì, S. & Breviario, D. Introns are key regulatory elements of rice tubulin expression. Planta218, 693–703 (2004).
Jeon, J. S. et al. Tissue-preferential expression of a rice alpha-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol.123, 1005–1014 (2000).
Liu, R. Y., Liu, F., Zhang, Z. Q. & Guan, C. Y. Functional analysis of BnFAD2-C5 promoter and intron at expression level in Brassica napus. Acta Agronomica Sin.42, 1471 (2016).
Li, S. H., Wang, F. R., Huang, R. F., Dong, J. G. & Wang, J. Function Analysis of VTC1 5’UTR Intron in Arabidopsis. J. Agric. Sci. Technol.18, 52–57 (2016).
Hormuzdi, S. G., Penttinen, R., Jaenisch, R. & Bornstein, P. A gene-targeting approach identifies a function for the first intron in expression of the alpha1(I) collagen gene. Mol. Cell. Biol.18, 3368 (1998).
Lee, S. et al. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol. Biol.57, 805–818 (2005).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol.16, 735–743 (1998).
Zhang, J. X. et al. Fine mapping and identification of candidate Br-or gene controlling orange head of Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol. Breed.32, 799–805 (2013).
Li, Y. & Zhang, Y. Study on agrobacterium tumefaciens-mediated transient transformation of tobacco by infiltration. Exp. Technol. Manag.27, 50–52 (2010). (in Chinese).
Giusti, M. M. & Wrolstad, R. E. Characterization and measurement of anthocyanins by UV–visible spectroscopy. Curr. Protoc. Food Anal. Chem.Unit F1.2, 1–13 (2001).
Lux, A., Morita, S., Abe, J. & Ito, K. An improved method for clearing and staining free-hand sections and whole-mount samples. Ann. Bot.96, 989–996 (2005).
Caraux, G. & Pinloche, S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics21, 1280–1281 (2005).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant No. 31672163) and the National Key Research and Development Program of China (grant No. 2017YFD0101802).
Author information
Authors and Affiliations
Contributions
L.Z. and Q.H. conceived and designed the experiment and wrote and revised the paper. Q.H. and W.Z. conducted the main experiments, such as the RNA and DNA template preparation, gene cloning, vector construction, genetic transformation, material planting, and determination of total anthocyanin contents. J.W. performed the fine mapping analysis, and Y.X. performed the cosegregation analysis. R.L. provided assistance during the planting of materials and anthocyanin extractions. All authors have read and approved the final paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, Q., Wu, J., Xue, Y. et al. The novel gene BrMYB2, located on chromosome A07, with a short intron 1 controls the purple-head trait of Chinese cabbage (Brassica rapa L.). Hortic Res 7, 97 (2020). https://doi.org/10.1038/s41438-020-0319-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-0319-z
This article is cited by
-
Mutation in BrFLS encoding flavonol synthase induced anthocyanin accumulation in Chinese cabbage
Theoretical and Applied Genetics (2024)
-
Genome-wide identification and expression analysis of the anthocyanin-related genes during seed coat development in six Brassica species
BMC Genomics (2023)
-
Identification of R2R3-MYB family in blueberry and its potential involvement of anthocyanin biosynthesis in fruits
BMC Genomics (2023)
-
Research progress and applications of colorful Brassica crops
Planta (2023)
-
Fine mapping of genes controlling pigment accumulation in oilseed rape (Brassica napus L.)
Molecular Breeding (2023)