Abstract
Vitis riparia, a critically important Native American grapevine species, is used globally in rootstock and scion breeding and contributed to the recovery of the French wine industry during the mid-19th century phylloxera epidemic. This species has abiotic and biotic stress tolerance and the largest natural geographic distribution of the North American grapevine species. Here we report an Illumina short-read 369X coverage, draft de novo heterozygous genome sequence of V. riparia Michx. ‘Manitoba 37’ with the size of ~495 Mb for 69,616 scaffolds and a N50 length of 518,740 bp. Using RNAseq data, 40,019 coding sequences were predicted and annotated. Benchmarking with Universal Single-Copy Orthologs (BUSCO) analysis of predicted gene models found 96% of the complete BUSCOs in this assembly. The assembly continuity and completeness were further validated using V. riparia ESTs, BACs, and three de novo transcriptome assemblies of three different V. riparia genotypes resulting in >98% of respective sequences/transcripts mapping with this assembly. Alignment of the V. riparia assembly and predicted CDS with the latest V. vinifera ‘PN40024’ CDS and genome assembly showed 99% CDS alignment and a high degree of synteny. An analysis of plant transcription factors indicates a high degree of homology with the V. vinifera transcription factors. QTL mapping to V. riparia ‘Manitoba 37’ and V. vinifera PN40024 has identified genetic relationships to phenotypic variation between species. This assembly provides reference sequences, gene models for marker development and understanding V. riparia’s genetic contributions in grape breeding and research.
Similar content being viewed by others
Introduction
Grapes (Vitis spp.), used for wine, juice, table grapes, raisins, and rootstocks are the most valuable fruit crop in the world. While the cultivated species Vitis vinifera is the predominant species used in the industry, other wild grape species are important contributors to commercial production. In particular, several North American species have been used by breeders to develop scion and rootstock cultivars that are disease, insect, and abiotic stress tolerant1. However, evidence from ongoing genome sequencing projects in grapevine demonstrates high variability between cultivars and that the wild grapevine species remain under-sampled for genomic data. With the advent of sequencing technologies for crop development, such as genotype by sequencing (GBS) and RNase H2-dependent amplicon sequencing (rhAmpSeq), many crop breeders are adopting larger genomic studies2,3. Thus, development of grape genomic resources is invaluable for ongoing crop improvement and ongoing gene annotation and gene function analyses.
Fossil evidence of Vitis seed from the Rocky Mountain region of the United States date back to the Plaocene era (65.5–55.8 Mya)4,5,6,7. Recent nuclear DNA analyses suggest that the most recent common Vitis ancestor for the existing global grape species originated in North America, diversifying from the rest of Vitaceae ~28 Mya (CI 41.2, 16.2 Mya), with Vitis and Muscadinia diverging ~18 Mya8. The Vitis genus, containing about 60 inter-fertile living species, is suggested to have diversified at 12–1.3 Mya8. The major cultivated species, V. vinifera is thought to have been domesticated from its wild ancestor V. sylvestris in the Mediterranean region of South and East Europe9. There are two major centers of wild grapevine species diversity, North America and East Asia with 28 and 30 species, respectively1,8. In the period following the retreat of the Wisconsin glaciation (~11–10,000 years ago), which radically altered the geography of central and eastern North America, it is likely that the receding glaciers and harsh conditions allowed the eastern North American grapevine species to expand their range north into more varied and marginal conditions8. In eastern North America, Vitis riparia, V. labrusca, V. aestivalis, and V. cinerea developed large overlapping distributions1. Hybridization of these species is common in the wild; however, distinct geographic, topographic, and climatic conditions have maintained the species diversification10. Many breeders have utilized the biotic and abiotic stress tolerance traits in these species to produce grapes that are sustainable for production in harsh climatic and biotic conditions, whereas V. vinifera would be killed outright or require extraordinary viticultural intervention to maintain production11,12.
One of the most commonly used species in abiotic and biotic stress tolerance breeding, V. riparia, has the largest continental distribution of the North American species. Its range stretches, from Texas in the south to the Riding Mountains (Manitoba, Canada) in the North and from the Rocky Mountains to the East Coast1. V. riparia genotypes have been utilized extensively in rootstock and scion breeding for its freezing tolerance, disease resistance (powdery mildew, downy mildew, and, botrytis), and phylloxera resistance11,12,13,14. V. riparia genotypes collected from the species’ northern range have been used to develop new cultivars. Presumably due to the capture of adaptive trait complexes that allow survival under harsh winter conditions. V. riparia based cultivars have contributed to the expansion of grapes in cold climate, resulting in over 2400 ha (hectares) of new production and 300 new wineries in the Northern United States and Southeastern Canada11,12,13,15,16. Mining the locally adaptive traits from V. riparia has resulted in new cultivars that incorporate the traits of early ripening, high sugar content and maximum freezing tolerance traits11,12,17. While breeding new cultivars using V. riparia has been successful, there are large gaps in our understanding of the genetic architecture of positive and negative traits that this species brings into the breeding programs.
To fill this gap, the genome of V. riparia ‘Manitoba 37’ a genotype from the Riding Mountains of Manitoba, Canada was sequenced for assembly, single nucleotide polymorphism mapping, pan-generic marker panel development and genetic analysis. V. riparia ‘Manitoba 37’ was obtained from the breeding program at the University of Minnesota, St. Paul, MN, USA and later placed in the USDA ARS Germplasm Repository at Geneva, New York under the identifier of PI588259. The strategy undertaken here was to use the highly accurate Illumina short-read sequencing in conjunction with mate pair libraries to produce an assembly with high fidelity that matches the greater heterozygosity of the V. riparia ‘Manitoba 37’ genome. This genotype has been used extensively in studies to develop improved cold hardiness, photoperiod responsiveness, identify the female allele of the Vitis sex locus, and break the linkage drag between high malic acid and soluble solids in segregating populations2,3,17,18,19. In addition to breeding, this genotype has also been used extensively for physiological, proteomic, transcriptomic, and metabolomic analyses and GBS mapping studies2,3,20,21,22.
Results
We present here, a detailed analysis of stress tolerant V. riparia ‘Manitoba 37’ genome assembly, using short-read Illumina data. First the genetic relationship of V. riparia ‘Manitoba 37’ and USDA ARS Germplasm Repository V. riparia materials, originally collected throughout V. riparia’s native range, was examined using informative SNPs. Secondly, a de novo assembly was developed for this heterozygous species using paired-end short-read libraries totaling 369.7X coverage. Gene prediction was conducted using RNASeq data from multiple tissues and experiments. Finally, the final assembly and gene models were compared with the reference genome V. vinifera PN40024 12X.2 and V3 annotation and other recent Vitis cultivar assemblies and gene conservation was analyzed. In addition to validating the quality of the V. riparia ‘Manitoba 37’ short-read assembly, we conducted analysis of the WRKY domain (WRKY), myelobsastosis (MYB), and ethylene response factor (ERF) transcription factor (TF) gene families and evaluated transposon composition in V. riparia versus V. vinifera genomes. Sequenced SNP markers from a F2 mapping population, identified by aligning genotype sequence data to V. vinifera, were used to test the fidelity and utility of this V. riparia genome2. The SNP markers were aligned to the V. riparia ‘Manitoba 37’ and V. vinifera genomes and quantitative trait loci regions over genic locations were mapped to the genomes using V. riparia phenotypes.
Genetic analysis of natively collected V. riparia
The genetic relationship of V. riparia ‘Manitoba 37’ relative to other V. riparia in the USDA ARS Germplasm Repository, Geneva, NY USA indicated that ‘Manitoba 37’ is representative of the V. riparia collected throughout its native range (Fig. 1a, b; Supplementary Fig. 1). Principal component analysis (PCA) of SNP data from 68 V. riparia samples demonstrated that V. riparia diversity is best described as two separate clusters. Both V. riparia ‘Manitoba 37’ and the important rootstock cultivar V. riparia ‘Gloire de Montpellier’ are in the primary cluster, and a secondary cluster is made up of species from the Northwestern edge of V. riparia’s range (Fig. 1a, b). This split is not observed in PCA2vsPCA3 (Supplementary Fig. 1). Further differentiation between ‘Manitoba 37’ and ‘Gloire de Montpellier’ and the secondary cluster can be observed in PCA1vsPCA4 (Fig. 1a, b).
a, b Principal component analysis of informative SNPs in 68 V. riparia individuals in the USDA ARS Geneva New York germplasm repository. Symbols represent sample origin by state (United States) and Canadian province as noted in the USDA Germplasm Resource Information Network. Open squares indicate individuals with unknown geographic origin. V. riparia ‘Manitoba 37’ and V. riparia’Gloire de Montpellier’ (unknown geographic origin) are represented by blue diamond and black square, respectively. SNP data was derived from GBS data that was previously reported in Klein et al.10. a Principal components one and two, 10.6% and 5.9% of variation, respectively. b Presents principal components one and four, 10.6% and 4.5% of variation, respectively. c K-mer analysis of V. riparia ‘Manitoba 37’ genome. The 19 k-mer was carried out with 262.3X coverage by Jellyfish and heterozygosity obtained by GenomeScope. The first peak located at coverage 89X corresponds to the heterozygous and the second peak at 184X corresponds to the homozygous peak. The genome contains 1.39% heterozygosity (identified with 262.3X coverage). d Dot plot of global alignment between V. vinifera PN40024 on the x-axis and V. riparia ‘Manitoba 37’ on the y-axis
De novo heterozygous assembly and validation
Using Illumina HiSeq (Illumina, USA) short reads and three mate-pair libraries of varying insert sizes, we generated 2295.4 M raw reads for the V. riparia diploid genome draft assembly. A 1.39% heterozygosity was estimated from the unprocessed short-reads with the Jellyfish plot showing the heterozygous peak slightly lower than the homozygous peak (Fig. 1c). The initial assembly had a N50 of 512,151 bp and 13.63% scaffold %N. After gap closing, the final V. riparia assembly was 494.6 Mb in 69,616 scaffolds, with an N50 of 518,740 bp and a scaffold N of 3.57% (Table 1 and Supplementary Table 1). The V. riparia ‘Manitoba 37’ and V. vinifera PN40024 alignment are shown as a dot plot (Fig. 1d).
In addition to the Assemblathon statistics, 96% of the filtered reads mapped back to the V. riparia ‘Manitoba 37’ genome assembly with zero mismatch. REAPR analysis of genome assembly accuracy using mate paired-end reads found evidence (low mate paired coverage) for potential mis-assembly in no more than 16% of the scaffolds (Supplementary Table 2a). Mapping of EST and BAC sequences indicated that 1935 of 1974 (98%) ESTs and 3811 of 4171 (91.3%) of the BACs mapped to the assembled genome (Supplementary Table 2b). Alignment of three different V. riparia de novo transcriptomes with the V. riparia genome assembly resulted in >99% mapping of the total transcripts for each transcriptome, with >93% transcripts mapping with >90% identity and >90% coverage (Supplementary Table 2b).
Alignment of V. riparia ‘Manitoba 37’ assembly with V. vinifera genomes
The V. riparia ‘Manitoba 37’ de novo scaffolds aligned well with V. vinifera ‘PN40024’. A total of 76% of the V. vinifera PN40024 scaffolds were aligned with >90% identity and >1000 bp identity and 9496 scaffolds had >95% identity with > 1000 bp alignment (Supplementary Fig. 2, Supplementary Table 3). A total of 1,607,090 high quality SNPs were called with a rate of 1 every 504 bases (Supplementary Fig. 2).
Similar alignment statistics were observed for other recent Vitis genomes. V. riparia ‘Manitoba 37’ scaffolds aligned with >90% identity and >1000 bp identity to primary-contig scaffolds of V. vinifera cultivars ‘Cabernet Sauvignon’, ‘Chardonnay’, and ‘Carménère’ and V. riparia ‘Gloire de Montpellier’ at 75%, 74%, 76.5%, and 90.6%, respectively (Supplementary Table 3). A zero-mismatch filtered reads mapping found only 89% of the reads mapping to ‘Gloire de Montpellier’ assembly, in contrast to 96% mapping to the ‘Manitoba 37’ assembly.
Repeat identification and de novo gene prediction
Repeat sequences were predicted to make up 46% of the V. riparia ‘Manitoba 37’ assembly (Supplementary Table 4a). These repeats are predominantly LTR regions (17.68%), long interspersed nuclear elements (LINE) (4.21%), DNA elements (2.06%) and Unclassified repeats (19.95%) (Supplementary Table 4a). The de novo gene prediction using V. riparia ‘Manitoba 37’ RNAseq data identified 40,019 putative coding sequences with the average size of predicted coding sequence (CDS) is 984.4 bp (Table 1). A total of 1548 (96%) Benchmarking Universal Single-copy Orthologs (BUSCO) were identified in the predicted genes (Supplementary Table 4b). Species distribution of BLASTX results showed most of the predicted V. riparia genes correspond with V. vinifera (Supplementary Table 4c). Over 89% of coding sequences were fully annotated using the V. vinifera ‘PN40024’ 41,733 CDS. A total of 41,189 (99%) V. vinifera ‘PN40024’ coding sequences aligned to the V. riparia ‘Manitoba 37’ assembly. Of these 33,370 (80%) V. vinifera CDS had >90% identity and >70% coverage (Supplementary Table 2b) with V. riparia predicted gene models. In total, 5,596 enzymes were identified from the six enzyme classes (Supplementary Table 4d). Gene ontology characterization of the V. riparia coding sequences of V. riparia functionally annotated the majority of the CDS (Biological Process (45,720), Cellular component (34,793), and Molecular Function (29,046)) (Supplementary Table 4e). All coding sequences were queried against the InterPro database and 35,643 sequences were annotated with 20,382 coding sequences containing GO identifiers. Analysis of KEGG annotations resulted in 133 different pathways covering 1,233 coding sequences (Supplementary Table 4f).
Comparative genome analysis
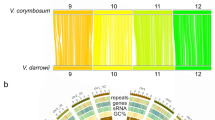
Synteny comparisons were made for V. riparia ‘Manitoba 37’, V. vinifera ‘PN40024’ and V. vinifera ‘Sultanina’ and genome assemblies in the Fabidae and Malvidae orders of the superrosids (Medicago trunculata, and Fragaria vesca and Populus trichocarpa and Arabidopsis thaliana respectively). The V. riparia scaffolds arranged in pseudo-molecules were aligned with each plant genome separately (Supplementary Table 5a, c, d). The greatest number of extended conserved syntenic blocks (>10 kb) was observed with the reference genome V. vinifera ‘PN40024’ and then V. vinifera ‘Sultanina’ (Fig. 2a; Supplementary Table 5c). Medicago had the highest number of syntenic blocks for the examined Rosid genomes, followed by Populus and Fragaria; the lowest synteny was observed with Arabidopsis thaliana (Supplementary Table 5b, c).
a V. riparia ‘Manitoba 37’ synteny with V. vinifera ‘PN40024’ and V. vinifera ‘Sultanina’. b Core orthologous genes present in V. vinifera ‘PN40024’, V. vinifera ‘Sultanina’ and V. riparia ‘Manitoba 37’. c Orthologous genes in V. riparia ‘Manitoba 37’, A. thaliana, F. vesca, and P. trichocarpa. d Neighbor-joining phylogenetic tree of V. riparia ‘Manitoba 37’ with other species based on the orthologous genes
A comparison of predicted V. riparia genes to multiple species was used to further characterize gene coverage. There were 13,964 common orthologous protein sequences shared by V. riparia, V. vinifera 12X.2 and V. vinifera ‘Sultanina’, and 9661 found in common with Rosid species (Arabidopsis thaliana, Fragaria vesca and Populus trichocarpa) (Fig. 2b, c, Supplementary Table 5d).
Plant transcription factor identification
Plant transcription factor analysis indicated that the V. riparia ‘Manitoba 37’ assembly contained representatives of all the transcription factor gene families found in V. vinifera ‘PN40024’ (Supplementary Table 6). A total of 1,723 transcription factors from 58 families were identified in the V. riparia assembly. There were 67 V. riparia WRKY transcription factors annotated and compared phylogenetically to the V. vinifera WRKY and other species (Fig. 3; Supplementary Table 6a–c; 7a, b). A putative novel WRKY domain (WVDTDKR) was identified in the V. riparia gene Vitri g36183.t1; however, this putative WRKY domain was not shared with any WRKY domain in V. vinifera ‘PN40024’ (Fig. 3; Supplementary Table 7a–c). There was a greater number of potential MYB family transcription factors identified in V. riparia (13 MYB genes and 35 MYB-related genes) than found in V. vinifera ‘PN40024’ (Supplementary Table 7d). In the 25 MYB family subgroups, only subgroup 6 had potential gene duplications in comparison to V. vinifera ‘PN40024’ (Supplementary Fig. 3). Comparison of the V. riparia Ethylene Response Factor transcription factor (ERF) genes with the V. vinifera ‘PN40024’ ERF groupings indicate all groups were represented in V. riparia; however, there were fewer ERF identified in V. riparia ‘Manitoba 37’ than in V. vinifera. (Supplementary Fig. 4; Supplementary Table 7e).
Phylogenetic relationship of WRKY TFs between the two species annotated by conserved group. Red dots represent V. riparia ‘Manitoba 37’ genes and blue dots represent V. vinifera ‘PN40024’ genes. Bootstrap values displayed are at nodes. Gene id and group identity are found in Supplementary Table 7b
Analysis of protein families from V. riparia and V. vinifera
The predicted V. vinifera ‘PN40024’, V. riparia ‘Manitoba 37’, and V. vinifera ‘Sultanina’ genes were aligned to the protein family database (PFAM). The V. riparia and the seedless cultivar ‘Sultanina’ had a lower copy number of predicted genes related to transposases and transposons than V. vinifera (Fig. 4a). These results are supported by annotation to GO terms for each family (Fig. 4b). A closer look at the RETROTRANS_GAG 2 (LTR) transposon family shows paralogous duplication of genes in each clade that most likely occurred after the species diverged geographically (Fig. 4c, Supplementary Table 7f). A comparison of LATERAL ORGAN BOUNDARY DOMAIN containing proteins (LBD) in V. vinifera and V. riparia ‘Manitoba 37’ found high conservation similarities between members of each subfamily from both genomes (Fig. 4d, Supplementary Table 7g).
Heatmap displaying the differences in numbers of genes per a protein family or b Gene ontology (GO) term. c Protein alignment of members of the “retrotrans_gag 2” family (LTR retrotransposons). Monophyletic clades containing genes from V. vinifera ‘PN40024’ and V. riparia ‘Manitoba 37’ were annotated and the node containing the most recent common ancestor was indicated by black triangle. Gene id and group are found in Supplementary Table 7f. d Protein alignment of LOB Domain containing (LBD) family proteins, bootstrap values are at nodes. Gene id and group are found in Supplementary Table 7g
Alignment of markers from an F2 mapping population to V. riparia and V. vinifera genome assemblies implies translocation events
SNP marker sequences for F2 mapping population derived from a single F1 (generated by crossing V. riparia ‘Manitoba 37’ (female; grandmother) and the cultivar ‘Seyval’ (male; grandfather)) previously identified from GBS SNP analysis against V. vinifera PN40024 12X.1 genome were used to evaluate the utility of the V. riparia genome as a reference. Alignment of the SNP marker sequence to V. riparia ‘Manitoba 37’ and V. vinifera ‘PN40024’ chromosomes indicated that 89.5% of the markers aligned to both species, while about 6.2% and 4.2% mapped uniquely to V. riparia or V. vinifera, respectively (Supplementary Fig. 5a, b, Supplementary Table 8). The number of markers that mapped was not proportional to genome or chromosome size between the two species indicating an even distribution (Supplementary Table 8). Putative rearrangements, needing further study to verify, were noted on sections of chromosomes 5, 6, and 8 appear between the two species, as well as between chromosomes 14 and 15 (Supplementary Fig. 6b).
Using the aligned SNP markers and phenotype data for flower sex and summer lateral shoot cessation photoperiod response, we identified and aligned genes between flanking markers of the respective QTL (Fig. 5a, b) in the V. riparia pseudo-chromosomes and the V. vinifera ‘PN40024’ chromosomes. Genes in common between the species with similar position alignment and markers with similar LOD score as well as differential marker LOD and gene distribution differences are apparent.
The QTL’s represented are for a summer lateral critical photoperiod growth cessation and b flower type. In the top–middle row, markers found within a genomic region of each genome between the two flanking markers are plotted by their relative position along the x axis and the LOD score along the y axis. The LOD score at each marker is normalized and represented as values between 0 and 1 of the maximum LOD score. F2 markers present in V. riparia are colored red and those present in V. vinifera are blue. In the two bottom panels, genes are annotated by PFAM name, and genes within the QTL in both genomes are colored orange. In the top row, marker positions within the F2 genetic map are connected to the same marker position on each genome. Gene id and PFAM names are found in Supplementary Table 9
Discussion
The first published Vitis reference genome for cultivated grapevine was produced using a highly homozygous inbred line derived from V. vinifera ‘Pinot noir’23. While immensely useful in early genomic studies in grape, this inbred individual does not reflect the high heterozygosity found in other grapevine species and cultivars. The principal component analysis showed that V. riparia ‘Manitoba 37’ is likely a good representative reference for the V. riparia genome as it clusters with most of the collected diversity of this species. Both V. riparia ‘Manitoba 37’ and the other sequenced V. riparia genome ‘Gloire de Montpellier’24 are part of this core diversity. When examined at other principal component levels, ‘Manitoba 37’ appears to be more genetically related to Midwest and Eastern collections of V. riparia than the V. riparia ‘Gloire de Montpellier’. Unfortunately, geographic passport data for the ‘Gloire’ variety is unknown and its representation of V. riparia may not be verifiable. The analysis presented here using only V. riparia data also suggests, for the first time, that there may be two clades of V. riparia, with the North Dakota and Montana genotypes forming the second subclade of genotypes. This result was not observed when examining GBS based relationship data as presented by Klein et al10, presumably due to the inclusion of a much wider collection of SNP data across the Vitis genus in that study. Our results demonstrate the potential for cryptic speciation within V. riparia for this Northwestern subclade of genotypes, or loss of diversity due to population extinction or poor representation in the USDA germplasm collection for the full diversity of the species.
Recently, an improved reference genome V. vinifera ‘PN40024’ was published with improved gene prediction and annotation25. Genomes for other V. vinifera cultivars, as well as the rootstock variety ‘Riparia Gloire de Montpellier’ have recently been produced as well24,26,27,28. However, there remains a dearth of genome data available for wild grapevine species, and those being used in breeding programs in particular. Illumina short-read are very accurate and preserve the heterozygosity of the genome sequence; however, they provide a more fractured assembly than long-read sequences. For example, the V. vinifera ‘Sultanina’ genome, Illumina sequences were used to construct a haploidified assembly29 and subsequently an improved assembly with higher fidelity to the genotypes heterozygous nature was developed using the PLATANUS short-read assembler30. With interest in developing an assembly that could be used for the development of molecular markers for V. riparia3 Illumina short-read paired-end sequencing and mate paired long insert libraries were employed in the present study to assemble the heterozygous, diploid V. riparia ‘Manitoba 37’ genome.
This draft genome sequence, assembled using high quality Illumina reads (>369X coverage), provides a valuable resource for marker development and breeding efforts using wild germplasm. The assembly of 495 Mb in 69,616 scaffolds, has an N50 of 518 kb which is greater than the N50 value reported for the V. vinifera ‘Sultanina’ genome assembled using Illumina data30. The closest Vitis representative to our draft genome is V. riparia ‘Riparia Gloire de Montpellier’, a widely used rootstock variety. Interestingly, the long-read genome assembly of the ‘Gloire’ variety reports 33.9% repetitive sequences, far less than the 46% repeat sequences we detected in ‘Manitoba 37’ (which is similar V. vinifera ‘PN40024’ (41.4%))24,25. Several Vitis genome assemblies have been developed recently using Illumina HiSeq, PacBio RSII or 10X Chromium Genomics24,26,27,28. The alignment results of V. riparia ‘Manitoba 37’ with primary contigs of V. vinifera cultivars was greater than 74% and as expected, the greatest alignment occurred with V. riparia ‘Riparia Gloire de Montpellier’ PACBio assembly24. Further evidence of quality can be seen in the similarity of V. riparia and V. vinifera genes identified in a common orthologous core across multiple genus, which indicate not only gene preservation across genetic distance, but provide evidence of assembly quality. In addition, assembly quality was indicated by the strong relationship found between the predicted V. riparia and V. vinifera genes. Indeed, the strong similarity of LBD and WRKY transcription factor families, between V. riparia and V. vinifera provide further evidence of assembly quality31,32. The validation, completeness and accuracy of V. riparia ‘Manitoba 37’ genome features indicate that using only short-read Illumina sequences a high quality V. riparia genome assembly was developed.
V. riparia is typically used in breeding programs to incorporate abiotic stress tolerance traits into new hybrid cultivars. Thus, particular attention was paid to examining the WRKY, MYB, and ERF transcription factors that influence gene regulation and have a strong role in abiotic stress tolerance phenotypes32,33,34. The V. riparia predicted WRKY coding sequences were consistent with the published V. vinifera ‘PN40024’ 12X.1 WRKY groups32 and the V3 annotation of 12X.225, showing a great level of conservation between the species. The MYB family of transcription factor genes was explored specifically because of their importance to color, flavor and chemistry in grapevine species. Phylogenetic reconstruction of MYB subgroups found that the subgroups (4, 13, and 24) that had potential paralogous duplications were involved in ABA response, flavanol synthesis, secondary growth and anther development35,36. The only homologues with deletions in V. riparia ‘Manitoba 37’ was MYB113 of subgroup 6 which has been shown to regulate the production of anthocyanins in a BASIS HELIX-LOOP-HELEX (bHLH) dependent manner36. We also noted an increased number of bHLH genes predicted in V. riparia ‘Manitoba 37’ genome assembly (by > 20 genes) which is known to produce high amounts of predominantly diglucoside derivatives as opposed to monoglucosides in V. vinifera37,38. This result adds genomic context to one of the key issues facing acceptance of hybrid derived grape products as the presence of diglucoside derivatives is considered an indication of low quality in hybrid wines39. Analysis of ERF genes using alignment and motif comparison between V. riparia and V. vinifera found functional similarities between members of each subfamily from both genomes. However, there were many instances of duplications present in V. vinifera ‘PN40024’ that were not present in our assembly, such as in subfamily IX where some duplications presented with different motifs.
Retrotransposon activity has long been associated with diversification of species clades. We observed a lower number of genes associated with transposases and retrotransposons in the V. riparia ‘Manitoba 37’ and V. vinifera ‘Sultanina’ than in the V. vinifera 12X.2. All three species seemed to share common ancestors for each LTR gene but experienced paralogous gene duplication at different rates in each clade. We posit that this change in retrotransposons could have played some impact on the divergent evolution of the species, as it has been found previously that Tvv1 transposon markers could accurately distinguish between North American species and V. vinifera cultivars40.
The SNP markers that were developed using GBS of the F2 population in comparison with V. vinifera ‘PN40024’ 12X.1 allowed further analysis of the V. riparia ‘Manitoba 37’ assembly. Aligning these SNP markers with the pseudochromosomes of the grandparent, V. riparia ‘Manitoba 37’ and the V. vinifera ‘PN40024’ 12X.2 chromosomes showed that the F2 population more closely modeled V. riparia ‘Manitoba 37’. This can be expected since informative SNPs were predicted using the F2 grandparents and the male parent ‘Seyval’ has a complex pedigree including V. vinifera and other species. The presence of markers that aligned to chromosome 20 of V. vinifera, un-assembled scaffolds, but to other chromosomes on V. riparia may give us a better indication of the actual genomic position of those scaffolds on V. vinifera ‘PN40024’ assembly. By using the markers that mapped to different chromosomes in female grandparent and V. vinifera as a representative portion of the male grandparent, we found evidence for potential large genomic alterations between these species that may have occurred during the evolution and geographic isolation 3.5–9.5 million years ago. When we look at areas of both genomes containing QTL’s we can see that large translocations between chromosomes shows missing genes found between flanking markers in V. riparia relative to V. vinifera, thus impacting the resulting observed phenotype. This shows the potential power of sequencing and assembling a genetic grandparent of a F2 population in identifying the genetic basis of QTL regions.
In conclusion, we present high coverage short-read draft genome sequence of the wild grapevine species V. riparia. This genome represents the second genome assembly of this critically important species and the first representative of a locally adapted stress tolerant genotype. The V. riparia ‘Manitoba 37’ genome assembly provides an important resource for comparative genomic and genetic marker studies. This V. riparia ‘Manitoba 37’ genome has already proven useful for the development of molecular markers in North American breeding programs and will serve as an important tool in the development of genomics-assisted selection for grapevine improvement, particularly for traits associated with abiotic and biotic stress resistance.
Materials and methods
V. riparia ‘Manitoba 37’ materials
V. riparia ‘Manitoba 37’ (identified as ‘PI588259’ in USDA Germplasm Repository, Geneva, NY, USA) was used for sequencing. The genetic relationship of V. riparia ‘Manitoba 37’ to other V. riparia genotypes was analyzed using a data set extracted from genotype data collected from multiple species housed at the Geneva USDA-ARS grape germplasm repository10. To identify highly specific SNPs, VCFtools filters were applied to 156,799 SNPs from 74 unique V. riparia genotypes, keeping those found in at least 50% of the individuals, resulting in 54,029 SNPs41. We then removed six V. riparia genotypes with missing data at greater than 30% of the total SNPs. A high stringency filter was applied to the remaining SNPs keeping all SNPs found in 95% or greater of the V. riparia providing 1485 highly specific SNPs. SNPRelate R package42 was used to calculate the principal components of the specific SNPs data set and plotted the 68 individuals using ggplot243.
DNA sequencing and pre-processing of reads
One centimeter diameter new leaves of greenhouse grown vines were used for DNA extraction and sequencing. A total of nine paired-end libraries were constructed with insert sizes of 346, 473, 478 by Illumina I and 250, 450, 600, 3–5 kb, 8–10 kb, 15–20 kb by Illumina Hiseq 2500 sequencer. In total, 2295.4 M raw reads were generated with 658.4X coverage and read length from 100 nt–260 nt (Supplementary Table 10a). Raw reads were then filtered and corrected using cutadapt, Trimmomatic, PEAR, FastUniq, Quake, and NGSTOOLKIT (Tool references are found in Supplementary Table 10b). The k-mer analysis was carried out with Jellyfish with 19 bp k-mers using only 262.3X coverage of filtered reads. The genome’s heterozygosity and other results were obtained with GenomeScope. All filtered reads used for de novo genome assembly were mapped back to our assembly using bowtie2. The SAM files of the bowtie2 mapping results were converted to BAM files using SAMtools, and then the alignment statistics were obtained using the flagstat option of SAMtools (Supplementary Table 10b).
V. riparia ‘Manitoba 37’ de novo heterozygous genome assembly and assembly evaluation
A total of 1313.7 M filtered reads were used for de novo genome assembly. The first assembly was obtained with PLATANUS44 by changing parameters in each of the three steps ((assemble: -u 0.2 -d 0.3), (scaffold: -s 20 -v 20 -u 0.2) and (gap close: -s 20 -vo 20 -vd 20 -ed 0.1)).This assembly was processed with GapCloser and the result of this assembly was then subjected to a second round of GapCloser to produce the final assembly (https://openprairie.sdstate.edu/vitis_riparia_VR37_PI588259) (All tool references are found in Supplementary Table 10b). The assembly was tested for contamination using DeconSeq. The assembly statistics were evaluated using an Assemblathon script and processed with reference genome V. vinifera ‘PN40024’ assembly25 486,205,130 bp. The quality of the assembly was further assessed by four independent methods. (1) The percentage filtered reads were mapped back to the V. riparia ‘Manitoba 37’ genome using a zero mismatch. (2) The quality of the assembly was further assessed by using REAPR program which measures the number of times that there is low mapped mate-paired-end read coverage of any specific site to predict potential errors in contig assembly. While we found few mistakes in assembly of reads (only 16% of contigs had errors), we did observe low incidence of error free bases (maximum 42.11%). The reason for this low rate is unknown, however, REAPR is a relatively new tool in plant genome development and may not be properly calibrated for the high heterozygosity of this genome. (3) The V. riparia ‘Manitoba 37’ assembly quality was further characterized by generating a dot plot of V. vinifera ‘PN40024’ and V. riparia assembly using the D-genie program which plotted a sorted and denoised global alignment of the two assemblies. 4) V. riparia ESTs and BAC sequences and three de novo V. riparia transcriptomes were aligned with the genome. GMAP with default parameters was used to map and EST sequences of V. riparia from NCBI to our assembly (SAMN00174930, SAMN00152554, SAMN00152555, SAMN00152556, and SAMN00150676). Three V. riparia de novo transcriptomes (PI588259, PI588271, and PI588587), were aligned with the V. riparia genome assembly using BLAT. We used the MUMmer package for alignment of V. riparia BAC sequences (BioProject PRJNA550997) and V. vinifera ‘PN40024’25. (1) BACs were aligned to each V. riparia scaffold using nucmer with -mum option. (2) The output results from nucmer were filtered using delta-filter with the -g option. (3) The filtered results were used in the show-coords program and the coordinates of the resulting alignments were obtained. (4) The alignments that represented the longest length (top-hit) for each BAC were summed (top-hits-length). The same steps were followed for mapping the de novo V. riparia ‘Manitoba 37’ assembly with the reference genome V. vinifera ‘PN40024’ (12X.2, V3), V. vinifera cv. Cabernet Sauvignon, V. vinifera ‘Chardonnay’, V. vinifera ‘Carménère’, and V. riparia ‘Riparia Gloire de Montpellier’.
V. riparia SNP calling
All filtered reads were aligned to the reference genome V. vinifera ‘PN40024’25 with bowtie2. The SAM files were converted to BAM files then repeats were removed by rmdup, followed by sorting in bowtie2. We called the SNPs using the mpileup of SAMtools with default parameters. Then, SNPs were filtered by VCFtools using a window of 10, a minimum depth of 8, and a minimum quality 40. SNP effect was predicted by using the SnpEff program (Supplementary Table 10c).
De novo gene prediction and functional annotation
The repeats were identified with RepeatModeler and then repeats were masked by RepeatMasker (All tool references used in gene prediction and annotation are shown in Supplementary Table 10d). The RNAseq data of V. riparia from our lab were mapped to the masked V. riparia genome assembly and all.bam files were used for de novo gene prediction with BRAKER-1. The assembly gene coverage was preliminarily assessed by BUSCO (version 4.0.5), after gene prediction, using the latest plant early release database (embryophyta_odb10) in genome mode. Coding sequences were further annotated using Blast2GO. BLASTX was performed using the nr database with parameters of: E value 1.0E−3; number of blast hits 1, word size 6, HSP length cutoff 33, and eukaryotes selected as taxonomy. The results from the BLASTX of the assembly was then searched for enzyme classification databases: InterPro, GO (gene ontology), and KEGG pathway analysis using Blast2GO.
Whole genome synteny analysis of V. riparia and V. vinifera
The de novo V. riparia assembly was aligned to reference genome V. vinifera ‘PN40024’25. Masking of repeat sequences and gene predictions from this chromosome-level assembly were obtained as described above for the scaffold assembly. Syntenic blocks between the genomes of V. riparia and other Vitis genomes24,25,26,27,28 were computed by SyMAP (v4.2). V. riparia ‘Manitoba 37’ genome and gene models were aligned to all other genomes and gene models separately (V. vinifera ‘PN40024’25, V. vinifera ‘Sultanina’30, Populus trichocarpa, Medicago truncatula, and Arabidopsis thaliana45, and Fragaria vesca46, using the promer option of the MUMmer program (Supplementary Table 5b–d). Syntenic blocks between V. riparia and all other genomes were obtained from a script in SyMAP (v4.2). (All tool references are in Supplementary Table 10e).
Analysis of orthologous genes
All the predicted protein sequences from V. riparia, V. vinifera ‘PN40024’, V. vinifera Sultanina, Populus, Fragaria, and Arabidopsis were analyzed using OrthoMCL with default settings (All tool references are in Supplementary Table 10e). OrthoMCL was performed step-by-step as described in the manual. (1) An all vs. all BLASTP was performed to identify best hit pairs between species (orthologs), as well as sets of genes more closely related within than between species (in-paralogs). (2) This best hit matrix was used for ortholog definition with the parameter (I) = 1.5. 3). The results from OrthoMCL were visualized by OrthoVenn. The single copy gene present in all six species was used to generate a phylogenetic tree in the program MEGA7. (1) All protein sequences were aligned with ClustalW using PAM weight matrix. (2) The multiple sequence alignments were then used for phylogenetic tree construction by Maximum Likelihood method using default parameters in MEGA7.
Plant transcription factors prediction and phylogenetic tree of gene families
Using all predicted protein sequences from V. riparia ‘Manitoba 37’ assembly and V. vinifera ‘PN40024’ (12X.1, V2 and 12X.2, V3) annotation25 we predicted Plant transcription factors with PlantTFDB (4.0)35. The V. vinifera ‘PN40024’ 12X.1, V247 annotation was also used so that comparisons could be made with earlier transcription factor characterization publications. Transcription factors were predicted together for all assemblies so that comparisons could be made directly in this study. The WRKY gene family results and protein sequences of V. riparia ‘Manitoba 37’ and V. vinifera ‘PN40024’. 12X.1, V1; 12X.2, V3 were retrieved from PlantTFDB (4.0)35 and classified into different groups based on a previous WRKY study32. We created a circular phylogenetic tree for WRKY in two steps, (1) WRKY protein sequences of V. riparia and V. vinifera ‘PN40024’ aligned together by ClustalW in MEGA7 (Supplementary Table 10b). The circular phylogenetic tree was then constructed by the neighbor-joining method using the complete deletion option and bootstrapping with 1000 replicates. Annotation of subgroups in MYB and ERF TFs was done through a BLAST alignment to classified Arabidopsis TFs in PlantTFDB48,49. The neighbor-joining tree for the MYB and ERF TFs was generated from a clustal alignment of all genes through R code ggtree and msa packages (Supplementary Table 10f).
Alignment of F2 GBS markers to V. riparia ‘Manitoba 37’ and V. vinifera ‘PN40024’ 12X.2
The predicted V. riparia ‘Manitoba 37’ gene set and V. vinifera ‘PN40024’ annotation were aligned to the PFAM database using HMMer (All tool references for QTL alignment are in Supplementary Table 10g). The matching annotations were used to divide genes into families or domains. Then the differences in gene number for each gene family at each position of the corresponding gene on the V. vinifera chromosome were plotted with RCircos. The GBS genetic markers from a F2 mapping population, derived from a self of an individual F1 genotype from a cross of V. riparia ‘Manitoba 37’ and hybrid cultivar ‘Seyval’2, were then aligned to V. vinifera ‘PN40024’ 12X.2 and a pseudo-chromosomal assembly of V. riparia ‘Manitoba 37’. The V. riparia pseudo-chromosomal assembly was based on its genomic alignment to V. vinifera35, using the Bowtie aligner (Supplementary Table 10c). R programming was used to find the common marker set and plot the markers mapping to chromosomes using RCircos (Supplementary Table 10g). The gene containing regions of V. vinifera 12X.2 and V. riparia ‘Manitoba 37’ were extracted from between QTL markers for previously predicted QTL flanking markers for female sex and summer lateral cessation in response to decreasing photoperiod phenotype on chromosome 2 and scaled to markers shared between the species2,18,19. The LOD score of the species-specific markers were then plotted using scaled LOD values to present species protein domain distribution.
Data availability
The raw reads of Illumina data and BAC sequences are found in the NCBI BioProject PRJNA550997. EST sequences were downloaded from existing NCBI nucleotide resources. The genome assembly, gene annotation, proteins, and other data are publically available at: https://openprairie.sdstate.edu/vitis_riparia_VR37_PI588259.
References
Reisch, B. J. & Pratt, C. in Fruit Breeding, Vine and Small Fruits, (eds Janick, J. & More, J. N.) Ch. 4, Vol. 2 (John Wiley and Sons, Inc., 1996).
Yang et al. Next generation mapping of enological traits in an F2 interspecific grapevine hybrid family. PLoS ONE 11, e0149560 (2016).
Zou, C. et al. Haplotyping the Vitis collinear core genome with rhAmpSeq improves marker transferability in a diverse genus. Nat. Commun. 11, 413 (2020).
Gong, F., Karsai, I. & Liu, Y.-S. Vitis Seeds (Vitaceae) from the late Neogene Gray fossil site, Northeastern Tennessee, U.S.A. J. Rev. Palaeobot. Palyno. 162, 71–83 (2010).
Graham, A. The role of land bridges, ancient environments, and migrations in the assembly of the North American flora. J. Sys. Evol. 56, 405–429 (2018).
Manchester, S. R. Revisions to Rowland Brown’s North American Paleocene flora. Acta Musei Nationalis Pragae, Ser. B – Historia Naturalis 70, 153–210 (2014).
Nie, Z. L. et al. Molecular phylogeny and biogeographic diversification of Parthenocissus (Vitaceae) disjunct between Asia and North America. Am. J. Bot. 97, 1342–1353 (2010).
Wan, Y. Z. et al. A phylogenetic analysis of the grape genus (Vitis L.) reveals broad reticulation and concurrent diversification during Neogene and Quaternary climate change. BMC Evol. Biol. 13, 141 (2013).
Zohary, D. & Hopf, M. Domestication of plants in the old world: The origin and spread of cultivated plants in West Asia, Europe and the Nile Valley. 3rd edn. (Oxford University, New York, 2000)
Klein, L. L. et al. High-throughput sequencing data clarify evolutionary relationships among North American Vitis species and improve identification in USDA Vitis germplasm collection. Am. J. Bot. 105, 215–216 (2018).
Hemstad, P. R. & Breeder, G. in Grapevine Breeding Programs for the Wine Industry (ed Reynolds, A.) 1st edn, Ch. 17 (Woodhead Publishing, 2015).
Hemstad, P. R. & Luby, J. J. Utilization of Vitis riparia for the development of new wine varieties with resistance to disease and extreme cold. (eds Bouquet, A. & Boursiquot, M. J.) VII international symposium on grapevine genetics and breeding; Acta Horticulturae 528, 487–490 (2000).
Luby, J. & Fennell, A. Fruit breeding for the northern great plains at the University of Minnesota and South Dakota State University. HortSci. 41, 25–26 (2006).
Lowe, K. M. & Walker, M. A. Genetic linkage map of the interspecific grape rootstock cross Ramsey (Vitis champinii) x Riparia Gloire (Vitis riparia). Theor. Appl. Genet. 112, 1582–1592 (2006).
Tuck, B., Gartner, W. & Appiah, G. Vineyards and grapes of the North. University of Minnesota. University of Minnesota Digital Conservancy, http://hdl.handle.net/11299/197814 (2016).
Tuck, B., Gartner, W., Appiah, G. Wineries of the North. University of Minnesota. Retrieved from the University of Minnesota Digital Conservancy, http://hdl.handle.net/11299/197823 (2016).
Fennell, A., Mathiason, K. & Luby, J. Genetic segregation for indicators of photoperiod control of dormancy induction in Vitis species. Acta Hort. 689, 533 (2005). 539.
Garris, A. et al. Mapping of photoperiod-induced growth cessation in the wild grape Vitis riparia. J. Am. Soc. Hort. Sci. 134, 261–272 (2009).
Fennell, A., Alahakoon, D., Luby, J. & Clark, M. Mapping the genetic architecture of grapevine bud growth-cycling quantitative traits. Acta Hort. 1248, 337–343 (2019).
Fennell, A. & Line, M. J. Identifying differential tissue response in grape (Vitis riparia) during induction of endodormancy using nuclear magnetic resonance imaging. J. Am. Soc. Hort. Sci. 126, 681–688 (2001).
Fennell, A. et al. Short day transcriptomic programming during induction of dormancy in grapevine. Front Plant Sci. 6, 834 (2015).
George, I. S., Fennell, A. Y. & Haynes, P. A. Protein identification and quantification from riverbank grape, Vitis riparia: Comparing SDS-PAGE and FASP-GPF techniques for shotgun proteomic analysis. Proteomics 16, 3061–3065 (2015).
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007).
Girollet, N. et al. De novo phased assembly of Vitis riparia grape genome. Sci. Data 6, 127 (2019).
Canaguier, A. et al. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genomics Data 14, 56–62 (2017).
Chin, C.-S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016).
Minio, A. et al. Diploid genome assembly of the wine grape Carménère. G3 9, 1331–1337 (2019).
Roach, M. J. et al. Population sequencing reveals clonal diversity and ancestral inbreeding in the grapevine cultivar Chardonnay. PLoS Genet. 14, e1007807 (2018).
Di Genova, A. et al. Whole genome comparison between table and wine grapes reveals a comprehensive catalog of structural variants. BMC Plant Biol. 14, 7 (2014).
Patel, S. et al. Comparison of three assembly strategies for a heterozygous seedless grapevine genome assembly. BMC Genomics 19, 57 (2018).
Grimplet, J., Pimentel, D., Agudelo-Romero, P., Martinez-Zapater, J. M. & Fortes, A. M. The lateral organ boundaries domain gene family in grapevine: genome-wide characterization and expresson analyses during developmental processes and stress responses. Sci. Rep. 7, 15968 (2017).
Wang, M. et al. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 1, 14016 (2014).
Eulgem, T., Rushton, P. J., Robatzek, S. & Somssich, I. E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206 (2000).
Licausi, F. et al. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics 11, 719 (2010).
Jin, J. et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045 (2017).
Gonzalez, A., Zhao, M., Leavitt, J. M. & Loyd, A. M. Regulation of the anthocyanin pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827 (2008).
Li, Z., Pan, Q., Jin, Z., Mu, L. & Duan, C. Comparison on phenoloic compounds in Vitis vinifera cv. Cabernet Sauvignon wines from five wine-growing regions in China. Food Chem. 125, 77–83 (2011).
Liang, Z. et al. Anthocyanin composition and content in grape berry skin in Vitis germplasm. Food Chem. 111, 837–844 (2008).
Manns, D. C., Lenerz, C. T. M. C. & Mansfield, A. K. Impact of processing parameters on the phenolic profile of wines produced from hybrid red grapes Maréchal Foch, Corot noir, and Marquette. J. Food Sci. 78, 696–702 (2013).
Sant’Ana, G. C. et al. Comparison of a retrotransposon-based marker with microsatellite markers for discriminating accessions of Vitis vinifera. Genet. Mol. Res. 11, 1507–1525 (2012).
Danecek, P. et al. The variant call format and VCFtools. Bioinform 27, 2156–2158 (2011).
Zheng, X. et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag, New York, 2016).
Kajitani, R. et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24, 1384–1395 (2014).
Ensembl Plants. Available at: http://plants.ensembl.org/index.html. (Accessed: 1st June 2019).
Sook Jung et al. 15 years of GDR: new data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 47, D1137–D1145 (2018).
Index of /DATA. (2016). Available at: http://genomes.cribi.unipd.it/DATA/. (Accessed: 25th July 2019).
Dubos et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581 (2010).
Nakano, T., Suzuki, K., Fujimura, T. & Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and Rice. Plant Physiol. 140, 411–432 (2006).
Acknowledgements
This material is based upon work supported by the National Science Foundation Award No. IIA1355423, USDA-NIFA SCRI Award No. 2011–51181–30635, and the state of South Dakota. We acknowledge Moore Brian, Kevin Brandt, Chad Julius, Alan Carter, and Fangping Mu for assistance with computational resources.
Author information
Authors and Affiliations
Contributions
S.P. processed sequences for V. riparia, assembled genome, conducted synteny analysis, gene prediction and annotation, and comparative genomics; M.R. and D.A. conducted marker and gene analysis; R.V.D. and P.S. prepared and uploaded sequences; A.F., S.P., M.R., J.L., D.A., and R.V-D. wrote and finalized the paper; all authors reviewed the final paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Patel, S., Robben, M., Fennell, A. et al. Draft genome of the Native American cold hardy grapevine Vitis riparia Michx. ‘Manitoba 37’. Hortic Res 7, 92 (2020). https://doi.org/10.1038/s41438-020-0316-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-0316-2