Abstract
Cold stress profoundly affects plant growth and development and is a key factor affecting the geographic distribution and evolution of plants. Plants have evolved adaptive mechanisms to cope with cold stress. Here, through the genomic analysis of Arabidopsis, three Brassica species and 17 other representative species, we found that both cold-related genes (CRGs) and their collinearity were preferentially retained after polyploidization followed by genome instability, while genome-wide gene sets exhibited a variety of other expansion mechanisms. The cold-related regulatory network was increased in Brassicaceae genomes, which were recursively affected by polyploidization. By combining our findings regarding the selective retention of CRGs from this ecological genomics study with the available knowledge of cold-induced chromosome doubling, we hypothesize that cold stress may have contributed to the success of polyploid plants through both increasing polyploidization and selectively maintaining CRGs during evolution. This hypothesis requires further biological and ecological exploration to obtain solid supporting evidence, which will potentially contribute to understanding the generation of polyploids and to the field of ecological genomics.
Similar content being viewed by others
Introduction
A now-classical idea with much circumstantial support is that polyploids adapt better than their diploid progenitors to environmental extremes1. At increasingly northerly latitudes, there is an increasing abundance of polyploid plants2. The frequency and level of polyploidy peak in arctic regions, where 73% of the flora are polyploids, approximately two-thirds of which have five or more subgenomes3. Stebbins reported a correlation between polyploid frequency and glaciation and proposed the hypothesis “second contact” between plant populations that have diverged in different refugia, resulting in hybridization and polyploidy to produce new races, some of which become adapted to different ecological conditions in regions vacated by ice4,5.
The success of polyploids is thought to result at least partly from the heterozygosity accompanying polyploidization6. It has been proposed that the evolutionary success of allopolyploids in the Arctic may be due to their fixed-heterozygous genomes, which buffer against inbreeding and genetic drift during periods of dramatic climate change2. The potential effects of polyploidy on ecological adaptation mediated by altered morphology and physiology remain poorly understood. Since the middle of the last century, many hypotheses have been proposed to explain the abundance and distribution of polyploids7,8,9. Many ecological characteristics have been attributed to polyploids, such as a higher colonization ability, higher tolerance, and greater ecological tolerance or niche breadth, which are associated with invasiveness in novel habitats10. However, most of this evidence is anecdotal or comes from observational studies.
Genomics provides valuable opportunities for studying polyploids by allowing deductions to be made about their distant evolutionary past11. The sequencing of model plants such as Arabidopsis and rice, has revealed recursive polyploidization even though these species often exhibit small genomes12. Rich genomic evidence shows that polyploidization could be a major driver of plant evolution correlated with the origination and rapid diversification of major plant taxa13. Based on phylogenetic and evolutionary analysis, genomic evidence has been suggested to link bursts of polyploidy to ecological catastrophes such as the Cretaceous-Tertiary (K-T) mass extinction14. The K-T extinction, which is generally thought to have been triggered by a massive comet or asteroid impact 66 Mya, resulted in catastrophic effects on global ecology, killing 75% or more species15. This event is thought to have caused lingering effects for years, with the persistence of environmental conditions under which it was impossible for plants to carry out photosynthesis. However, the link between mass extinction and polyploidization is weakened by uncertainty in the estimation of the ages of polyploidization events16. Moreover, polyploidization events were by no means scarce either before or after the K-T extinction, with some such event being quite recent17. Nevertheless, these facts do not eliminate the possibility of a relationship between polyploidization events and ecological changes.
Temperature profoundly influences the metabolism of plants and is thus a key factor determining their geographic distribution and growing season18,19. Cold stress, including chilling and/or freezing, adversely affects plant growth, development, and crop productivity20. Plants have evolved a repertoire of adaptive mechanisms for coping with cold temperature, including seed and bud dormancy, vernalization, photoperiod sensitivity, supercooling, and especially cold acclimation19,21. Most plants can undergo cold acclimatization and acquire tolerance to extracellular ice formation in their vegetative tissues22. The exploration of the freezing tolerance of these species has contributed greatly to the understanding of cold stress21,23.
All species are the products of ecological systems and are continuously affected by ecological changes. Considering that selection may leave genomic signatures, we investigated whether there is nonrandom enrichment of CRGs after polyploidization in the present study. Cold stress induces the expression of transcription factors (TFs) such as C-repeat binding factors (CBFs), which can activate the expression of downstream cold-responsive (COR) genes24. CBF genes are regulated by several upstream TFs, including the basic helix-loop-helix (bHLH) and inducer of CBF expression 1 (ICE1) proteins, among others20.
In this study, we conducted comprehensive comparative analyses of CRGs to explore their retention, positive selection, expression and regulatory networks after polyploidy. By studying tens of sequenced plant genomes affected by multiple polyploidization events, we sought to minimize bias due to lineage-specific characteristics, elucidating general principles of the relationship between polyploidy and ecological adaptation.
Results
Recursive polyploidization and accumulation of CRG analogs in Arabidopsis
First, in the angiosperm model Arabidopsis thaliana, we systematically collected 115 CRGs based on previous reports (Table S1, Arabidopsis CRGs list). Then, using these 115 genes, we identified 420 CRG analogs via BlastP searches (E-value <1 × 10−10, amino acid identity >60%: Table S2). To determine whether the 420 CRG analogs could be related to ancestral polyploidization events, a program from the MCScanX packages that could detect different types of gene duplication was implemented as reported25. All CRGs were classified into five groups: singletons (5, 1.19%), dispersed genes (42, 10.00%), proximal duplicates (3, 0.71%), tandem duplicates (17, 4.05%), and polyploidy-related genes (353, 84.05%) (Figure S1a, Table S3-4). Colinear genes between homoeologous regions are those whose ancestral locations are preserved, which are often used to infer paleo-polyploidy events6. Here, polyploidy-related genes were inferred based on collinearity persisting in the regions surrounding the selected genes, albeit noting that the result may be an underestimate due to the erosion of gene collinearity and gene losses after polyploidization26. Compared with the total of 27,417 genes annotated in Arabidopsis classified into the above five types, CRGs were extremely overrepresented (Chisq-test P-value = 1.92E−151) in polyploidy-related groups and underrepresented in all other groups (Table S4). In addition, we used the Gene Ontology (GO) database to identify CRGs, and the results were similar to those of the above analysis (Table S5).
Recursive polyploidization contributed to the expansion of rice CRGs
Using the sequences from the CRG library of Arabidopsis and rice, we identified 430 homologs by BLAST searches in rice, a model monocot plant (Table S2). The classification of rice cold-related homologs into singletons (5), dispersed genes (101), proximal or tandem duplicates (23), and polyploidy-related genes (301) showed significant overrepresentation in the polyploidy-related group (P-value ~ 0) (Figure S1, Table S3-4) and underrepresentation in other groups (Table S4). Specifically, ~70% (301 of 430) of cold-related colinear genes were associated with recursive polyploidization according to our previous classification27 (Table 1, Table S6). These findings suggested that polyploidization might be correlated with the accumulation of cold-related homologs in rice and possibly other grasses.
Polyploidization contributed to the expansion of CRGs in other species
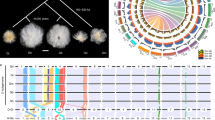
In addition to Arabidopsis and rice, we investigated the relationship between the accumulation of CRGs and polyploidization in a total of 21 species, including model plants such as Zea mays, a basal angiosperm (Amborella trichopoda), a lycophyte (Selaginella moellendorffii), a moss (Physcomitrella patens) and other land plants (Fig. 1, Table S2, Table S7). There were more cold-related homologs in Brassica napus (1520) than in any other plant, followed by Gossypium hirsutum (1210) and Brassica oleracea (852). In contrast, S. moellendorffii exhibited the fewest CRG homologs (124) (Table S6, Fig. 1). At the whole-genome level, 1.83% (750) of Brassica rapa genes belonged to the cold-related group, followed by the proportions in Musa acuminata (1.78%, 652) and G. hirsutum (1.72%), while only 0.56% of S. moellendorffii genes belonged to this group (Table S3).
Information regarding polyploidization was obtained from the Plant Genome Duplication Database (PGDD). The percentages of cold-related genes and all genes in colinear regions are displayed in parallel, gradient rectangles, where C: cold-related genes (CRGs), and A: all genes in each genome; each gene classification is displayed with solid circles, with significance levels relative to genome-wide averages shown in different colors: enriched (orange, P < 0.01), depleted (blue, P < 0.01), or not significantly different (gray)
In 20 of the 21 species, with the exception of S. moellendorffii, cold-related homologs were significantly enriched (P < 0.05) for polyploidy-related genes at the whole-genome level (Table 1, Table S6). In 17 of the 21 plant species, polyploidization was the predominant mechanism by which CRG homologs formed, accounting for 46.32–96.00% of these homologs in each genome (Figure S1, Table S6). CRGs were also enriched for dispersed duplications in three plants (A. trichopoda, P. patens, and S. moellendorffii) and proximal duplications in one plant (M. truncatula). CRGs were underrepresented (P < 0.05) among singleton genes in all 21 species (Fig. 1, Table S4). Notably, nucleotide-binding site (NBS) genes showed a trend that was precisely opposite that of CRGs. The most NBS genes were found in M. truncatula (565, 1.28%), followed by P. trichocarpa (563, 1.36%) and O. sativa (500, 0.90%), while NBS genes represented <0.50% of the gene sets of B. napus, G. hirsutum, B. rapa, and M. acuminata (Table S8). These phenomena indicated that the expansion and evolutionary mechanisms were different between CRGs and NBS genes. Whole-genome duplication/triplication (WGD/T) and segmental duplication played a much greater role in the expansion of CRGs than NBS genes. Overall, these findings suggested that the expansion of CRGs in a wide range of plants might be correlated with polyploidization.
Hierarchical analyses of recursive polyploidies and CRG expansion
The 21 plants in which we investigated the accumulation of CRGs have experienced several independent polyploidizations (Fig. 2). Using a hierarchical and event-related approach by which we deciphered gene collinearity in many other genomes28,29, we identified colinear genes produced by each of the WGD/T events. CRGs were significantly increased in colinear regions in all species except for S. moellendorffii. The most recent events generally contributed the most to the enrichment of CRGs (Fig. 2, Table 2).
A blue four-pointed star represents tetraploidy events, including β, α, τ, σ, ρ and some θ events. A red six-pointed star represents hexaploidy events, including γ and some θ events. A yellow circle represents decaploidy events, which occurred only in Malvaceae. The number on the branch represents the number of CRGs included in each species related to each duplication event. Venn diagrams show the numbers of common and unique CRGs in each examined species
For example, Arabidopsis has been affected by one polyploidization common to core eudicots and two shared with other Brassicaceae plants30,31. By checking the gene collinearity inferred above and polyploidization-produced paralogous regions reported previously32, 31.43, 54.52, and 54.29% of the 420 cold-related homologs could be connected to the respective events in temporal order. Genes associated with an older event were recounted if they were reduplicated in a younger event. This general pattern was found for nearly all events affecting the other plants under study (Table 2), consistent with the fact that gene collinearity is better preserved from recent than ancient events.
In contrast, colinear regions were poor in NBS genes in nearly all species (Table S6). The ratio of NBS genes in colinear regions was significantly lower than that at the whole-genome level in almost all species, except for M. truncatula, A. trichopoda, and S. moellendorffii (Table S9). NBS genes largely expanded by proximal and tandem duplications through a rapid birth-and-death process.
Regulatory network analyses
Networks constructed from all candidate CRGs
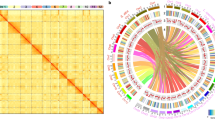
By using the Arabidopsis Interactions Viewer with data obtained from several methods, such as gene coexpression analysis (see Methods), we constructed an interaction network for all 420 inferred CRGs of Arabidopsis. The software queries a database of 70,944 predicted and 39,505 confirmed interacting proteins in Arabidopsis. For other species, we also characterized protein–protein interactions among putative orthologs of the Arabidopsis CRGs using the established Arabidopsis interaction network and predictions from the method of equivalent replacement. The inferred interaction relationships were illustrated using Gephi software33.
We found that taxa that have experienced more duplication events generally exhibited much larger interaction networks. For example, Brassica plants (B. rapa, B. oleracea) have undergone triplication since divergence from Arabidopsis34,35,36, and B. napus (AACC genome) originated via hybridization between B. rapa (AA genome) and B. oleracea (CC genome)31. The interaction network of CRGs was much larger in B. rapa and B. oleracea than in Arabidopsis and was larger still in B. napus. For Arabidopsis, 136 of 420 CRGs were involved in the network, among which 102 could be related to polyploidy events, with 3, 17, and 16 related to γ, β, and α polyploidizations, respectively (Fig. 3, Table S10). For B. rapa, 39.7% (298 of 750) of CRGs were involved in the network, and 278 were related to polyploidy events, with 0, 4, 0, and 66 genes that were specific to γ, β, α, and θ (the latest) events, respectively (Fig. 3, Table S10). For B. oleracea, 330 of 852 CRGs were involved in the network, among which 312 were related to polyploidy, with 0, 5, 0, and 65 being specific tο γ, β, α, and θ events, respectively. For B. napus (Fig. 3, Table S10), 589 of 1520 CRGs were involved in the network, with 293 and 294 coming from subgenomes A and C, respectively. Among these genes, 455 genes were related to polyploidy, with 0, 5, 3, and 104 that were specific to γ, β, α, and θ events.
The network of CRGs in Arabidopsis was constructed using Arabidopsis Interactions Viewer (v2.0). The networks associated with the Arabidopsis orthologs of CRGs in the other three species were constructed using Gephi (v0.8.2) software. The pie charts show the number of CRGs located in the network and outside of the network. WGD (tetraploidy) events include β, α, and θ events, while γ represents a WGT (hexaploidy) event. Venn diagrams show the numbers of CRGs common or specific to different polyploidization events (α, β, γ, and θ). Dots represent α (blue), β (purple), γ (green), and θ (orange) events, respectively
Genes from plants that have experienced more polyploidization exhibited more interactions. Among these four networks, the maximum edge number for a single Arabidopsis gene was only 39 (At2g43790), vs. 356, 262, and 676 for the B. rapa, B. oleracea, and B. napus genes, respectively (Figure S2a, Table S11). Likewise, the total edge number was highest for B. napus (7887), followed by B. oleracea (2722), B. rapa (2165), and Arabidopsis (213) (Figure S2b).
The complexity of the regulatory networks was likely related to transcription factors (TFs). More than half of the CRGs were putative TFs in Arabidopsis and Brassica (Figure S3a, Tables S12, 13). A total of 16 types of TFs were identified among all CRGs, including ethylene-responsive factor (ERF), myeloblastosis (MYB), APETALA2 (AP2), related to ABI3/VP1 (RAV), cysteine 2/histidine 2 (C2H2), NAM, ATAF1/2, and CUC2 (NAC) TFs, with ERF and MYB being significantly more abundant than other types of TFs (Figure S3b, Table S12), which indicated that these two kinds of TFs might play important roles in cold regulation networks in the studied plants.
Networks constructed with reported CRGs
We investigated previously reported cold regulation networks in Arabidopsis19,20 and selected 20 key CRGs for further analyses. The interaction networks among putative orthologs of the Arabidopsis CRGs in other plants were inferred using the above methods (Table S14) (Fig. 4a). The published data on the expression of these CRGs37 came from two tissues, shoots and roots, under different durations of cold treatment (0, 0.5, 1.0, 3.0, 6.0, 12.0, and 24.0 h) (Table S15). Most of the examined CRGs were upregulated after cold treatment, with only high expression of osmotically responsive genes 2 (HOS2) being downregulated after 24 h of treatment (Fig. 4b). For MYB15, expression was downregulated in some time periods but upregulated after cold treatment for 1–24 h. These expression changes indicated that these genes play important roles in the cold response of Arabidopsis. Furthermore, we studied the expression of these genes in seven different tissues, including the roots, stems, leaves, shoots, flowers, pollen, and seeds. Some genes showed tissue-specific expression; for example, MYB15 exhibited much higher expression levels in the roots than in other tissues (Figure S4, Table S16).
a An interaction network of key CRGs in Arabidopsis was constructed based on Arabidopsis Interactions Viewer. Dots represent α (blue), β (purple), γ (green), and θ (orange) events. b Expression cluster analyses were performed for key CRGs under different durations (0, 0.5, 1.0, 3.0, 6.0, 12.0 and 24.0 h) of cold treatment of the shoots (S) and roots (R) in Arabidopsis. The expression values were log2 transformed for heatmap cluster analyses. Red and green colors represent up- and downregulated genes, respectively. c The networks associated with the Arabidopsis orthologs of key CRGs in B. napus were constructed using Gephi software. Dots represent α (blue), β (purple), γ (green), and θ (orange) events, respectively. d Copy numbers of key CRGs from Arabidopsis in B. napus. Venn diagrams indicate the numbers of CRGs that are common or specific to different events in B. napus
Among these networks, the maximum edge number for a single gene (zinc finger of Arabidopsis thaliana 10, ZAT10) was 60 in B. napus (Table S17). The total edge number was highest for B. napus (1022), followed by B. oleracea (418) and B. rapa (300) (Figure S2c d, Table S18). The cold regulation networks of several species, such as B. napus, B. oleracea, B. rapa, and M. acuminata, were much larger than those of Arabidopsis (Fig. 4a, c, Figure S5). The ratios of edge number to node number in these four species were higher than in Arabidopsis (3.35) and other examined species. The highest ratio was detected in B. napus (10.76), followed by B. oleracea (7.46) and B. rapa (5.45) (Table S18). Among these key CRGs, one or more copies were identified in each of B. napus, B. oleracea, and B. rapa, except for SIZ1 in B. rapa (Fig. 4d).
Similarly, we identified more copies of the CBF1-3, MYB15, related to AP2 1 (RAP2.1), and ICE1 genes in most species than were found in Arabidopsis (Fig. 5). There were 42 and 37 MYB15 copies in G. hirsutum and G. max, respectively (Table S14, Fig. 5). However, we failed to detect an ortholog of Arabidopsis COR15-2 in all species. For COR15-1, COR47, and COR6.6, orthologous genes were only detected in B. napus, B. rapa, and B. oleracea, while none were found in all other species (Figure S6, Table S14). In P. patens, we detect no copies of 14 of these genes, one copy of 4, and two copies only for RAP2.1, which could reflect sequence variation, high structural variation or deletions.
The regulatory network of key CRGs was drawn, mainly in reference to a previous report20. The key orthologous CRGs in B. napus and other examined species were identified using the OrthoFinder program
There were no B. napus CRGs specific to γ, β, or α polyploidization events, while 25 were specific to the most recent event (θ) (Fig. 4d). In addition, we detected 11 genes that were duplicated in all four genome duplication events (γ, β, α, and θ). For B. oleracea, 0, 2, 0, and 15 key CRGs were specific to γ, β, α, and θ events, respectively (Figure S5), with 5 genes involved in all four types of genome duplication events. For B. rapa, no CRG was specific to γ, β, or α events, but 9 were specific to the most recent polyploidization (θ) (Figure S5), and 14 were duplicated in all four types of genome duplication events.
Strengthened CRG regulatory networks after polyploidization
The robustness of CRG regulatory networks was enriched with extra homologs after polyploidization. We evaluate the robustness value (R) of a network, reflecting the size of the largest linked subgraph after randomly removing certain numbers of nodes from an initial network or graph (see Methods for details). We used two approaches to compare the robustness values of Arabidopsis and Brassica plants. First, the same number of nodes was removed from each network, comparing the corresponding networks (Table 3). For the networks constructed from the key CRGs, when 5 genes were removed from each network, the Arabidopsis network presented a robustness value of 0.967, which was lower than those for the Brassica plants. The Arabidopsis network robustness was still lower even after the removal of 10 nodes of Brassica plants (≥0.998). Second, the same percentages of nodes were removed. When 5% of nodes were removed in the network constructed with key CRGs, the Arabidopsis network presented a robustness value of 0.992, compared with 0.995, 1.000, and 1.000 for B. rapa, B. oleracea, and B. napus, respectively, and its robustness value remained lower even after 20% of nodes were removed from the B. oleracea and B. napus networks. Robustness was also checked with the networks constructed from all candidate CRGs, which resulted in the same conclusion (Table 3).
Positive selection analyses of key CRG families
To explore the selective pressures acting on genes involved in the cold regulation network, we collected and selected key CRGs according to previous reports19,20. The best BlastP matches or likely orthologs from all plants were used to perform positive selection analyses (Figure S7). A total of 118 branch nodes showing positive selection were detected in 16 phylogenetic trees (Table S19). The greatest number of branch nodes with positive selection (16) was detected in the COR15 gene family, followed by CBF (12) and COR47 (10) (Figure S8). In contrast, there were few nodes showing positive selective in the COR6.6 (3), LOS2 (4), MYB15 (4), and salt-induced zinc finger protein 1 (SIZ1) (4) gene families (Table S19).
Dosage effect analysis
We identified B. rapa genes that were colinear with key Arabidopsis CRGs. The results showed that 1, 5, 8, and 5 key Arabidopsis CRGs exhibited 0–3 colinear orthologs in B. rapa (Figure S9a, Table S20). The expression patterns of five groups with three colinear orthologs were studied in six tissues of B. rapa. We found that several colinear genes exhibited similar expression trends among tissues (Figure S9b, c), although the expression levels were slightly different. These findings indicated that these colinear homologs may play similar or related roles after duplication.
We found that in several cases, colinear orthologs presented divergent expression patterns, showing likely dosage differentiation or compensation effects (Figure S9d–f). For example, Bra002527 exhibited much higher expression in almost every tissue than its two colinear homologs. A similar phenomenon was found for Bra023742, which presented significantly higher expression in roots than the other two colinear homologs. These findings might also indicate that polyploidy-produced homologs may have diverged in function, possibly resulting in subfunctionalization or neofunctionalization. In addition, we found that some genes exhibited tissue specificity in their expression. Bra020284 and Bra023742 were highly expressed in roots but not in other tissues.
Discussion
Numerous lines of evidence suggest that polyploidization is associated with the expansion of gene families related to the cold temperature response, an association that may be related to the direct effects of cold on gamete formation38. Cold stress can alter the formation of radial microtubule arrays at telophase II and lead to the formation of diploid and polyploid pollen grains, which will ultimately increase the possibility of polyploidization39. Genes including those encoding transcription factors are involved in determining the sensitivity of male gametogenesis to cold stress40. Thus, cold temperatures may lead to the formation of polyploidy, and cold tolerance may be important for the survival of newly formed polyploids. High temperature can also induce the production of unreduced pollen in Populus41 and in Rosa sp.42. It is likely that temperature stresses disrupt the normal process of cell division and can cause chromosome number increases in plants. Previous analyses demonstrated that extreme cold temperatures may represent an absolute boundary for species survival, whereas warm temperatures do not43. The observation of more polyploids in higher-latitude regions in the Northern Hemisphere agrees with this inference2. Here, the analysis of the abundance and expansion of CRGs in dozens of land plants provided further support for this hypothesis.
Experimental evidence suggests that both CRGs and heat tolerance genes are often members of the temperature stress-responsive or thermostress-responsive (TSR) gene families. For example, the transgenic overexpression of a Primula forrestii heat-induced gene, PfHSP17.2, in Arabidopsis increased cold tolerance44. The overexpression of a zinc finger transcription factor (TaZnF) from wheat conferred tolerance to both heat and cold stresses in Arabidopsis45. Even in insects, most heat shock protein (HSP) genes are induced or repressed by thermal stress (40 °C) and cold treatment (4 °C)46. Strong evidence led to the conclusion that certain temperature stress-responsive gene families, or even individual members of these families, may be involved in the responses to both high- and low-temperature stresses. Therefore, it was no surprise that we identified the preferential retention of cold-related family genes as an outcome of paleopolyploidization in plants from both cold region and warm regions. We conducted analyses of heat shock transcription factors (Hsfs) among 21 representative plants (Table S21). The repeat types were identified for these Hsf genes and compared with those of cold-related genes in each species. Six species showed more and three species showed fewer WGD-type duplications than non-WGD-type duplications (Table S21). On average, 50.80% of Hsf genes belonged to WGD events (including segmental duplications), confirming that some heat-related genes were also enriched by WGD, even though the number was smaller than that of cold-related genes. We are not aware of any reports that heat induces polyploidy.
Plants with doubled genomes have been suggested to present a greater chance of survival in dramatically altered environments14. Polyploidization was previously related to the occurrence of mass extinctions, albeit with some problems16. Catastrophic events such as volcanic eruptions and widespread global cooling (ice ages) could have triggered mass extinctions47. However, we must note that icy or cold days may occur nearly every year and in nearly all regions on Earth, thereby affecting plant cell division, and a single undivided cell may ultimately grow into a polyploid plant15. Previous research has shown that angiosperms had to achieve new structural and functional trait solutions before making forays into cold environments48. Our findings suggest that the expansion of cold-related genes in a wide range of plants might be correlated with polyploidization. CRGs were significantly more often retained in polyploids compared with the genome-wide average. Moreover, CRGs were enriched in genomic regions produced by recent WGD/T events, suggesting that recurrent polyploidization may enrich the repertoire of CRGs. One exception, S. moellendorffii, was inferred to present the fewest CRG homologs, possibly due to its fragmented genome assembly49.
Moreover, previous studies have indicated that polyploid plants exhibit tolerance to a wider range of environmental conditions than their diploid relatives50. This is thought to occur because gene duplication promotes diversification and evolutionary success through the emergence of novel gene functions51. The overrepresentation of CRGs in the polyploidy-related gene group suggests that polyploidization has contributed significantly more to the expansion of CRGs than the other four gene duplication types. Moreover, if all genes were equally affected by collinearity erosion, newer polyploidization events should have made the largest contributions not only to the CRG reservoir but also to other gene families, as those associated with older events decrease over time. This is clearly not a general characteristic of all large gene families; for example, NBS genes are predominantly clustered, and ectopic duplication is the major cause of their expansion52. Although CRGs, among which the NBS genes are a major component, have expanded via various mechanisms other than WGD53, the present study confirmed this phenomenon in a diverse group of taxa and provided a direct opposite example to the group of CRGs (Tables S8, 9, 22). In addition, we identified drought-related genes in Arabidopsis that were not significantly enriched during polyploidization.
Many WGD/T events are distributed across angiosperms and might lead to gene expression changes and epigenetic remodeling47, which may provide variation that allows rapid adaptation to novel environments54. Altered reproductive modes, increased phenotypic variability, and subfunctionalization may allow polyploids to survive in conditions that are beyond the adaptive range of their diploid progenitors55. For example, arctic allopolyploids have been particularly efficient in invading deglaciated (relatively warmer) areas during periods of dramatic climate change2.
The plasticity of expression patterns might confer polyploids with a wider phenotypic range than their diploid progenitors. Based on various environmental stresses, duplicated genes could become subfunctionalized by partitioning ancestral expression patterns, which might improve adaptation to different ecological conditions56. In addition, several key CRGs, such as COR15 and CBF genes, show evidence of positive selection in their evolutionary history. Most CRGs belong to transcription factor families, and their regulatory networks are much larger in plant genomes affected by more polyploidization events. These results suggest that polyploidy plays an important role in resisting the stress imposed by a cold external environment. These findings may contribute to understanding the formation of polyploids and contribute new knowledge to the field of ecological genomics.
Materials and methods
Retrieval of sequences
The B. napus, B. oleracea, and B. rapa genome sequences were retrieved from the BRAD database57. The Arabidopsis and rice genome sequences were downloaded from the TAIR (https://www.arabidopsis.org) and RGAP (http://rice.plantbiology.msu.edu) databases, respectively. The A. trichopoda genome sequences were obtained from the Amborella Genome Database58. The S. lycopersicum genome sequences were downloaded from the Sol Genomics Network (https://solgenomics.net). The G. hirsutum, G. arboretum, and G. raimondii genome sequences were downloaded from CottonFGD (http://www.cottonfgd.org). The M. acuminata and M. domestica genome sequences were downloaded from the Banana Genome Hub (https://banana-genome-hub.southgreen.fr) and GDR (https://www.rosaceae.org), respectively. The genome sequences of the other nine examined species (M. truncatula, G. max, P. trichocarpa, V. vinifera, Z. mays, A. comosus, E. guineensis, S. moellendorffii, and P. patens) were downloaded from JGI Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html). These species are representative of different branches in the plant phylogenetic tree, and the WGD/WGT information was obtained through searches in the plant genome duplication database59.
Identification and characterization of CRGs and NBS genes
First, 115 CRGs of Arabidopsis were employed in searches against rice protein sequences using the Blastp program with an E-value threshold of 1 × 10−10. The top-ranked rice hit was used to construct the CRG library combined with 115 Arabidopsis CRGs. Then, homologous CRGs in other species were identified through comparison with the Arabidopsis and rice CRG library by using the Blastp program (E-value < 1 × 10−10, identity > 60%) according to a previous report60. The Pfam (v32.0, http://pfam.xfam.org) database and a related Perl script were used to identify the NBS gene family61. Pfam accession number PF00931.18 was used to identify the NB-ARC domain. The genes were further verified with the SMART program62.
Identification of gene collinearity and specific duplication events
BLAST and the Multiple Collinearity Scan toolkit (MCScanX) were used for gene collinearity analysis with default parameters according to previous reports25,63. Venn diagrams were drawn by using the R (v3.5.0) program VennDiagram. A program (duplicate_gene_classifier) in MCScanX was used to infer different types of duplicated genes. To identify the cold-related genes associated with a specific polyploidization event, we inferred colinear genes. First, BlastP was used to search for potential anchors (E-value < 10–5; top five matches) between each possible pair of chromosomes in multiple genomes. A loose E-value threshold accommodates the highly divergent evolutionary rates of duplicated genes produced by polyploidization millions of years ago.
The protein sequences from each species were searched against its own or other genome sequences. The best, second best, and other gene pairs were indicated by red, blue, and gray dots, respectively. Then, the homologous blocks within each genome and between different species (maximal gap ≤ 50 genes; P-value < 0.05) were determined using ColinearScan64. The key parameter of the maximum gap between neighboring genes along a chromosome showing collinearity with genes along the counterpart chromosome sequence was set to 50 intervening genes, which was proven to be successful in previous studies29.
Orthologous and paralogous CRG identification
Orthologous and paralogous CRGs in B. napus and other species were identified using OrthoFinder (v2.2.7, https://github.com/davidemms/OrthoFinder/releases). The relationships between orthologous and paralogous genes were plotted using Circos (v1.0)65. Clustering was conducted by using the Markov cluster algorithm (MCL, v1.0, −I > 1.5).
Gene expression analyses of CRGs
We conducted CRG expression analyses in B. rapa using RNA-Seq data reported previously (Tong et al.66). These data came from six tissues, including the roots, stems, leaves, flowers, siliques, and calli, and fragments per kilobase of transcript per million fragments mapped (FPKM) values were used for comparative analyses. The expression values of these CRGs in Arabidopsis were obtained from the eFP browser (http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi) according to a previous report37; these values came from two tissues, the shoots and roots, under different cold treatments (0, 0.5, 1.0, 3.0, 6.0, 12.0, and 24.0 h). The expression values of these CRGs in seven different tissues, including the roots, stems, leaves, shoots, flowers, pollen, and seeds, were also downloaded from the Arabidopsis eFP browser. The heat maps of hierarchical clustering according to gene expression were plotted using Tree View software (http://jtreeview.sourceforge.net/).
Construction of regulatory networks
The interaction network of CRGs in Arabidopsis was constructed by using Arabidopsis Interactions Viewer (v2.0) (http://bar.utoronto.ca/interactions/cgi-bin/arabidopsis_interactions_viewer.cgi). The interaction networks related to orthologs of Arabidopsis genes in other species were constructed with Gephi (v0.8.2) software using the Force-based algorithm ForceAtlas33. The plug-ins of Multi Color Renderer were used to illustrate the WGD/WGT events (α, β, γ, and θ). The key CRG network for Arabidopsis was constructed by using the String databases (v10.5)67.
Evaluation of network robustness
The robustness criterion is defined as R = C/(N−Nr), where N is the size of a linked group or network defined by the number of nodes within it; Nr is the number of removed nodes; and C is the size of the largest linked group after the removal of those Nr nodes68. In our implementation, certain numbers or percentages of nodes were randomly removed from a certain network, and R was then calculated. This process was implemented 100 times, and an average R value was used to describe the robustness of the network.
Selective pressure estimation
To estimate the divergence times of the colinear CRG pairs, the protein sequences were aligned with the ClustalW2 program and translated into the coding sequence alignment. The synonymous rate (Ks), nonsynonymous rate (Ka), and evolutionary constraint (Ka/Ks) between colinear gene pairs were calculated using the method of Nei and Gojobori, implemented in KaKs_Calculator (v2.0)69. The divergence times of our selected species were collected and integrated from several previous reports70,71,72. We employed the Codeml program from PAML (v4.9) and analyzed 16 key CRG families to calculate ω, which is the ratio of nonsynonymous to synonymous distances73, and we checked positive selection by employing a likelihood ratio test between the M7 and M8 models. Maximum-likelihood phylogenetic trees were constructed with the PhyML program (v3.0)74.
References
Van de Peer, Y., Maere, S. & Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732 (2009).
Brochmann, C. et al. Polyploidy in arctic plants. Biol. J. Linn. Soc. 82, 521–536 (2004).
Rice, A. et al. The global biogeography of polyploid plants. Nat. Ecol. Evol. 3, 265–273 (2019).
Stebbins, G. L. Polyploidy, hybridization and the invasion of new babits. Ann. Mo. Botanical Gard. 72, 824–832 (1985).
Ramsey, J. & Ramsey, T. S. Ecological studies of polyploidy in the 100 years following its discovery. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130352 (2014).
Soltis, P. S., Marchant, D. B., Van de Peer, Y. & Soltis, D. E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 35, 119–125 (2015).
Jiao, Y. & Paterson, A. H. Polyploidy-associated genome modifications during land plant evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130355 (2014).
Brochmann, C. Reproductive strategies of diploid and polyploid populations of arcticDraba (Brassicaceae). Plant Syst. Evolution 185, 55–83 (1993).
Hsu, T. C. & Moorhead, P. S. Chromosome anomalies in human neoplasms with special reference to the mechanisms of polyploidization and aneuploidization in the HeLa strain. Ann. N. Y Acad. Sci. 63, 1083–1094 (1956).
Blaine Marchant, D., Soltis, D. E. & Soltis, P. S. Patterns of abiotic niche shifts in allopolyploids relative to their progenitors. N. Phytol. 212, 708–718 (2016).
Soltis, D. E., Visger, C. J. & Soltis, P. S. The polyploidy revolution then…and now: Stebbins revisited. Am. J. Bot. 101, 1057–1078 (2014).
Paterson, A. H., Bowers, J. E. & Chapman, B. A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA 101, 9903–9908 (2004).
Soltis, D. E. et al. Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 98, 704–730 (2011).
Fawcett, J. A., Maere, S. & Van de Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl Acad. Sci. USA 106, 5737–5742 (2009).
Renne, P. R. et al. Time scales of critical events around the Cretaceous-Paleogene boundary. Science 339, 684–687 (2013).
Wang, X. et al. Genome alignment spanning major poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol. Plant 8, 885–898 (2015).
Soltis, D. E. & Burleigh, J. G. Surviving the K-T mass extinction: new perspectives of polyploidization in angiosperms. Proc. Natl Acad. Sci. USA 106, 5455–5456 (2009).
Kaplan, F. et al. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 136, 4159–4168 (2004).
Chinnusamy, V., Zhu, J. K. & Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639, 39–55 (2010).
Chinnusamy, V., Zhu, J. & Zhu, J. K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451 (2007).
Kang, J. et al. Natural variation of C-repeat-binding factor (CBFs) genes is a major cause of divergence in freezing tolerance among a group of Arabidopsis thaliana populations along the Yangtze River in China. N. Phytol. 199, 1069–1080 (2013).
Yu, C. et al. Molecular breeding of water lily: engineering cold stress tolerance into tropical water lily. Hortic. Res. 5, 73 (2018).
Chai, F. et al. Comparative metabolic profiling of Vitis amurensis and Vitis vinifera during cold acclimation. Hortic. Res. 6, 8 (2019).
Zhu, J. K. Abiotic stress signaling and responses in plants. Cell 167, 313–324 (2016).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012).
Freeling, M. et al. Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15, 9 (2012).
Sun, S. et al. Alignment of common wheat and other grass genomes establishes a comparative genomics research platform. Front Plant Sci. 8, 1480 (2017).
Wang, J. et al. An overlooked paleo-tetraploidization in Cucurbitaceae. Mol. Biol. Evolution 35, 16–26 (2018).
Wang, J. et al. Hierarchically aligning 10 legume genomes establishes a family-level genomics platform. Plant Physiol. 174, 284–300 (2017).
Jiao, Y. et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13, R3 (2012).
Chalhoub, B. et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953 (2014).
Bowers, J. E., Chapman, B. A., Rong, J. & Paterson, A. H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438 (2003).
Jacomy, M., Venturini, T., Heymann, S. & Bastian, M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE 9, e98679 (2014).
Wang, X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039 (2011).
Liu, S. et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930 (2014).
Cheng, F., Wu, J. & Wang, X. Genome triplication drove the diversification of Brassica plants. Hortic. Res 1, 14024 (2014).
Kilian, J. et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363 (2007).
te Beest, M. et al. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 109, 19–45 (2012).
De Storme, N., Copenhaver, G. P. & Geelen, D. Production of diploid male gametes in Arabidopsis by cold-induced destabilization of postmeiotic radial microtubule arrays. Plant Physiol. 160, 1808–1826 (2012).
Liu, B., De Storme, N. & Geelen, D. Cold interferes with male meiotic cytokinesis in Arabidopsis thaliana independently of the AHK2/3-AHP2/3/5 cytokinin signaling module. Cell Biol. Int. 41, 879–889 (2017).
Zhao, S. et al. Development and application of KASP marker for high throughput detection of AhFAD2 mutation in peanut. Electron. J. Biotechnol. 25, 9–12 (2017).
Pecrix, Y. et al. Polyploidization mechanisms: temperature environment can induce diploid gamete formation in Rosa sp. J. Exp. Bot. 62, 3587–3597 (2011).
Vetaas, O. R. Realized and potential climate niches: a comparison of four Rhododendron tree species. J. Biogeogr. 29, 545–554 (2002).
Zhang, L., Hu, W., Gao, Y., Pan, H. & Zhang, Q. A cytosolic class II small heat shock protein, PfHSP17.2, confers resistance to heat, cold, and salt stresses in transgenic Arabidopsis. Genet. Mol. Biol. 41, 649–660 (2018).
Agarwal, P. & Khurana, P. Characterization of a novel zinc finger transcription factor (TaZnF) from wheat conferring heat stress tolerance in Arabidopsis. Cell Stress Chaperones 23, 253–267 (2018).
Wang, X. R. et al. Genome-wide identification and characterization of HSP gene superfamily in whitefly (Bemisia tabaci) and expression profiling analysis under temperature stress. Insect Sci. 26, 44–57 (2019).
Adams, K. L. & Wendel, J. F. Novel patterns of gene expression in polyploid plants. Trends Genet. 21, 539–543 (2005).
Zanne, A. E. et al. Three keys to the radiation of angiosperms into freezing environments. Nature 506, 89–92 (2014).
Banks, J. A. et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 (2011).
Zhao, Z. et al. Genome-wide expression analysis of salt-stressed diploid and autotetraploid Paulownia tomentosa. PLoS ONE 12, e0185455 (2017).
Taylor, J. S. & Raes, J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev. Genet. 38, 615–643 (2004).
Mun, J. H. et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10, R111 (2009).
Panchy, N., Lehti-Shiu, M. & Shiu, S. H. Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316 (2016).
Rieseberg, L. H. et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301, 1211–1216 (2003).
Hegarty, M. & Hiscock, S. Polyploidy: doubling up for evolutionary success. Curr. Biol. 17, R927–R929 (2007).
Force, A. et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999).
Cheng, F. et al. BRAD, the genetics and genomics database for Brassica plants. Bmc Plant Biol. 11, 136 (2011).
Albert, V. A. et al. The Amborella genome and the evolution of flowering plants. Science 342, 1467 (2013).
Lee, T. H., Tang, H., Wang, X. & Paterson, A. H. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–D1158 (2013).
Song, X. et al. Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom. Sci. Rep. 5, 14631 (2015).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Letunic, I., Doerks, T. & Bork, P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305 (2012).
Song, X. et al. Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol. Genet. Genomics 289, 541–551 (2014).
Yun, W. et al. Defective carbohydrate metabolism in mice homozygous for the tubby mutation. Physiol. Genomics 27, 131 (2006).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Tong, C. et al. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics 14, 689 (2013).
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015).
Wei, D., Cai, M. & Du, H. Criteria to evaluate the robustness of networks and the application. Xi’ Jiaotong Univ. 44, 7 (2010).
Wang, D., Zhang, Y., Zhang, Z., Zhu, J. & Yu, J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteom. Bioinforma. 8, 77–80 (2010).
Li, Z. et al. Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell 28, 326–344 (2016).
Van de Peer, Y., Fawcett, J. A., Proost, S., Sterck, L. & Vandepoele, K. The flowering world: a tale of duplications. Trends Plant Sci. 14, 680–688 (2009).
Jiao, Y., Li, J., Tang, H. & Paterson, A. H. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 26, 2792–2802 (2014).
Yang, Z., Nielsen, R., Goldman, N. & Pedersen, A. M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449 (2000).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31801856 to X.S., 2016YFD0101001 to X.W.), the Hebei Province Higher Education Youth Talents Program (BJ2018016 to X.S.), and the Natural Science Foundation of Hebei (C2017209103 to X.S.).
Author information
Authors and Affiliations
Contributions
X.S. and X.W. planned and designed this research. X.S., J.W., P.S., X.M., Q.Y., J.H., S.S., Y.L., J.Y., S.F., Q.P., T.Y., N.Y. and Y.L. contributed to the data collection and bioinformatics analysis. X.W., X.S., X.L. and A.H.P. participated in preparing and writing the paper. All authors contributed to revising the paper. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, XM., Wang, JP., Sun, PC. et al. Preferential gene retention increases the robustness of cold regulation in Brassicaceae and other plants after polyploidization. Hortic Res 7, 20 (2020). https://doi.org/10.1038/s41438-020-0253-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-0253-0
This article is cited by
-
Large-scale analysis of the ARF and Aux/IAA gene families in 406 horticultural and other plants
Molecular Horticulture (2024)
-
Whole-genome and dispersed duplication, including transposed duplication, jointly advance the evolution of TLP genes in seven representative Poaceae lineages
BMC Genomics (2023)
-
The fate of drought-related genes after polyploidization in Arachis hypogaea cv. Tifrunner
Physiology and Molecular Biology of Plants (2022)
-
Chloranthus genome provides insights into the early diversification of angiosperms
Nature Communications (2021)
-
Molecular Evolution and Local Root Heterogeneous Expression of the Chenopodium quinoa ARF Genes Provide Insights into the Adaptive Domestication of Crops in Complex Environments
Journal of Molecular Evolution (2021)