Abstract
Reporters have been widely used to visualize gene expression, protein localization, and other cellular activities, but the commonly used reporters require special equipment, expensive chemicals, or invasive treatments. Here, we construct a new reporter RUBY that converts tyrosine to vividly red betalain, which is clearly visible to naked eyes without the need of using special equipment or chemical treatments. We show that RUBY can be used to noninvasively monitor gene expression in plants. Furthermore, we show that RUBY is an effective selection marker for transformation events in both rice and Arabidopsis. The new reporter will be especially useful for monitoring cellular activities in large crop plants such as a fruit tree under field conditions and for observing transformation and gene expression in tissue culture under sterile conditions.
Similar content being viewed by others
Introduction
Various genetically encodable reporters have been developed to monitor gene expression, protein subcellular localization, protein stability, hormonal signaling, and impacts of environmental signals. The green fluorescent protein (GFP) and its derivatives such as RFP, mCherry, and YFP have many applications as reporters for gene expression or as fusion proteins1,2. Although GFP is easy to use, it needs light sources to visualize the fluorescence signals. The β-glucuronidase (GUS) reporter has been widely used in plants for monitoring gene expression patterns and as a reporter for hormonal signaling3. For example, DR5-GUS transgenic lines are commonly used to monitor auxin distribution and auxin signaling4. Luciferase is another broadly used reporter in both animals and plants5. Both GUS and luciferase require the addition of expensive substrates X-Gluc (5-Bromo-4-chloro-1H-indol-3-yl β-d-glucopyranosiduronic acid) and luciferin, respectively. Whereas the traditional reporters have been very useful, they have limitations. Fluorescent proteins are often monitored under a microscope, rendering it less useful in analyzing plants in natural growing fields or analyzing large samples such as a tree. GUS-staining is invasive and often requires sacrifice of the plants. Luciferase can be used noninvasively, but it requires a special camera and spraying the expensive substrate. It is also not very practical to use them in fields. GUS and luciferase may not be optimal for sterile conditions such as tissue culture because addition of substrates increases the chance for contamination of microbes. Therefore, there is a need to develop new reporter systems that can be widely used to monitor cellular activities noninvasively, continuously, and cost-effectively. For the past few years, gene editing has been widely used in basic research and crop improvement. A visible marker for transgenes will greatly accelerate the isolation of edited plants that no longer harbor the gene editing machinery6,7.
Plants produce many colorful compounds that potentially can serve as reporters. For example, anthocyanins display bright red-blue colors and anthocyanin-producing rice plants have been used to generate interesting patterns in rice field. However, synthesis of anthocyanins requires multiple enzymes and varies greatly among different plants8,9. It is difficult to use anthocyanin biosynthesis pathways as a universal visible reporter. Betalains are a class of plant natural products derived from the amino-acid tyrosine10,11. The bright red color seen in beets, dragon fruit, Swiss chard, and other plants is resulted from accumulation of betalains. Biosynthesis of betalains has been well studied and only needs three enzymatic reactions to convert tyrosine into betalain (Fig. 1a)12. Tyrosine is first hydroxylated on the benzene ring, resulting in l-3,4-dihydroxyphenylalanine (l-DOPA) (Fig. 1a). The reaction is catalyzed by the P450 oxygenase CYP76AD1 (Fig. 1a). l-DOPA can be further oxidized into cyclo-DOPA by CYP76AD1 (Fig. 1a). Alternatively, l-DOPA is catalyzed by l-DOPA 4,5-dioxygenase (DODA) into betalamic acid, which is subsequently condensed with cyclo-DOPA into betanidin. The condensation reaction does not require an enzyme (Fig. 1a). Finally, a sugar moiety is added to betanidin by a glucosyltransferase to generate the colorful betalain (Fig. 1a). Betalain has a very bright red color, which potentially can serve as a reporter to track gene expression or to visualize transgenic events. Because every cell contains the amino-acid tyrosine, exogenous application of tyrosine to tissues may not be required. We hypothesized that betalain would be a more convenient reporter than the aforementioned reporters. It is visible to naked eyes without any needs for special equipment. It does not require processing samples and it allows continuously monitoring events throughout the life cycle of an organism. Moreover, it is applicable to large plants grown under normal field conditions. Herein, we synthesize an artificial open reading frame named RUBY that when expressed can produce all of the enzymes required for betalain biosynthesis. We show that RUBY is a very effective marker for noninvasively selecting transformation events in both rice and Arabidopsis. Moreover, we show that RUBY can be used to visualize gene expression without any chemical treatments or special equipment, providing useful tools for visualizing gene expression in large plants under natural field growth conditions.
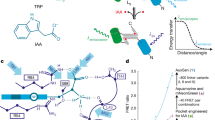
a Chemical reactions for converting tyrosine into betalain, which has a red color. Tyrosine is first oxidized by the P450 CYP76AD1 into l-3,4-dihydroxyphenylalanine (l-DOPA), which can be further converted into cyclo-DOPA by P450 CYP76AD1. In the presence of l-DOPA 4,5-dioxygenase (DODA), DOPA is oxidized and circularized into betalamic acid. Cyclo-DOPA condenses with betalamic acid, a non-enzymatic reaction, to produce betanidin. Glucoyslastion of betanidin generates the red color betalain. b A strategy for expressing the whole betalain biosynthetic pathway in a single cassette. The three betalain biosynthetic genes were fused into a single open reading frame, which can be expressed using a single promoter and terminator. Between the genes, sequences that encode 2A peptides were inserted. The 2A peptides undergo self-cleavage, thus releasing the individual enzymes for betalain biosynthesis. The betalain synthesis unit can be placed under the control of a promoter of interest. The terminator used here was the Arabidopsis HSP18.2 terminator. The open reading frame of 2A-linked betalain biosynthesis genes is named RUBY.
Results and discussion
Construction of a single open reading frame for the betalain biosynthetic pathway
Heterologous expression of CYP76AD1, DODA in tobacco, and other plants demonstrated that the betalain biosynthetic pathway can be re-constituted in plant cells13,14. In order to use betalain as a visual reporter, we need to effectively co-express the entire pathway using a single promoter. We organized CYP76AD1, DODA, and Glucosyltransferase into a single open reading frame (Fig. 1b) (Supplemental Fig. 1). The stop codons of CYP76AD1 and DODA were removed. The three genes were linked by sequences that encode 2A peptides (Fig. 1b) (Supplemental Fig. 1)15,16. Upon transcription, the single transcript, which includes the coding regions of the three enzymes, produced the three separate enzymes through either 2A-mediated self-cleavage or ribosomal “skipping” (Fig. 1b)17. The 2A system enables the expression of multiple proteins under the control of a single promoter. We name the 2A-linked unit of CYP76AD1, DODA, and Glucosyltransferase RUBY (Fig. 1b). RUBY can be expressed when a promoter is placed in front of it. The expression pattern and level of a particular gene may be inferred from the red color of betalain if the gene’s promoter is used to drive RUBY expression.
RUBY is capable of synthesizing betalain in tobacco
We first placed RUBY under the control of Cauliflower Mosaic Virus (CaMV) 35S promoter, which is a widely used constitutively strong promoter18. To test whether RUBY can produce functional enzymes for betalain synthesis, we infiltrated tobacco leaves with Agrobacteria that contain RUBY-expressing plasmid (Supplemental Fig. 2). Transient expression of RUBY led to the production of betalain in tobacco leaves, suggesting that the synthetic open reading frame RUBY can produce the functional enzymes for the synthesis of betalain. Moreover, we observed that betalain was not transported from the spots of Agrobacterium-infiltration spots to other leaves of the plant (Supplemental Fig. 2).
Synthesis of betalain by RUBY in Arabidopsis
We transformed the 35S:RUBY construct into Arabidopsis using Agrobacterium-mediated floral dipping19. Two days after floral dipping, we noticed that the transformed plants displayed patches of red color (Supplemental Fig. 3a), indicating that the RUBY cassette was functionally expressed and that RUBY may be used to monitor transient Arabidopsis transformation. Once the seeds from the Agrobacterium-dipped plants were harvested, transgenic seeds could be easily differentiated from non-transgenic seeds (Supplemental Fig. 3b). The transformed seeds had a dark red color (Supplemental Fig. 3b), demonstrating that RUBY can be used as a visual selection marker for transgenic events in Arabidopsis. We previously used mCherry as a very effective marker to select transgenic events (Gao et al., 2016), which requires a dissecting microscope with fluoresence capability. RUBY is a better option because it does not require special equipment.
The 35S:RUBY plants produced sufficient amount of betalain to become visually evident (Fig. 2a). Consistent with previous reports that CaMV 35S promoter is constitutively active, we observed red color in all tissues throughout the plant life cycle (Fig. 2a) (Supplemental Fig. 3c, d). We also expressed RUBY reporter under the control of the Maize UBIQUITIN promoter, which has been widely used to overexpress genes in monocots20. Similar to 35S:RUBY plants, UBQ:RUBY plants were also visibly red in leaves, stem, and flowers (Fig. 2b). These results clearly demonstrated that RUBY could be expressed in Arabidopsis and that our RUBY reporter was able to functionally re-constitute the betalain biosynthetic pathway.
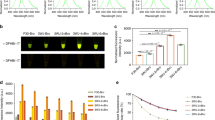
a RUBY expression driven by the CaMV 35S promoter led to a red Arabidopsis plant (right) compared with the WT plant (left). b UBIQUITIN promoter was effective in driving RUBY expression throughout the plant (right). Non-transgenic WT was shown left. c The seed specific promoter At2S3 did not lead to RUBY expression in leaves and stems. The transgenic plant (right) and non-transgenic plant (left) were indistinguishable. The siliques of At2S3:RUBY were similar to those of WT, however, when the silique was opened, the seeds of At2S3:RUBY were clearly red. Moreover, it was obvious that transgenic and no-transgenic seeds were segregating in a silique from a T1 At2S3:RUBY plant. d YUC4:RUBY plants displayed patches of red at the tip of leaves and apical region of a gynoecium. e DR5:RUBY was expressed in rice calli. The red color can be used to distinguish transgenic (red) and non-transgenic calli (white). DR5:eGFP has been used in rice calli f, but it was much more difficult to distinguish transgenic from non-transgenic using DR5:eGFP compared with RUBY. g Roots of DR5:RUBY and DR5eGFP rice plants, which had similar patterns. The three DR5:eGFP pictures were generated with the same root: bright field (left), 488 nm fluorescence field (middle), and the merged (right). h Activation of DR5 promoter in DR5:RUBY leaves was easy to observe whereas DR5:eGFP was much more difficult to detect: bright field (left), 488 nm fluorescence field (middle), and the merged (right).
We expressed RUBY using the seed specific At2S3 promoter, which we previously used to drive mCherry expression in Arabidopsis to facilitate the selection of transgenes6. As shown in Fig. 2c, the transgenic plants were indistinguishable from wild type plants. When we checked the seeds in a silique from an At2S3:RUBY T1 plant, RUBY-expressing seeds displayed strong red color, whereas the non-transgenic seeds were green (Fig. 2c). RUBY can be conveniently used to select single T-DNA insertion events by analyzing the ratio of red seeds to green seeds, which should be ~3:1 for single insertions. The At2S3:RUBY results demonstrate that RUBY could be an effective marker for Arabidopsis transformation. Furthermore, betalain was not widely transported from the sites of synthesis to other tissues as we did not see any red color in leaves (Fig. 2c). We also expressed RUBY under the control of the Arabidopsis YUC4 promoter (Fig. 2d). YUC4, which encodes a key enzyme in auxin biosynthesis, was shown to express in small regions of embryos, leaves, and flowers21,22. GUS signals were observed in leaf tips and apical region of a gynoecium in YUC4 promoter:GUS transgenic plants. We observed similar patterns of betalain production in YUC4:RUBY lines (Fig. 2d).
RUBY synthesizes betalain in rice
Unlike Arabidopsis, rice and many other plants are transformed through tissue culture and the formation of calli, which are often mosaic. A visible marker for transformation at tissue culture stage will be very useful. We placed RUBY under the control of DR5, an auxin-responsive synthetic promoter4, which was used to monitor stem cell initiation during tissue culture. We also used the rice ACTIN1 promoter, which is considered a strong promoter, to control RUBY expression. We transformed the DR5:RUBY (Fig. 2e) and OsACTIN1: RUBY (Supplemental Fig. 4) into rice calli. RUBY expression rendered calli vividly red. The presence of the RUBY gene corelated to the observed RUBY expression in rice calli (Supplemental Fig. 4). We also observed a positive correlation between the brightness of the calli the expression levels of RUBY gene (Supplemental Fig. 4). Introduction of RUBY made it easier to distinguish the transformed calli from untransformed calli (Fig. 2e). RUBY enables selection of calli that have better expression of transgenes among the hygromycin-resistant calli in the same tissue block (Supplemental Fig. 4). In comparison, DR5:eGFP has been used to visualize transgenic calli (Fig. 2f). Our RUBY system is clearer and much more convenient to operate during tissue culture condition. The DR5:RUBY and DR5:eGFP in rice roots showed similar patterns (Fig. 2g), but DR5:RUBY was easier to observe without any treatments or a change in light conditions. The advantages of DR5:RUBY over DR5:eGFP was also obvious in rice leaves (Fig. 2h). Moreover, RUBY as a visible marker was very useful for monitoring transgenes in intact adult rice plants (Supplemental Fig. 5).
We demonstrate that our synthetic cassette of betalain biosynthetic genes was able to produce betalain in Arabidopsis and in rice, providing a visible color for monitoring gene expression and plant transformation. We believe that RUBY will be very useful in large plants such as fruit trees and in field conditions. Because RUBY does not require either special equipment or expensive substrates, RUBY provides a cost-effective reporter and RUBY is a convenient alternative to the existing reporters. We envision that RUBY can be adapted for applications in some microbes and animals because the substrate tyrosine exists in all cells. For example, RUBY may provide a more convenient marker than β-galactosidase (LacZ) in yeast two-hybrid screens. Betalain is a natural product and was shown to have health benefits. Using RUBY as a reporter has less environmental and health concerns compared with antibiotic and/or herbicide resistance markers.
Materials and methods
Arabidopsis constructs and transformation
The backbone of the RUBY constructs for Arabidopsis is the plasmid pHDE, which was described previously6. We took advantage of the P2A peptide, which has the sequence of (GSGATNFSLLKQAGDVEENPGP), to link the CYP76AD1, DODA, and glucosyltransferase (GT) coding regions. The transcriptional terminator used was the HSP18.2 terminator from Arabidopsis. The 2A-linked CYP76AD1, DODA, and GT unit was named RUBY, which was cloned into pHDE along with the HSP terminator by Gibson assembly at the XbaI23. The entire sequence of RUBY and terminator was shown in Supplemental Fig. 1.
Various promoters can be cloned into the PmeI site of pHDE-RUBY to drive RUBY expression. Promoters were amplified by PCR and the primers used in this study were listed in Supplemental Table 1. We expressed RUBY using CaMV 35S, Maize UBIQUITIN promoter, At2S3 promoter, and YUC4 promoter to test whether RUBY can serve as an effective reporter. The constructs were transformed into Arabidopsis Columbia plants by Agrobacterium-mediated floral dipping19. Transgenic seeds for 35S:RUBY, UBQ:RUBY, and At2S3: RUBY were easily identified by red color. Transgenic plants for YUC4:RUBY were selected on MS medium containing hygromycin (16.7 μg/ml).
Rice constructs and transformation
The auxin-responsive promoter (DR5) was composed of a synthetic promoter with 16 repeats of core auxin response element (AuxRE) sequence (TGTCTC) linked with CaMV minimal 35S promoter4. The DR5 promoter was synthesized and then cloned into pDX218124 between BamH I and Pst I sites by Gibson assembly with primer pair Oligo15 and Oligo16 (Supplemental Table 1), resulting in the plasmid pDR5:eGFP.
To construct the pDR5:RUBY plasmid, we linked the three betalain biosynthetic genes through 2A peptides. Here, we used the F2A peptide, which has the following peptide sequence: QLLNFDLLKLAGDVESNPGP. The RUBY cassette replaced the eGFP in the pDR5:eGFP plasmid, resulting in the pDR5:RUBY. The rice version of RUBY was also detailed in the Supplemental Fig. 1. Both the DR5:RUBY and DR5:eGFP plasmids were transformed into XiaoweiNIP25 through Agrobacterium-mediated plant transformation following a protocol that was previously described26.
Quantitative analysis the expression level of RUBY
The relative expression levels of RUBY in the transgenic calli of OsACTIN1: RUBY were determined by reverse transcription quantitative PCR using the primer pair Oligo26/Oligo27. The primer pair Oligo28/ Oligo29 was specific for the rice UBIQUITIN (UBQ) gene, which served as the endogenous reference gene27. The primers were listed in Supplemental Table 1.
Data availability
The plasmids will be available at Addgene.
References
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W. & Prasher, D. C. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994).
Heim, R., Cubitt, A. B. & Tsien, R. Y. Improved green fluorescence. Nature 373, 663–664 (1995).
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987).
Sabatini, S. et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472 (1999).
Contag, C. H. & Bachmann, M. H. Advances in in vivo bioluminescence imaging of gene expression. Annu. Rev. Biomed. Eng. 4, 235–260 (2002).
Gao, X., Chen, J., Dai, X., Zhang, D. & Zhao, Y. An effective strategy for reliably isolating heritable and Cas9-free arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 171, 1794–1800 (2016).
He, Y. & Zhao, Y. Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants. aBIOTECH 1, 88–96 (2020).
Misyura, M., Colasanti, J. & Rothstein, S. J. Physiological and genetic analysis of Arabidopsis thaliana anthocyanin biosynthesis mutants under chronic adverse environmental conditions. J. Exp. Bot. 64, 229–240 (2013).
Li, P. et al. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. N. Phytol. 210, 905–921 (2016).
Strack, D., Vogt, T. & Schliemann, W. Recent advances in betalain research. Phytochemistry 62, 247–269 (2003).
Xu, J.-J., Fang, X., Li, C.-Y., Yang, L. & Chen, X.-Y. General and specialized tyrosine metabolism pathways in plants. aBIOTECH, https://doi.org/10.1007/s42994-019-00006-w (2019).
Polturak, G. & Aharoni, A. Advances and future directions in betalain metabolic engineering. N. Phytol. 224, 1472–1478 (2019).
Polturak, G. et al. Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. N. Phytol. 210, 269–283 (2016).
Polturak, G. et al. Engineered gray mold resistance, antioxidant capacity, and pigmentation in betalain-producing crops and ornamentals. Proc. Natl Acad. Sci. USA 114, 9062–9067 (2017).
Liu, Z. et al. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 7, 2193 (2017).
Wang, J. & Chen, H. A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH 1, 6–14 (2020).
Sharma, P. et al. 2A peptides provide distinct solutions to driving stop-carry on translational recoding. Nucleic Acids Res. 40, 3143–3151 (2012).
Benfey, P. N. & Chua, N. H. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250, 959–966 (1990).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Cornejo, M. J., Luth, D., Blankenship, K. M., Anderson, O. D. & Blechl, A. E. Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Biol. 23, 567–581 (1993).
Cheng, Y., Dai, X. & Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799 (2006).
Cheng, Y., Dai, X. & Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439 (2007).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Ye, R., Zhou, F. & Lin, Y. Two novel positive cis-regulatory elements involved in green tissue-specific promoter activity in rice (Oryza sativa L ssp.). Plant Cell Rep. 31, 1159–1172 (2012).
Hu, S. et al. Xiaowei, a new rice germplasm for large-scale indoor research. Mol. Plant 11, 1418–1420 (2018).
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994).
He, Y. et al. PINOID is required for formation of the stigma and style in rice. Plant Physiol. 180, 926–936 (2019).
Acknowledgements
This work was partially supported by grants from the National Transgenic Science and Technology Program (2019ZX08010-003; 2019ZX08010-001) to Y.H. and H.Z. T.Z. is a TIGS postdoctoral fellow. We thank Mr. Vitor Pinoti for helping the tobacco transformation.
Author information
Authors and Affiliations
Contributions
Y.H. and Y.Z. conceived the idea. Y.H. and H.S. conducted the rice experiments. T.Z. performed the Arabidopsis experiments. Y.H., T.Z., and Y.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, Y., Zhang, T., Sun, H. et al. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic Res 7, 152 (2020). https://doi.org/10.1038/s41438-020-00390-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-00390-1
This article is cited by
-
An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity
Nature Chemical Biology (2024)
-
Monitoring genetic transformation with RUBY visible reporter in Nicotiana tabaccum L.
Plant Cell, Tissue and Organ Culture (PCTOC) (2024)
-
Efficient genetic transformation of rice using Agrobacterium with a codon-optimized chromoprotein reporter gene (ChromoP) and introducing an optimized iPCR method for transgene integration site detection
Plant Cell, Tissue and Organ Culture (PCTOC) (2024)
-
The RUBY reporter for visual selection in soybean genome editing
aBIOTECH (2024)
-
Establishment and Advances of Third-Generation Hybrid Rice Technology: A Review
Rice (2023)