Abstract
The carotenoid isomerase gene (BoaCRTISO) of Chinese kale was targeted and edited using the CRISPR/Cas9 system in the present study. The results showed a high mutation rate (81.25%), and 13 crtiso mutants were obtained. Only two types of mutations, insertions and replacements, were found. Both the total and individual carotenoid and chlorophyll concentrations of the biallelic and homozygous mutants were reduced, and the total levels declined by 11.89–36.33%. The color of the biallelic and homozygous mutants changed from green to yellow, likely reflecting a reduction in the color-masking effect of chlorophyll on carotenoids. The expression levels of most carotenoid and chlorophyll biosynthesis-related genes, including CRTISO, were notably lower in the mutants than in the WT plants. In addition, the functional differences between members of this gene family were discussed. In summary, these findings indicate that CRISPR/Cas9 is a promising technique for the quality improvement of Chinese kale and other Brassica vegetables.
Similar content being viewed by others
Introduction
Chinese kale (Brassica oleracea var. alboglabra) is a member of the Brassicaceae family, and the main edible parts of this plant are the highly nutritious tender leaves and bolting stems1. Our previous studies have shown that Chinese kale is rich in antioxidants and anticancer compounds, including vitamin C, glucosinolates, and carotenoids2,3. Carotenoids are a class of natural pigments that play important roles in photoprotection and antioxidant processes4. Their presence affects the color of many vegetables, fruits, and flowers, including those of tomato5, carrot6, cauliflower7, watermelon8, citrus9, and lily10.
Carotenoid isomerase (CRTISO), an enzyme that acts before the bifurcation point in the carotenoid biosynthetic pathway, is responsible for catalyzing the conversion of lycopene precursors to lycopene11. Previous studies have shown that the loss of function of CRTISO results in a yellow color in several crop species, including tomato12, rice13, and Chinese cabbage14. The rice zebra mutant accumulates lycopene precursors when growing in the dark and exhibits a “zebra” phenotype when growing in the light because the expression of the CRTISO gene ZEBRA2 is reduced13. Su et al. found that the loss of function of the BrCRTISO gene caused an accumulation of lycopene precursors in and an orange phenotype of Chinese cabbage14.
Crop quality improvement is a topic of perennial importance in both food and agricultural research. However, traditional techniques for quality improvement, such as hybridization and chemical spraying, can be time consuming and inefficient. Because of its efficiency and convenience, the CRISPR/Cas9 system has been successfully used on many horticultural crop species to enhance plant resistance and improve yields. For instance, citrus resistance to canker was significantly improved by editing CsLOB1 with the CRISPR/Cas9 system15. Targeted editing of SlNPR1 was performed to alter the drought resistance of tomato16. Moreover, the heading stage of rice was accelerated, and the yield of oilseed rape was increased by editing Hds17 and ALC18, respectively. Recently, targeted editing of five tomato genes (SGR1, LCY-B1, LCY-B2, LCY-E, and Bic) were performed by Li et al.5 which blocked the biosynthesis of α- and β-carotene and increased the lycopene content by more than fivefold. Consequently, the nutritional quality of the edited tomato fruits improved. However, the application of CRISPR/Cas9 technology to the quality improvement of horticultural crops remains limited.
Recently, we established a CRISPR/Cas9 gene editing system in Chinese kale19 and used it to demonstrate functional differences among members of the PDS family, which are important genes involved in the carotenoid biosynthetic pathway20. Chinese kale is not typically colorful: the edible organs of most varieties are green, except for a few varieties that have red bolting stems21. Thus, the purpose of the present study was to perform targeted editing of BoaCRTISO using the CRISPR/Cas9 system to change the color and pigment concentrations of Chinese kale.
Material and methods
Plant materials and cultivation conditions
The typical white-flowered Sijicutiao cultivar of Chinese kale was used in this study. Sterile seedlings were grown on half-strength Murashige and Skoog (1/2 MS) media consisting of 0.8% Phytagar in a tissue culture chamber with 25/20 °C (day/night) temperatures, a 16/8 h (day/night) photoperiod, and a light intensity of 36 μmol m−2 s−1. After 7 d, cotyledons with 1–2 mm long petioles were cut and used as explants for Agrobacterium-mediated transformation.
Plasmid construction
The primers used for plasmid construction are listed in Supplementary Table S1. The sequence of the BoaCRTISO gene (GenBank accession MN810158) was analyzed by using an online analysis tool (http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR), and a target site located on exon 11 with the sequence ACGTATGGACCAATGCCAAGAGG was selected; the underlined portion of the sequence is the protospacer adjacent motif of the target. We used a vector described in a previously published study22. CRTISO-CRISPR-F and CRTISO-CRISPR-R primers were obtained by removing AGG at the end of the target site, adding ATTG at the 5′ end of the target site, and adding AAAC at the 3′ end of the target site. The primers pairs CRTISO-CRISPR-F and CRTISO-CRISPR-R were then synthesized. To produce a vector that expressed both the Cas9 protein and guide RNAs, the primers pairs CRTISO-CRISPR-F and CRTISO-CRISPR-R were annealed, after which the primers and BsaI-digested pAtU6-26-sgRNA-SK plasmids were mixed together to give rise to entry plasmids. The resulting recombinant plasmid rendered the sgRNA cassettes released by NheI and SpeI digestion. The cassettes were cloned into a pCAMBIA1300-pYAO:Cas9 binary vector previously digested by SpeI, followed by dephosphorylation. The resulting plasmids were used for transformation of wild-type (WT) plants (Supplementary Fig. S1).
Agrobacterium-mediated transformation of Chinese kale plants
The transformation of Chinese kale was performed as described in our previous study19. Explants were precultured for 3 days in MS media consisting of 0.5 mg L−1 2,4-D and 0.8% Phytagar, after which the explants were infected with the Agrobacterium strain GV3101 by immersion for 1–2 min. The explants were cocultivated with Agrobacterium for 3 d in MS media consisting of 0.03 mg L−1 naphthaleneacetic acid (NAA), 0.75 mg L−1 boric acid (BA), and 0.8% Phytagar. The explants were then transferred to MS media supplemented with 0.03 mg L−1 NAA, 0.75 mg L−1 BA, 0.8% Phytagar, 325 mg L−1 carbenicillin, and 325 mg L−1 timentin for 7 d. Hygromycin-resistant shoots that regenerated on the same media supplemented with 12 mg L−1 hygromycin B were transferred to tissue culture bottles that contained the subculture media. After 3 months, hygromycin-resistant plantlets were obtained, after which they were transplanted into trays containing a mixture of peat and vermiculite (3:1) within an artificial climate chamber with a light intensity of 80 μmol m−2 s−1, a temperature of 25/20 °C (day/night), a 12/12 h (day/night) photoperiod, and 75% relative humidity.
Determination of transformation efficiency
Genomic DNA of both hygromycin-resistant and WT plants was extracted by the standard cetyltrimethylammonium bromide (CTAB) method23, and Hyg-F and Hyg-R specific primers (Supplementary Table S1) were designed based on the hygromycin gene sequence with the vector. Genomic DNA of each transgenic plant was used as a template for PCR-based amplification to screen the positive transgenic plants. Approximately 10 monoclones of each mutant were detected by sequencing. The PCR procedure was as follows: denaturation at 95 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 30 s. The transformants with the hygromycin gene were used for further analysis19.
Detection of mutations
To evaluate mutations introduced into the CRISPR/Cas9 transgenic plants, the genomic DNA of each positive transgenic shoot was amplified using the specific primers CRTISO-CRISPR-test-F/R (Supplementary Table S1), which were designed to amplify the 730-bp flanking regions of the target site. The PCR products were then sequenced for genotypic analysis of the transgenic plants. The mutation rate was calculated, and all sequencing data were collected to analyze the mutation type19. Amino acid changes in the mutants were analyzed by DNAMAN 6.0 (Lynnon Biosoft, California, USA).
Color measurements
The colors of the mutants were determined using an NR110 colorimeter (3nh, Shenzhen, China). The colorimeter was calibrated via a standard white plate according to the manufacturer’s instructions. Three positions on the leaves and the bolting stems of each mutant were randomly selected, and the L*, a*, and b* color values were obtained. The chromaticity (L*) represents the brightness (0–100:0 = black, 100 = white). Negative chromaticity (a*) indicates green, and positive chromaticity (a*) indicates red. Similarly, negative chromaticity (b*) stands for blue, and positive chromaticity (b*) stands for yellow. The a* and b* values are balanced toward 0, which represents white.
Chlorophyll and carotenoid assays
Chlorophyll and carotenoid concentrations were determined using the methods of Shi et al.24. Two hundred milligrams of leaves or bolting stems was ground and extracted with 25 mL of acetone. The samples were sonicated for 20 min and then centrifuged at 4000 × g at room temperature for 5 min. The supernatant was filtered through 0.22 μm cellulose acetate filters and then analyzed by high-performance liquid chromatography (HPLC). HPLC analysis of the carotenoids and chlorophyll was carried out using an Agilent 1260 instrument equipped with a VWD detector. Samples (10 μL) were separated at 30 °C on a Waters C18 column (150 × 3.9 mm id; 3 μm particle size) using isopropanol and 80% acetonitrile-water at a flow rate of 0.5 mL min−1; the absorbance was measured at 448 and 428 nm.
RNA extraction and qPCR expression analysis
The CTAB method25 was used to extract the total RNA from the leaves and bolting stems of mutant and WT plants. First-strand cDNA was synthesized by reverse transcription using a PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time, Takara). The qPCR primers used for the genes involved in the biosynthesis and degradation of carotenoids and chlorophyll in Chinese kale were designed based on Brassica oleracea primer sequences retrieved from the qPCR primer database (https://biodb.swu.edu.cn/qprimerdb/), except for the CLH2, PPH, and NYC genes26 (Supplementary Table S1). qPCR was performed following the procedures and recommended conditions of the TB Green Premix Ex Taq™ II qPCR Kit (Tli RNaseH Plus, Takara). Finally, fold changes in gene expression were calculated in terms of threshold cycles using the 2−ΔΔCT method27, with the housekeeping gene β-actin28 serving as an internal reference. The expression level of BoaCRTISO in the WT bolting stems was set to one for genes that encode carotenoid biosynthesis enzymes. The expression levels of genes encoding carotenoid-degrading enzymes and chlorophyll biosynthesizing- and degrading-enzymes were calculated based on the respective expression levels of the respective genes in WT bolting stems.
Statistical analysis
All the results are shown as the means ± standard deviations (SDs) of three replicates. Statistical analysis was performed using SPSS version 18 (SPSS Inc., Illinois, USA). The data were analyzed using one-way analysis of variance, and differences were compared using the least significant difference test at a significance level of 0.05.
Results
Analysis of BoaCRTISO mutations
Twenty-three hygromycin-resistant plants were obtained from ~2000 explants. No hygromycin target bands were observed in 7 out of the 23 plants (lines 1, 2, 5, 17, 18, 22, and 23) (Supplementary Fig. S2), indicating that the target expression cassette was transferred into the other 16 lines, and the transgenic efficiency was 69.57%.
The sequencing results showed two mutation types, insertions and replacements, whereas no deletions were found. Combinatorial mutagenesis (32-bp insertions and 27-bp replacements occurring simultaneously) was predominant in these mutants, accounting for 97.40% of the total mutations, and other mutations (1- and 3-bp replacements) accounted for only 2.60% (Fig. 1a, Supplementary Fig. S3).
a CRISPR/Cas9-induced mutations in Chinese kale. The target sequence is indicated in blue, the PAM sequence (NGG) is underlined in red, the mutated bases are indicated in red font, and the asterisks indicate spacing between bases. WT wild-type plant, M # number of mutants, i # number of base insertions, r # number of base replacements. M1 is a biallelic mutant with two kinds of sequences. M3, M6, and M16 are homozygous mutants with one kind of sequence. b Number of different mutant types. c Frequency of different mutant types
Thirteen out of the 16 transgenic plants harbored mutations, including one biallelic mutant that accounted for 6.25% of the total number of mutations among the transgenic plants, five homozygous mutants that accounted for 31.25%, six heterozygous mutants that accounted for 37.5%, and one chimeric mutant that accounted for 6.25%. Thus, biallelic and homozygous mutants accounted for 37.5% of the transgenic plants, and the percentage of transgenic plants with mutations was as high as 81.25% (Fig. 1). The base mutations caused several changes in the translation of the amino acids (Supplementary Fig. S4). The biallelic mutant M1 and the homozygous mutants M3, M6, and M16 were selected for subsequent studies.
Color of crtiso mutants
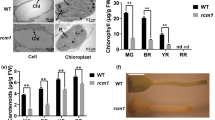
Marked differences in color were observed between the crtiso mutants and WT plants. The leaves and bolting stems of the four tested mutants were yellow, with M6 being the most yellow (Fig. 2). The CILAB color parameters of the leaves and bolting stems of the WT and mutant plants were measured, the results of which are shown in Fig. 2b. The variation in chromatic parameters in the leaves and bolting stems of the four mutants was consistent. Specifically, the values of L* and b* were significantly higher than those of WT plants, while the values of a* were significantly lower. For example, the yellowness parameters (b*) of the leaves and bolting stems of M6 were 2.69- and 2.07-fold higher than those of WT plants (Fig. 2b).
a Phenotype of the crtiso mutants and wild-type plants at 6 weeks after transplanting. WT wild-type plant, M1 biallelic plant, M3, M6, M16 homozygous plants. b Color parameters of the crtiso mutants and wild-type plants at 6 weeks after transplanting. The data are expressed as the means of three replicates. The same letter in the same column indicates no significant differences among values (p < 0.05) according to the LSD test
Pigment concentrations of crtiso mutants
Six individual pigments were measured: four carotenoids (β-carotene, violaxanthin, neoxanthin, and lutein) and two types of chlorophyll (chlorophyll a and chlorophyll b). The concentrations of individual and total carotenoids and chlorophyll in the leaves were significantly higher than those in the bolting stems. Chlorophyll a was the predominant pigment, while lutein was the major carotenoid in both the leaves and bolting stems (Fig. 3). The total carotenoid and chlorophyll concentrations in the mutants were significantly lower than those in the WT plants: the concentrations were more than 16% and 18% lower in the leaves, respectively, and more than 11% and 29% lower in the bolting stems, respectively. In particular, the M6 plant with the most obvious yellow phenotype had an ~25% lower total carotenoid concentration and a more than 30% lower total chlorophyll concentration in both the leaves and bolting stems.
a Concentrations of carotenoids and chlorophyll in the leaves of the mutants and WT plants at 6 weeks after transplanting. b Concentrations of carotenoids and chlorophyll in the bolting stems of the mutants and WT plants at 6 weeks after transplanting. WT wild-type plant, M1 biallelic mutant, M3, M6, M16 homozygous mutants. The data are expressed as the means ± SDs. The same letter in the same histogram indicates that there is no significant difference between the values tested, according to the LSD (p < 0.05)
The patterns of variation of individual pigments differed among the different organs. All pigments in the M6 leaves significantly decreased by more than 20% compared with those in the WT leaves. In particular, the violaxanthin concentration in the M6 leaves was 0.64 mg g−1 DW, 40% of that in the WT leaves. The concentrations of most pigments in the leaves of M1, M3, and M16 were also significantly lower than those in the WT leaves, although the differences in violaxanthin, β-carotene, and neoxanthin were not significant. The concentrations of lutein, chlorophyll a, and chlorophyll b in the bolting stems of the mutants were significantly lower than those of WT plants (by 16–32%, 23–36%, and 21–36%, respectively), whereas there were no significant differences in bolting stem concentrations of β-carotene, neoxanthin, or violaxanthin.
Carotenoid- and chlorophyll-related gene expression levels in crtiso mutants
In WT plants, the expression levels of carotenoid biosynthesis-related genes in the leaves were substantially higher than those in bolting stems. The genes with the highest and lowest expression levels in the leaves were ZEP1 and NXS, respectively, and they differed in their expression by ~50-fold. The genes with the highest and lowest expression levels in the bolting stems were ZEP2 and VDE, respectively, and they differed in their expression by ~350-fold. Moreover, marked differences in gene expression were found among different members of the same gene family. The members with the highest expression levels in the PSY, PDS, and LCYe gene families were PSY1, PDS1, and LCYe2, respectively (Fig. 4).
The leaves and bolting stems of mutants and WT plants were sampled 6 weeks after transplanting. The main axis represents the amount of gene expression in the leaves, and the secondary axis represents the amount of gene expression in the bolting stems. WT wild-type plant, M1 biallelic mutant, M3, M6, M16 homozygous mutants. GGPP geranylgeranyl diphosphate, PSY phytoene synthase, PDS phytoene desaturase, ZDS ζ-carotene desaturase, Z-ISO ζ-carotene isomerase, CRTISO carotenoid isomerase, LCYe lycopene ε-cyclase, LCYb lycopene β-cyclase, ε-OHase ε-carotene hydroxylase, β-OHase β-carotene hydroxylase, VDE violaxanthin de-epoxidase, ZEP zeaxanthin epoxidase, NXS neoxanthin synthase, CCD carotenoid cleavage dioxygenase
After the targeted editing, the CRTISO gene expression in the leaves of the crtiso mutants was consistently downregulated. CRTISO expression was lowest in the M6 leaves, equal to 40.81% of that in the WT leaves. The CRTISO expression in the bolting stems of the mutants was also downregulated; M1 had the lowest CRTISO expression in the bolting stems—only 35% of that in the WT bolting stems (Fig. 4).
The expression levels of all carotenoid biosynthesis-related genes were measured, and the results indicated that the BoaCRTISO mutation led to reduced expression of most carotenoid biosynthesis-related genes in the leaves and bolting stems, although the degree of reduction varied widely (Fig. 4). Take M6 as an example. The expression of ZEPs in M6 leaves was downregulated by more than 90% compared with that in WT leaves, and the expression levels of most carotenoid biosynthesis-related genes (PSY1, PSY2, PDS1, ZDS, CRTISO, LCYb, LCYe1, LCYe2, β-OHase, ε-OHase, and VDE) were downregulated by 60–80%, whereas the expression levels of PDS2 and NXS were downregulated by 30.62% and 40.51%, respectively. On the other hand, the expression of some genes whose products act upstream of the carotenoid biosynthetic pathway were upregulated rather than downregulated in the leaves and bolting stems of the mutants, although the upregulated genes were not identical in the leaves or bolting stems of a given mutant. Specifically, the expression levels of PSY1 (M3), PSY3 (M3 and M6), and PDS2 (M1, M3, and M16) were clearly upregulated in the leaves, whereas the expression levels of PSY3 (M1, M3, and M16) and ZDS (M6) were upregulated in the bolting stems. The expression level of CCD1 in the crtiso mutant decreased significantly, except for the upregulation in M6 bolting stems. Nevertheless, the expression levels of CCD4 were too low to be detected in both the WT and mutants.
The expression levels of chlorophyll biosynthesis-related genes in the leaves and bolting stems of crtiso mutants were generally downregulated (Fig. 5). The expression of chlorophyll biosynthesis-related genes in the leaves of M6 was reduced by more than 65%. However, some chlorophyll biosynthesis-related genes were upregulated. These genes included ALAD and ChlI in M1 leaves; ALAD, ChlI, and CS in M3 leaves; and ChlI in M6 bolting stems. The expression levels of genes encoding chlorophyll-degrading enzymes were also analyzed. The expression levels of CLH1 in the leaves and bolting stems of all the mutants were upregulated, and the expression levels of NYC in the leaves and bolting stems were downregulated and upregulated, respectively. The expression levels of other genes encoding chlorophyll-degrading enzymes in the leaves and bolting stems varied from mutant to mutant (Fig. 5).
The leaves and bolting stems of mutants and WT plants were sampled 6 weeks after transplanting. The main axis represents the amount of gene expression in the leaves, and the secondary axis represents the amount of gene expression in the bolting stems. WT wild-type plant, M1 biallelic mutant, M3 M6, M16 homozygous mutants. ALAD 5-aminolevulinic acid dehydratase, HemE1 glutamyl tRNA reductase, ChlI magnesium-chelatase I, ChlD magnesium-chelatase D, ChlH magnesium-chelatase H, CS chlorophyll synthase, CLH chlorophyllase, PaO pheide a oxygenase, PPH pheophytinase, RCCR red Chl catabolite reductase, NYC nonyellow coloring
In summary, the results can be presented as a pattern, as shown in Fig. 6. The inhibition of BoaCRTISO gene expression in the crtiso mutants led to the downregulation of expression levels of carotenoid and chlorophyll biosynthesis-related genes, as well as carotenoid and chlorophyll contents, which led to the yellowing of the crtiso mutants.
The solid frames indicate the presence of the substance in Chinese kale, and the dashed frames indicate the absence of the substance in Chinese kale. The blue genes were downregulated in the mutants, the red genes were upregulated in the mutants, and the black genes did not change significantly in terms of their expression. The down arrow next to a pigment indicates a decrease in its content. ⊥ indicates suppression
Discussion
In this study, the carotenoid isomerase gene from Chinese kale (BoaCRTISO) was targeted and edited using the CRISPR/Cas9 system and Agrobacterium-mediated stable transformation. As expected, 13 Chinese kale mutants were obtained, and the transgenic efficiency and mutation rates were 69.57% and 81.25%, respectively. The efficiency was relatively high compared with that in other plant species, such as tomato16, rice18, and watermelon29. The percentage of biallelic and homozygous mutants, the two most expected types of CRISPR/Cas9 mutants, reached 37.5%, which was substantially higher than that in previous reports16,29. Furthermore, BoaCRTISO was knocked down rather than knocked out in our study. Carotenoids are precursors of abscisic acid, which is essential for plant growth and stress resistance. Therefore, the knockdown of BoaCRTISO may be a better outcome than its knockout, as the latter may severely affect plant growth, development, and resistance to abiotic and biotic stress. These findings suggest that the CRISPR/Cas9 system is a promising and efficient technique for quality improvement in Chinese kale.
The concentrations of carotenoids and chlorophyll decreased in the crtiso mutants. The mutants also exhibited a yellow color, especially M6 (Fig. 2). In a previous study, the chlorophyll content of cucumber mutants decreased significantly, resulting in yellow leaves, which is consistent with our results30. We speculate that this is due to reduced chlorophyll concentrations and a concomitant reduction in their masking effect on carotenoid color. The color of a horticultural commodity is a primary characteristic perceived by consumers31,32. Vegetables with a higher chroma value and more vivid colors are typically judged to be fresher and of better quality than vegetables with dull colors33. The perception of color also affects other sensory perceptions, including taste34. Therefore, crtiso mutants of Chinese kale with a new color will likely have better market prospects in the future.
Carotenoids are crucial components of light-harvesting antenna complexes and play important roles in photosynthesis and photoprotection, especially the photoprotection of chlorophyll35. Our results showed that trends in chlorophyll reduction in the mutants were consistent with those of carotenoid reduction and were similar to those observed following virus-induced gene silencing of LCYe in tobacco plants24. This result may reflect photooxidative damage to chlorophyll caused by the reduction in the carotenoid content35. The chlorophyll content is influenced by the expression levels of genes related to chlorophyll biosynthesis and degradation36. In this study, the expression levels of chlorophyll biosynthesis-related genes significantly decreased, and the expression levels of several genes encoding chlorophyll-degrading enzymes were upregulated or unchanged in the crtiso mutants. In particular, the expression level of CLH1, a key gene that encodes a chlorophyll-degrading enzyme26, was notably induced in the mutants compared to the WT plants. All these changes resulted in a decrease in chlorophyll content.
CRTISO has emerged as a key regulatory step in the carotenoid biosynthetic pathway. When CRTISO expression is altered, the transcript level of other carotenoid genes can also be regulated37. In our study, the expression of most carotenoid biosynthesis-related genes in the crtiso mutants was downregulated. Similar results were found in the “zebra” mutant of rice: reduced CRTISO expression resulted in relatively low expression of all carotenoid genes except VDE13. CCD1 and CCD4 are the key genes that encode carotenoid-degrading enzymes in Brassica38. In the present study, the expression level of the CCD1 gene was also significantly reduced, which may be a response to the decrease in the expression levels of carotenoid biosynthesis genes. In the crtiso mutants of Chinese kale, some genes differed significantly in their expression between the leaves and bolting stems. For example, the expression of ZEP1 in the mutants was downregulated in the leaves but upregulated in the bolting stems. In the Chinese cabbage crtiso mutants, gene expression patterns in the inner, middle and outer leaves were not completely consistent14. These results suggest that the expression and regulatory patterns of genes differ among tissues.
The members of a gene family, having significant similarities in both structure and function, encode similar protein products. However, they are usually located at different positions on the same chromosome or on different chromosomes, each with different regulatory patterns and functions39. In our previous study, the two members of the BoaPDS gene family in Chinese kale (BaPDS1 and BaPDS2) played different and indispensable roles, and their functions were partially complementary20. In this study, knockdown of BoaCRTISO resulted in downregulation of most carotenoid biosynthesis-related genes. However, some genes, such as PSY3 and PDS2, had higher expression levels in the mutants. These results suggest that when the expression of one gene family member is inhibited, another member may be induced as a compensatory mechanism. However, the functional differences between these gene family members have not been determined and require further study.
In conclusion, this study used the CRISPR/Cas9 system to edit the BoaCRTISO gene of Chinese kale, resulting in the production of 13 mutants of the biallelic, homozygous, heterozygous, and chimeric types. Only insertion and replacement mutations were observed. The expression levels of carotenoid and chlorophyll biosynthesis-related genes in the crtiso mutants were significantly downregulated. Inhibition of CRTISO expression affected both the carotenoid and chlorophyll pathways, leading to decreased carotenoid and chlorophyll concentrations and creating a new yellow color of Chinese kale, with improved market prospects. The CRISPR/Cas9 system is therefore a promising technique for crop quality improvement.
References
Sun, B. et al. Molecular cloning and expression analysis of the ζ-carotene desaturase gene in Chinese kale (Brassica oleracea var. alboglabra Bailey). Hortic. Plant J. 4, 94–102 (2018).
Chen, J. et al. Assessment of glucosinolates in Chinese kale by near-infrared spectroscopy. Int. J. Food Prop. 17, 1668–1679 (2014).
Sun, B., Yan, H. Z., Liu, N., Wei, J. & Wang, Q. M. Effect of 1-MCP treatment on postharvest quality characters, antioxidants and glucosinolates of Chinese kale. Food Chem. 131, 519–526 (2012).
Guo, X. Y., Song, C. K., Ho, C. T. & Wan, X. C. Contribution of L-theanine to the formation of 2, 5-dimetheylpyrazine, a key roasted peanutty flavor in Oolong tea during manufacturing processes. Food Chem. 263, 18–28 (2018).
Li, X. D. et al. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9, 559 (2018).
Ma, J., Li, J. W., Xu, Z. S., Wang, F. & Xiong, A. S. Transcriptome profiling of genes involving in carotenoid biosynthesis and accumulation between leaf and root of carrot (Daucus carota L.). Acta Biochim. Biophys. Sin. 50, 481–490 (2018).
Lu, S. et al. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18, 3594–3605 (2006).
Lv, P., Li, N., Liu, H., Gu, H. H. & Zhao, W. E. Changes in carotenoid profiles and in the expression pattern of the genes in carotenoid metabolisms during fruit development and ripening in four watermelon cultivars. Food Chem. 174, 52–59 (2015).
Kato, M. et al. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 134, 824–837 (2004).
Yamagishi, M., Kishimoto, S. & Nakayama, M. Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed. 129, 100–107 (2010).
Yuan, H., Zhang, J., Nageswaran, D. & Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2, 15036 (2015).
Isaacson, T., Ronen, G., Zamir, D. & Hirschberg, J. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 14, 333–342 (2002).
Chai, C. L. et al. ZEBRA2, encoding a carotenoid isomerase, is involved in photoprotection in rice. Plant Mol. Biol. 75, 211–221 (2011).
Su, T. B. et al. Loss of function of the carotenoid isomerase gene BrCRTISO confers orange color to the inner leaves of Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol. Biol. Rep. 33, 648–659 (2015).
Jia, H. G. et al. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823 (2016).
Li, R. et al. CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 19, 38 (2019).
Braatz, J. et al. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174, 935–942 (2017).
Li, X. F. et al. High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genom. 44, 175–178 (2017).
Sun, B. et al. CRISPR/Cas9-mediated mutagenesis of homologous genes in Chinese kale. Sci. Rep. 8, 16786 (2018).
Sun, B. et al. Functional differences of BaPDS1 and BaPDS2 genes in Chinese kale. R. Soc. Open Sci. 6, 190260 (2019).
Lei, J. J., Chen, G. J., Chen, C. M. & Cao, B. H. Germplasm diversity of Chinese kale in China. Hortic. Plant J. 3, 101–104 (2017).
Yan, L. H. et al. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823 (2015).
Doyle, J. J. & Doyle, J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15 (1987).
Shi, Y. M. et al. Molecular cloning and functional characterization of the lycopene ε-cyclase gene via virus-induced gene silencing and its expression pattern in Nicotiana tabacum. Int. J. Mol. Sci. 15, 14766–14785 (2014).
Chen, Q. et al. An alternative cetyltrimethylammonium bromide-based protocol for RNA isolation from blackberry (Rubus L.). Genet. Mol. Res. 11, 1773–1782 (2011).
Hasperué, J. H., Gómez-Lobato, M. E., Chaves, A. R., Civello, P. M. & Martínez, G. A. Time of day at harvest affects the expression of chlorophyll degrading genes during postharvest storage of broccoli. Postharvest Biol. Technol. 82, 22–27 (2013).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Büchert, A. M., Civello, P. M. & Martínez, G. A. Effect of hot air, UV-C, white light and modified atmosphere treatments on expression of chlorophyll degrading genes in postharvest broccoli (Brassica oleracea L.) florets. Sci. Hortic. 127, 214–219 (2011).
Tian, S. W. et al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 36, 399–406 (2017).
Miao, H. et al. Changes of the photosynthetic pigment and differential expression of the correlated genes in a chlorophyll-deficient cucumber mutant (Cucumis sativus L.). Sci. Agric. Sin. 43, 4027–4035 (2010).
Gu, H. H. et al. Characterization of the appearance, health-Promoting compounds, and antioxidant capacity of the florets of the loose-curd cauliflower. Int. J. Food Prop. 18, 392–402 (2015).
Guiné, R. P. F. & Barroca, M. J. Effect of drying treatments on texture and color of vegetables (pumpkin and green pepper). Food Bioprod. Process. 90, 58–63 (2012).
Lee, S. M., Lee, K. T., Lee, S. H. & Song, J. K. Origin of human colour preference for food. J. Food Eng. 119, 508–515 (2013).
Manninen, H., Paakki, M., Hopia, A. & Franzén, R. Measuring the green color of vegetables from digital images using image analysis. LWT Food Sci. Technol. 63, 1184–1190 (2015).
Nisar, N., Li, L., Lu, S., Khin, N. C. & Pogson, B. J. Carotenoid metabolism in plants. Mol. Plant 8, 68–82 (2015).
Koichi, K. & Tatsuru, M. Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 1811, 1–17 (2016).
Soufflet-Freslon, V. et al. Functional gene polymorphism to reveal species history: the case of the CRTISO gene in cultivated carrots. PloS ONE 8, e70801 (2013).
Zhang, B. et al. Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. N. Phytol. 206, 1513–1526 (2015).
Di, F. F. et al. Genome-wide analysis of the PYL gene family and identification of PYL genes that respond to abiotic stress in Brassica napus. Genes 9, 156 (2018).
Acknowledgements
We are grateful to Prof. Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for kindly providing the plasmid used in this study. This work was supported by the National Natural Science Foundation of China (31500247), the Ministry of Agriculture of China (2016ZX08009003-001), the Technology Innovation Program of Zhejiang Province (ZJWR0102001), and Sichuan Science and Technology Program (2018NZ0081).
Author information
Authors and Affiliations
Contributions
Q.-M.W., H.-R.T., F.Z., and B.S. conceived and designed the experiments; M.J., Y.J., W.-L.H., Q.Y., Q.C., and Y.W. performed the experiments; H.Z., A.-H.Z., Y.-T.Z., Y.-X.L., and X.-R.W. analyzed the data; and B.S., M.J., H.Z., and F.Z. wrote the paper. All the authors have given final approval for publication.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, B., Jiang, M., Zheng, H. et al. Color-related chlorophyll and carotenoid concentrations of Chinese kale can be altered through CRISPR/Cas9 targeted editing of the carotenoid isomerase gene BoaCRTISO. Hortic Res 7, 161 (2020). https://doi.org/10.1038/s41438-020-00379-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-00379-w
This article is cited by
-
CRISPR-Cas technology secures sustainability through its applications: a review in green biotechnology
3 Biotech (2023)
-
The application of CRISPR/Cas technologies to Brassica crops: current progress and future perspectives
aBIOTECH (2022)
-
An apple (Malus domestica) AP2/ERF transcription factor modulates carotenoid accumulation
Horticulture Research (2021)
-
Comparative transcriptomic and metabolomic analyses of carotenoid biosynthesis reveal the basis of white petal color in Brassica napus
Planta (2021)
-
Improvement of glucosinolates by metabolic engineering in Brassica crops
aBIOTECH (2021)