Abstract
The plant immune system is divided into two branches; one branch is based on the recognition of pathogen-associated molecular patterns (PAMP-triggered immunity), and the other relies on pathogenic effector detection (effector-triggered immunity). Despite each branch being involved in different complex mechanisms, both lead to transcription reprogramming and, thus, changes in plant metabolism. To study the defense mechanisms involved in the Brassica oleracea–Xanthomonas campestris pv. campestris (Xcc) interaction, we analyzed the plant transcriptome dynamics at 3 and 12 days postinoculation (dpi) by using massive analysis of 3′-cDNA ends. We identified more induced than repressed transcripts at both 3 and 12 dpi, although the response was greater at 12 dpi. Changes in the expression of genes related to the early infection stages were only detected at 12 dpi, suggesting that the timing of triggered defenses is crucial to plant survival. qPCR analyses in susceptible and resistant plants allowed us to highlight the potential role of two calcium-signaling proteins, CBP60g and SARD1, in the resistance against Xcc. This role was subsequently confirmed using Arabidopsis knockout mutants.
Similar content being viewed by others
Introduction
Plant leaves are relatively isolated from the environment by physical barriers (i.e., the cuticle) that prevent desiccation and the penetration of phytopathogens. However, leaf metabolism requires gas and water interchange with the environment through pores that interrupt this barrier surface. These pores (stomata or hydathodes on the leaf margin), together with wounds in leaf tissues, are the entry points of pathogenic bacteria to the intercellular space where they can proliferate.
Unlike metazoans, plants lack mobile defender cells and an adaptive innate immune system, so plant defense relies on the capacity of individual cells to sense and respond to pathogens by reprogramming cell metabolism to induce the expression of defense genes1. Jones and Dangl2 model the plant cell immune system in a zigzag response organized at two different levels. The early basal response or PTI (Pathogen-associated molecular pattern (PAMP)-triggered immunity) is triggered upon the recognition of PAMPs on the cell surface and can prevent pathogen colonization of the cell3. However, compatible pathogens can overcome this first barrier of the immune system and trigger a second level of defense called ETI (effector-triggered immunity), mediated by NB-LRR proteins, which occurs mainly intracellularly2,4. The burst of these mechanisms of defense has also been associated with the activation of HR (hypersensitive response) and SAR (systemic acquired resistance)5. Several studies have revealed a high percentage of overlapping networks between the PTI and ETI phases6,7 to the extent that most authors consider the PTI a weak variant of ETI8. However, Pombo et al.9 reported that as little as 14% of the transcriptomic changes occur in response to the attack of Pseudomonas syringae on tomato plants are common between PTI and ETI responses. This apparent contradiction could be explained by the use of different plant-pathogen systems. Tsuda and Katagari3 reviewed the plant mechanisms in the response to different bacterial PAMPs and effectors, concluding that different PAMPs trigger the PTI response through common signaling pathways, whereas the cellular response to different pathogen effectors diverges among microbial types.
The immune response burst is driven by extensive transcriptional reorganization. Early response to pathogen recognition involves an increase in cytosolic secondary messengers to amplify the immune response through the activation of transcription factors. Increased intercellular Ca+2 levels along with ROS production play a pivotal role in this response. Many Ca+2 sensors have been described in land plants that perceive changes in cytosolic Ca+2 levels and transduce it into a downstream signaling response10. Intracellular Ca+2 sensors are represented by three families, i.e., calmodulin (CaM) and calmodulin-like proteins (CMLs), Ca+2-dependent protein kinases (CPKs) and Ca+2 and calmodulin dependent protein kinases (CCaMK)11. These three families can be grouped into two types: sensor relays (CaM and CMLs) and sensor responders (CPKs and CCaMK). Sensor relays function through bimolecular interactions, whereas sensor responders function through intramolecular interactions12. However, the mechanism by which plant sensors modulate the plant response to different stimuli remains elusive. The specificity of the Ca+2-mediated response can be reached at several levels (for review see ref. 13), which includes the spatial and temporal transcriptional regulation of genes involved in the Ca+2 downstream signaling network.
With the aim of deciphering the molecular mechanisms involved in the response to bacterial pathogenesis, we investigated the transcriptome dynamics of Brassica oleracea in response to Xanthomonas campestris pv. campestris (Xcc) race 1 infection at 3 and 12 days after inoculation.
Results
Global changes in the B. oleracea transcriptome after Xcc infection
To identify the genes that were upregulated and downregulated after Xcc infection, we performed a MACE analysis. We selected two different time points (3 and 12 dpi) based on previous observations. At 3 dpi, small necrotic lesions were observed at the inoculation points, whereas at 12 dpi, lesions reached the leaf edge and the midrib (Fig. S1). Statistical analysis was carried out within each time point to compare the transcriptomic expression of inoculated vs. control B. oleracea plants. An average of 217,132 tag sequences was read from each sample, and 77,637 of them were unambiguously annotated to the databases. We identified a higher transcriptomic response at 12 than at 3 dpi, albeit in both harvest times, the number of induced transcripts was higher than the number of repressed transcripts (Fig. 1a). Among the differentially expressed transcripts between the conditions, 37 were coregulated at both times (Fig. 1b). We identified three major functional groups among these genes. The major group corresponded to transcripts involved in phytohormone metabolism, which included important genes in the synthesis (lipoxygenases (LOX) or allele oxide synthase (AOS)) and perception (JAZ-proteins) of jasmonic acid (JA) and two methyl-transferases involved in the synthesis of methyl salicylate, an active form of salicylic acid (SA). It is commonly accepted that phytohormones are rapidly and transiently activated after a pathogenic attack; however, we found marked levels of these transcripts at 12 dpi. The other two groups encompassed typical genes involved in plant defense mechanisms (pathogenesis-related (PR) proteins and secondary metabolism activation, such as GSLs and flavonoids).
Altered pathways during early bacterial infection
The transcriptomic response during the first days after bacterial inoculation was characterized by the activation of defense genes (Fig. 2). Eight plant PR proteins were activated. Six of them belonged to the proteinase inhibitor class (PIs, PR-6 family), and the other two were plant defensins (PR-12 family). Likewise, Xcc infection caused a vast array of molecular reactions, including the accumulation of plant defensive secondary metabolites. In this case, different genes implicated in GSL production/degradation were upregulated. Some of these genes were involved in the biosynthesis of the core GSL structure, such as CYP79B3 and CYP79B2, which encode the enzymes responsible for the step from l-tryptophan to indole-3-acetaldehyde oxime during the synthesis of glucobrassicin (GBS) and its derivatives. Furthermore, one gene encoding a myrosinase-associated protein was overexpressed, suggesting that the GSL hydrolysis system was activated.
Transcriptional reprogramming at 12 dpi
Twelve days after Xcc colonization, plants exhibited cell death at the infection site followed by the spread of chlorosis and secondary necrosis to the surrounding uninfected tissue. In addition to the processes triggered at 3 dpi, a broad range of other complex mechanisms were activated at 12 dpi (Fig. 3). It is commonly accepted that to trigger the basal defense mechanisms, plants need to detect the presence of the pathogen or the damage produced due to pathogen activity. Our results showed that B. oleracea activated the transcription of several kinds of receptors at 12 dpi. Among the different receptor-like genes overexpressed, ~40 belonged to the receptor-like kinase (RLK) gene family, one of the largest gene families encoded by plant genomes14. In addition to RLK induction, other related perception mechanisms were activated at this infection point. The intracellular nucleotide-binding/leucine-rich-repeat (TIR-NB-LRR) proteins are involved in the recognition of pathogen effectors or their activity, and therefore, subsequent ETI activation15. Surprisingly, whereas the NB-LRR at1g72870-homolog gene was significantly overexpressed, NB-LRR at5g18350-homolog was repressed. These results indicate a simultaneous induction of factors related to the two branches of plant immunity.
Our results showed that the transcription of different genes encoding redox state-related enzymes was modified by Xcc infection, such as several thioredoxin transferases and peroxidases (class III peroxidases or ascorbate peroxidases). Both ascorbate peroxidase genes detected were repressed. In addition, 17 genes encoding calcium-signaling proteins were overexpressed. Most of these proteins are calcium sensors, which are essential factors for Ca+2 transport11. Members of the two calcium sensor types (sensor relays and sensor responders) were upregulated during the infection progression.
Analysis of the calcium-signaling response to Xcc infection
To further investigate calcium signaling in the B. oleracea-Xcc compatible interaction, we evaluated the relationship between the calcium-signaling proteins induced at 12 dpi by using the webtool STRING (v10.5) and setting other known calcium sensors in Arabidopsis as queries (Fig. 4a). The protein association network obtained showed a main functional module that formed a tightly connected cluster. Most of the calcium-signaling proteins differentially expressed between conditions (11 of 17 genes) were responsible for the formation of this major functional module, suggesting that they are part of the same highly connected signaling pathway. To confirm the MACE results, quantitative reverse-transcription-PCR (RT-qPCR) was employed to analyze the expression patterns of 10 of these 17 genes. This selection encompassed members of all known calcium sensor families and other proteins directly related to them. We obtained consistent results for eight of them, and only three genes presented significant differences between conditions, all of which showed a clear tendency (Fig. 4b).
a Protein association network built with all induced calcium-signaling proteins and the known calcium sensors in Arabidopsis. Bolded names are the differentially expressed genes detected at 12 dpi. Line thickness indicates the strength of the data support, which is based on experimental results and coexpression analysis. b Relative expression patterns of different calcium-signaling-related genes from the “Early Big” inbred line at 12 dpi. Data are the average of three biological replicates ± SE. *P-value < 0.05 (Student’s t test, Control vs. Inoculated)
In addition, the MACE results showed that the expression of four genes classified as CaM-binding proteins changed at 12 dpi. Among them, CAM-BINDING PROTEIN 60 g (CBP60g) and SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) are highlighted since they are two master transcription factors (TFs) of plant immunity. According to the literature, these genes have a partially redundant role16.
Expression of SARD1 and CBP60g in compatible and incompatible Xcc–B. oleracea interactions
To further study the roles of CBP60g and SARD1 in resistance to Xcc pathogenesis, we investigated their expression patterns in two different B. oleracea genotypes, one compatible (“Early Big”, used for the MACE analyses) and one incompatible (Badger Inbred-16), at 1, 2, 3, and 12 dpi (Fig. 5). The SARD1 qPCR results showed a clear difference between the two genotypes. The susceptible line showed a unique overexpression peak at 12 dpi, while the expression level in the resistant line was higher and constant, showing even a slight decrease at 12 dpi. The comparison of CBP60g expression between lines offered a striking image. The expression patterns formed a specular image in both genotypes. The expression of CBP60g was low during all days postinoculation (dpi) analyzed in the susceptible line, while Badger Inbred-16 maintained a constant and higher level of CBP60g expression. Therefore, it appears that the susceptible line is not able to activate CBP60g expression or not at a sufficient amount, and therefore, this calcium-signaling branch cascade is interrupted.
Relative expression patterns of SARD1 and CBP60g in two different B. oleracea lines (“Early Big” and Badger Inbred-16, susceptible and resistant to Xanthomonas campestris pv. campestris, respectively) at four different dpi. Data are the average of three biological replicates. FC was calculated as inoculated/control. Propagation of error was used to calculate uncertainty
Role of SARD1 and CBP60g in Xcc resistance
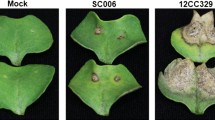
To determine whether the role of SARD1 and CBP60g is essential to promote plant resistance to Xcc, we evaluated the response of two Arabidopsis mutants, sard1-1 and cbp60g-1, to infection. We carried out the inoculation with the Xcc race 3 strain HRI5212 since the Arabidopsis plants did not show any symptoms to the infection with Xcc race 1 (even at the susceptible ecotype Sf2)17 (data not shown). Typical Xcc necrotic symptoms were clearly visible on the mutant leaves 7 days post infection, whereas Col-0 (wt) barely showed necrotic lesions (Fig. 6a). The area of the infected region was measured using ImageJ software. The statistical analysis of these data confirmed that the necrotic lesions were significantly larger in the two mutants than in wt (Fig. 6b). This result clearly supports our hypothesis that the SARD1 and CBP60g genes play essential roles in plant resistance to Xcc.
Discussion
To investigate the dynamics of the transcriptional response of B. oleracea plants challenged with Xcc, we used MACE technology, a high-resolution and cost efficient RNA-seq variant. Although extensive transcriptomic plant reorganization has been demonstrated to take place a few hours after pathogen perception, we observed a higher response at 12 dpi than at 3 dpi. This could be explained since the plant genotype used in this analysis is a compatible host for Xcc infection. Although susceptible hosts possess basal defense mechanisms, this response could be delayed in comparison with the response of an incompatible host. Generally, this delay is produced by the host’s inability to immediately recognize the invader and therefore produce the proper response18. Thus, 3 dpi may not be enough to observe a complete elicited response in our host-pathogen system. This is supported by the fact that we did not observe the activation of some common genes typically involved in immune system activation (i.e., factors responsible for direct or indirect pathogen perception or members implicated in cell-to-cell signaling pathways) during the first days after bacterial infection.
In our conditions, the basal defensive response to the attack of Xcc was characterized by the activation of PR proteins belonging to two major protein families (PR-6 and PR-12). The PR-6 proteins are involved in the reduction of the ability of the pathogen to complete its replication cycle, whereas the role of PR-12 remains unclear19. The accumulation of PR proteins is a typical response in plants exposed to both biotic and abiotic stress20. Likewise, Xcc infection causes the accumulation of transcripts of plant defensive secondary metabolites. Among these transcripts, those involved in GSL core structure biosynthesis were notable. Specifically, two of these transcripts encode important enzymes involved in the synthesis of GBS. Several authors have studied the role of GSLs or their autolytic breakdown products (isothiocyanates, ITCs) in the defense against Xcc21,22. These studies have shown that different ITCs and GSLs, such as gluconapin, sinigrin and sinalbin, had an antibiotic effect against Xcc; however, the GBS biocide effect against Xcc was weaker than that observed in other GSLs. Moreover, the quantification of the content of GSLs in our samples did not show significant differences between the control vs. inoculated plants.
This basal immune response seems to be mediated by phytohormones. It is well known that proper regulation of the immune response is essential to maintain an appropriate energy balance to reduce the inherent fitness cost of being well defended, which is precisely the main role of the phytohormones23. Generally, the SA pathway is related to plant defenses against biotrophic pathogens, whereas ethylene (ET) and JA are required to fight against necrotrophic pathogens. However, some authors debate whether pathogens are not often readily classifiable as purely biotrophic or necrotrophic, and the antagonistic or synergistic interactions between SA and JA/ET pathways depend on the specific pathogen and its lifestyle24,25. Our results showed an upregulation of genes related to the biosynthesis of these phytohormones, which reinforces the hypothesis that Xcc is not a total biotrophic or necrotrophic bacterium. Markedly, a higher number of transcripts involved in phytohormone metabolism and perception were activated at 12 dpi than at 3 dpi. The role of phytohormones in the long-term immune response has been neglected in the literature.
The plant immune system is composed of different phases in which several factors collaborate together with the aim of curbing pathogen progression. This system is conventionally divided into two interconnected branches: an earlier step called the PTI and the ETI, a later and amplified response that results in a hypersensitive cell death response (HR) at the infection site2. Despite this conventional zigzag model presenting ETI and PTI as two well-differentiated branches of plant immunity, recent evidence indicates that they have more in common than previously thought. It appears that depending on the specific plant-pathogen interaction, pattern recognition receptors (PRRs) and effector receptors can swap roles, and most defense mechanisms triggered by plants, such as oxidative burst, hormonal changes, HR or transcriptional reprogramming, can be produced by both responses8. Approximately 40 of the pathogen receptors that we identified at 12 dpi belong to the RLK gene family. Most of the members of the RLKs act as PRRs involved in the recognition of PAMPs in the plasma membrane, which is essential to trigger PTI26. We found PRR genes with different kinds of ectodomains, the epidermal growth factor-like domain, lectin and lysine motifs or LRRs, each involved in the recognition of specific PAMPs during pathogen invasion. Thus, although Xcc infection was well established and plants presented high levels of damage, plants were still triggering the mechanisms related to pathogen perception, which are processes generally associated with the early stages of basal immunity.
The perception of pathogen invasion produces the activation of multifaceted intracellular signaling pathways that initiate defense responses. The role of hydrogen peroxide (H2O2) as a second messenger is well known; if the organism is subjected to increasing levels of H2O2, as observed during pathogen attack, this signal must be propagated to trigger the appropriate responses27. Generally, this oxidative burst is accompanied by changes in the intracellular environment, such as fluxes in Ca+2 28, which acts as a pleiotropic second messenger to trigger numerous physiological processes. However, the mechanism of this essential signaling pathway remains unclear due to the cellular location and nature of these calcium signals that differ across both host species and pathogenesis, which probably reflects mechanistically distinct processes29.
CMLs, which are solely present in the plant kingdom, display a strong affinity for calcium ions. The interaction between a Ca+2 ion and one CML induces conformational changes that trigger its association with downstream target proteins11. CML targets include protein kinases, metabolic enzymes, transporters and transcription factors. Although the role of most of these CMLs remains unclear, some CMLs are stress responsive. For example, CML37 showed dual roles in biotic and abiotic responses, acting as a positive regulator of defense against herbivores and a negative regulator during drought stress tolerance11. CML42 is both a negative regulator of insect herbivory-induced defense and drought-induced ABA levels and a positive regulator of UV stress. Our in silico study focused on members of the calcium-signaling pathway suggests, for the first time, the possible implication of CML30, CML37, CML40, CML43, CML45, and CML47 in the response against pathogenesis.
Since these proteins appear to act as Ca+2 flow transmitters, most of the studies are focused on the identification and characterization of their downstream targets30. Among these genes, we identified two master transcription factors of plant immunity, CBP60g and SARD1. A study performed by Wang et al.31 using Arabidopsis cbp60g mutants showed that this protein contributes to PAMP-triggered SA accumulation, which enhances resistance against P. syringae. In addition to SA-dependent defense pathway activation, both CBP60g and SARD1 activate SA-independent defense mechanisms through the regulation of WRKY70 expression32. In fact, in this work, we reported the overexpression of WYRK70 at 12 dpi, which is further indication of the action of these transcription factors. Interestingly, in light of our results, the expression patterns of CBP60g and SARD1 are opposing. Despite these genes belonging to the same protein family, it has been shown that their regulation is different. CBP60g necessarily requires CML binding, and SARD1 provides a similar role in a Ca+2-independent manner33. Therefore, strictly speaking, SARD1 should not be considered a CaM-binding protein and indicates that the functions CBP60g and SARD1 are carried out in parallel32. This fact could explain their different expression patterns during Xcc infection.
The comparison of the expression of these genes between a compatible and an incompatible genotype suggests an essential role in the resistance against Xcc. The fact that Arabidopsis knockout mutants of these genes are more susceptible to Xcc infection than wt plants confirms this role. Apparently, both SARD1 and CBP60g are important during defense against Xcc. A time-course analysis of the expression of these genes suggests that SARD1 is induced too late in the compatible genotype when the pathogen is already spread through the whole plant. In addition, the incompatible genotype may not need a sharp increase in SARD1 expression, since this genotype presents a higher basal expression level.
In summary, the MACE results provide a complete view of the variation in the expression of genes potentially involved in the B. oleracea–Xcc interaction. In general, we identified more induced than repressed transcripts at both dpi analyzed, albeit the response was greater at 12 dpi. In addition, in contrast to expectations, changes in the expression of genes related to early infection stages, such as PAMP perception or ROS burst and Ca+2 flux signaling, were only detected at 12 dpi. Our results suggest that several CMLs could have an important role during Xcc pathogenesis and that the genes CBP60g and SARD1 act as downstream factors of Ca+2 signaling.
Material and methods
Plant material
The doubled haploid broccoli line “Early Big” (B. oleracea var. italica) was used for transcriptomic analysis. The Badger Inbred-16 line was subsequently used as a tester of resistance. This line presents a partial black rot-resistance, which is controlled by a quantitative trait locus (QTL). Plants were sown in plastic pots containing sphagnum peat (GRAMOFLOR GmbH & Co, Vechta, Germany) in a greenhouse with a minimum temperature of 20 °C during the day and 15 °C during the night, venting at 25 °C and 60% humidity.
Inoculation of B.oleracea
Xcc race 1 strain HRI3811 was provided by Warwick HRI (Wellesbourne, UK). Bacterial cultures were grown in screening media 523 (Sigma-Aldrich, St. Louis, MO, USA) at 30 °C in a rotary incubator at 100 rpm for 48 h. An aliquot was diluted in sterile water to reach a final absorbance of 0.5, which corresponds to a concentration of 5 × 108 cfu/ml. The turbidity of the suspension was measured with a microplate spectrophotometer (Spectra MR; Dynex Technologies, Chantilly, VA, USA) at a wavelength of 600 nm. Plants at the six-leaf stage were inoculated at the third leaf following the method described by Lema et al.34. Briefly, a sterilized “florists’ frog” (i.e., multiple needles mounted in circles) was used for inoculation by pressing through the leaf onto a sponge submerged in the inoculum at the edge of the distal side of the leaf. Control plants were mock inoculated following the same procedure. After inoculation, greenhouse conditions were changed to maintain a minimum temperature of 18 °C and an 80% relative humidity. The whole inoculated leaf was from three independent biological replicates of the control, and inoculated plants were collected at 3 and 12 dpi in liquid nitrogen and conserved at −80 °C until processed.
Inoculation of Arabidopsis thaliana
Arabidopsis plants were grown on sterilized peat in a growth chamber under fluorescent light (228 µmol m−2 s−1) in short-day conditions (8 h light/16 h darkness) and watered as needed. A constant day/night temperature was set at 20 °C. Xcc race 3 strain HRI5212 was provided by Warwick HRI. Bacterial cultures were prepared as described above. Fully expanded leaves of 6-week-old plants were inoculated as described in Meyer et al.17. Briefly, inoculation was performed using a sterilized inoculation needle dipped on bacterial culture. Three inoculation points were established in the midrib of each leaf. After inoculation, plants were covered with a plastic bag to maintain nearly 100% relative humidity. At 7 dpi, inoculated leaves were collected and digitalized with a scanner at 300 dpi resolution. The lesion area of each leaf was calculated using ImageJ software35.
RNA isolation, library preparation, and sequencing
Individual sample tissues were ground in liquid nitrogen, and total RNA from three biological replicates of each treatment was extracted using the SpectrumTM Plant Total RNA kit (Sigma-Aldrich, MO, USA). To remove any traces of genomic DNA, the RNA was treated with DNase according to the manufacturer’s instructions. Massive analysis of 3′-cDNA ends (MACE) was performed for each sample (GenXPro GmbH, Frankfurt am Main, Germany) as described by Zawada et al.36. Briefly, poly-adenylated mRNA was isolated from 1 μg of total RNA, and cDNA was produced by first- and second-strand synthesis using the SuperScript® III First-Strand Synthesis System (Invitrogen, Waltham, MA, USA) with modified barcoded poly-T adapters that are biotinylated at the 5′ end. After cDNA random-fragmentation, the 3′-ends were captured by streptavidin beads, and 5′ ends of ≈ 67 bp long fragments from the 12 barcode samples were sequenced (single-read) using an Illumina HiSeq 2000 version 4 chemistry (Illumina, Inc., San Diego, CA, USA), with 1 × 125 bps (6 bps were used for a barcode). To eliminate PCR-based copies from the generated dataset, GenXPro’s “TrueQuant” method was applied. This method identifies and eliminates copies with an identical barcode-sequence combination37. The average raw count of each gene within a library was normalized by dividing by the geometric mean of all counts in all samples, and the median of the quotients was calculated per library. Each raw count was then divided by the library-specific median value. Statistical analysis was performed using the DEseq R package according to Anders and Huber recommendations38.
Transcript annotation and functional analysis
Putative functions were assigned to the resulting transcripts by mapping in silico their translated protein sequences to the UniProtKB/Swiss-Prot, UniProtKB and Ref-Seq protein databases using the BLASTX algorithm available at NCBI in hierarchical manner, using an e-value of 10−5 as a threshold for considering them as homologs. Transcripts without homology to these sequences were subsequently annotated to the nonredundant nucleotide NCBI database (nr) by BLASTN using an e-value of 0.001 as a threshold.
Transcripts with a false discovery rate <0.05 and −1 < log2 fold change (FC) > 1 were considered to be differentially expressed between the control and inoculated samples. The functional classification of the differentially expressed genes was performed with MapMan 3.6.0RC1 software by using A. thaliana homolog genes as input. This tool allows the data to be organized according to Gene Ontologies (GO) and displayed by the user in the context of preexisting biological knowledge39. Furthermore, the webtool STRING (v. 10.5) was used to study the interconnections between the selected genes. This resource, in addition to the well-supported protein–protein interactions experimentally observed, includes indirect and predicted interactions on top40.
The transcriptomic data can be found in the Gene Expression Omnibus repository with the accession number GSE107720.
Quantitative reverse-transcription-PCR (RT-qPCR) validation
Plant from the inbred lines “Early Big” and Badger Inbred-16 were grown in the greenhouse as described above. Leaves from three biological replicates of each line were gathered at different infection points (1, 2, 3, and 12 dpi). The procedure for RNA extraction was the same as that followed in the MACE analysis. Three technical replicates were performed for each biological replicate. All primer pairs used are listed in Table S1. One microgram of total RNA was reverse transcribed using the GoScript™ reverse-transcription system and oligo (dT20) (Promega, Madison, WI, USA). RT-qPCR was performed in a 20 μl reaction with the Fast Start Universe SYBR Green Master (ROX) mix (Roche Molecular Systems Inc, Pleasanton, CA, USA), following the manufacturer’s instructions. The glyceraldehyde-3-phosphate-dehydrogenase transcript was used as housekeeping gene41. RT-qPCRs were carried out on a 7500 Real-Time PCR System (Applied Biosystem, Forster City, CA, USA), and primer efficiency was calculated using the LingRegPCR software42. Efficiencies were used to calculate relative gene expression using the ΔΔCt method43. Statistical significance was calculated using a two-tailed Student’s t test to compare the relative gene expression in the control vs. inoculated plants.
References
Agrawal, A. A. Induced responses to herbivory and increased plant performance. Science 279, 1201–1202 (1998).
Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Tsuda, K. & Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465 (2010).
Cui, H., Tsuda, K. & Parker, J. E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511, https://doi.org/10.1146/annurev-arplant-050213-040012 (2015).
Henry, E., Yadeta, K. A. & Coaker, G. Recognition of bacterial plant pathogens: local, systemic and transgenerational immunity. New Phytol. 199, 908–915, https://doi.org/10.1111/nph.12214 (2013).
Tao, Y. et al. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15, 317–330, https://doi.org/10.1105/tpc.007591 (2003).
Navarro, L. et al. The transcriptional innate immune response to flg22. interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128, https://doi.org/10.1104/pp.103.036749 (2004).
Thomma, B. P. H. J., Nürnberger, T. & Joosten, M. H. A. J. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15, https://doi.org/10.1105/tpc.110.082602 (2011).
Pombo, M. A. et al. Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector-triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome Biol. 15, https://doi.org/10.1186/s13059-014-0492-1 (2014).
Day, I. S., Reddy, V. S., Ali, G. S. & Reddy, A. S. N. Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 3, RESEARCH0056, https://doi.org/10.1186/gb-2002-3-10-research0056 (2002).
Ranty, B. et al. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 7, https://doi.org/10.3389/fpls.2016.00327 (2016).
Sanders, D., Pelloux, J., Brownlee, C. & Harper, J. F. Calcium at the crossroads of signaling. Plant Cell 14, S401–S417, https://doi.org/10.1105/tpc.002899 (2002).
McAinsh, M. R. & Pittman, J. K. Shaping the calcium signature. New Phytol. 181, 275–294, https://doi.org/10.1111/j.1469-8137.2008.02682.x (2009).
Kemmerling, B., Halter, T., Mazzotta, S., Mosher, S. & Nurnberger, T. A genome-wide survey for Arabidopsis leucine-rich repeat receptor kinases implicated in plant immunity. Front. Plant Sci. 2, https://doi.org/10.3389/fpls.2011.00088 (2011).
Narusaka, M. et al. Interfamily transfer of Dual NB-LRR genes confers resistance to multiple pathogens. Plos ONE 8, https://doi.org/10.1371/journal.pone.0055954 (2013).
Wang, L. et al. CBP60g and SARD1 play partially redundant critical roles in 552 salicylic acid signalings. Plant J. 67, 1029–1041, https://doi.org/10.1371/journal.pone.0055954 (2013).
Meyer, D., Lauber, E., Roby, D., Arlat, M. & Kroj, T. Optimization of pathogenicity assays to study the Arabidopsis thaliana-Xanthomonas campestris pv. campestris pathosystem. Mol. Plant Pathol. 6, 327–333 (2005).
O’Donnell, P. J. et al. Susceptible to intolerance - a range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 33, 245–257 (2003).
Agarwal, P. & Agarwal, P. K. Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Mol. Biol. Rep. 41, 599–611, https://doi.org/10.1007/s11033-013-2897-4 (2014).
Sels, J., Mathys, J., De Coninck, B. M. A., Cammue, B. P. A. & De Bolle, M. F. C. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950 (2008).
Aires, A. et al.Initial in vitro evaluations of the antibacterial activities of glucosinolate enzymatic hydrolysis products against plant pathogenic bacteria. J. Appl. Microbiol. 106, 2096–2105 (2009).
Sotelo, T., Lema, M., Soengas, P., Cartea, M. E. & Velasco, P. In vitro activity of glucosinolates and their degradation products against brassica-pathogenic bacteria and fungi. Appl. Environ. Microbiol. 81, 432–440 (2015).
Denance, N., Sanchez-Vallet, A., Goffner, D. & Molina, A. Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front Plant Sci. 4, https://doi.org/10.3389/fpls.2013.00155 (2013).
Mur, L. A. J., Kenton, P., Atzorn, R., Miersch, O. & Wasternack, C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262 (2006).
Adie, B. A. T. et al. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant cell 19, 1665–1681, https://doi.org/10.1105/tpc.106.048041 (2007).
Macho, A. P. & Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol Cell 54, 263–272 (2014).
Passaia, G. & Margis-Pinheiro, M. Glutathione peroxidases as redox sensor proteins in plant cells. Plant Sci. 234, 22–26 (2015).
Davies, D. R., Bindschedler, L. V., Strickland, T. S. & Bolwell, G. P. Production of reactive oxygen species in Arabidopsis thaliana cell suspension cultures in response to an elicitor from Fusarium oxysporum: implications for basal resistance. J. Exp. Bot. 57, 1817–1827, https://doi.org/10.1093/jxb/erj216 (2006).
Zipfel, C. & Oldroyd, G. E. D. Plant signalling in symbiosis and immunity. Nature 543, 328–336, https://doi.org/10.1038/nature22009 (2017).
Zhu, X. Y., Dunand, C., Snedden, W. & Galaud, J. P. CaM and CML emergence in the green lineage. Trends Plant Sci. 20, 483–489 (2015).
Wang, L. et al. Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. Plos Pathog 5, https://doi.org/10.1371/journal.ppat.1000301 (2009).
Sun, T. J. et al. ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nature Commun. 6, https://doi.org/10.1038/ncomms10159 (2015).
Truman, W. et al. The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol. 163, 1741–1751, https://doi.org/10.1104/pp.113.227108 (2013).
Lema Marquez, M., Terán, H. & Singh, S. P. Selecting common bean with genes of different evolutionary origins for resistance to Xanthomonas campestris pv. phaseoli. Crop Sci. 47, 1367–1374, https://doi.org/10.2135/cropsci2006.12.0769 (2007).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 18, 529, https://doi.org/10.1186/s12859-017-1934-z (2017).
Zawada, A. M. et al. Massive analysis of cDNA ends (MACE) and miRNA expression profiling identifies proatherogenic pathways in chronic kidney disease. Epigenetics 9, 161–172, https://doi.org/10.4161/epi.26931 (2014).
Yakovlev, I. A. et al. Temperature-dependent differential transcriptomes during formation of an epigenetic memory in Norway spruce embryogenesis. Tree Genet. Genomes 10, 355–366, https://doi.org/10.1007/s11295-013-0691-z (2014).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106–R106 (2010).
Thimm, O. et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37, 914–939 (2004).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368, https://doi.org/10.1093/nar/gkw937 (2017).
Duan, M. et al. Identification of optimal reference genes for expression analysis in radish (Raphanus sativus L.) and its relatives based on expression stability. Front. Plant Sci. 8, https://doi.org/10.3389/fpls.2017.01605 (2017).
Ramakers, C., Ruijter, J. M., Deprez, R. H. L. & Moorman, A. F. M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66, https://doi.org/10.1016/s0304-3940(02)01423-4 (2003).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
This research was supported by project AGL2015-66256-C2-R and RTI2018-096591-B-I00, by the European Regional Development Fund (ERDF) and by A Xunta de Galicia (IN607A 2016/13). M. T. acknowledges the FPI fellowship from the Ministry of Economy and Competitiveness (BES-2013-065744). We thank Professor Jane Glazebrook from the University of Minnesota for providing sard1 and cbp60g Arabidopsis mutants.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tortosa, M., Cartea, M.E., Velasco, P. et al. Calcium-signaling proteins mediate the plant transcriptomic response during a well-established Xanthomonas campestris pv. campestris infection. Hortic Res 6, 103 (2019). https://doi.org/10.1038/s41438-019-0186-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-019-0186-7
This article is cited by
-
Molecular basis for host responses to Xanthomonas infection
Planta (2022)
-
Status and advances in mining for blackleg (Leptosphaeria maculans) quantitative resistance (QR) in oilseed rape (Brassica napus)
Theoretical and Applied Genetics (2021)