Abstract
SCR/SP11 encodes the male determinant of recognition specificity of self-incompatibility (SI) in Brassica species and is sporophytically expressed in the anther tapetum. Based on dominance relationships in pollen and nucleotide sequence similarity, the S haplotypes in Brassica have been classified as class I or class II, with class-I S haplotypes being dominant over class-II S haplotypes. Here, we revealed that S-22 in B. rapa belonging to class I is recessive to class-II S-44 and class-I S-36 in pollen, whereas it is dominant over S-60, S-40, and S-29 based on pollination tests. SCR/SP11 of S-22 (SCR-22) was sequenced, revealing that the deduced amino-acid sequence of SCR-22 has the longest C-terminal domain among the SCR/SP11 sequences. The expression of SCR-22 was found to be suppressed in S-22/S-44 and S-22/S-36 heterozygotes. Normal transcription of SCR-44 was considered to be due to the transcription suppression of Smi sRNA of the S-22 haplotype and a very low methylation state of the SCR-44 promoter region in the tapetum of S-22/S-44 heterozygotes. In SCR-22, only the cytosine residue located at the –37 bp position of the promoter region was hypermethylated in the tapetum of S-22/S-44 heterozygotes, and few methylated cytosines were detected in the promoter and coding regions of SCR-22 in S-22/S-36 heterozygotes. SCR-22 was also expressed in microspores in S-22 homozygotes but not in S-22/S-44 and S-22/S-36 heterozygotes. These results suggest that a mechanism different from class-II SCR/SP11 suppression may operate for the suppression of recessive class-I SCR-22 in S heterozygotes.
Similar content being viewed by others
Introduction
Self-incompatibility (SI) is a genetic mechanism exploited by many angiosperm species to prevent inbreeding and to promote outcrossing. In most species, SI is controlled by a single S locus with a large number of haplotypes. The SI response occurs when the S haplotype of the pollen is the same as that of the pistil. In Brassica, three genes located at the S locus have been characterized, namely, S-receptor kinase (SRK), a female determinant of recognition specificity;1,2 S-locus cysteine-rich protein/S-locus protein 11 (SCR/SP11), a male determinant;3,4 and S-locus glycoprotein (SLG), which is highly similar to the extracellular domain (S-domain) of SRK5.
SCR/SP11 is a small cysteine-rich protein with ca. 50 amino-acid residues with 8 conserved cysteine residues. The sequence of mature SCR/SP11 protein is highly polymorphic, with less than 50% amino-acid similarity among different S haplotypes within the same species3,6,7,8,9. In most SCR/SP11 variants, only a few amino acids are conserved, such as the eight cysteines, a glycine between the first and second cysteines, and an aromatic amino-acid residue between the third and fourth cysteines3,6,7,10. SCR/SP11 proteins are mainly produced in the anther tapetum and are then transferred to the surface of mature pollen3,10,11,12. Therefore, the SI phenotype in pollen is consistent with the dominant S haplotypes carried by S heterozygous plants.
Based on dominance relationships relative to the other alleles in S-heterozygous plants and the nucleotide sequence similarity of S-locus genes, the S haplotypes in Brassica are classified into two groups: the pollen-dominant S haplotypes termed class I and the pollen-recessive S haplotypes termed class II13. The class-I S haplotypes are always dominant over the class-II S haplotypes in pollen. In Brassica rapa, SCR/SP11 sequences of four class-II members, namely, S-44, S-60, S-40, and S-29, have been identified, and a linear dominance relationship has been demonstrated among them14,15. The expression patterns of class-I and class-II SCR/SP11 alleles are slightly different. Class-II SCR/SP11 alleles are expressed only in the anther tapetum, whereas class-I SCR/SP11 alleles are expressed not only in the tapetum but also in microspores3,10,16. Phylogenetic analysis has revealed that class-II SCR/SP11 alleles form a distinct group separate from class-I SCR/SP11 alleles15. The promoter sequences of SCR/SP11 alleles show little similarity between the two classes, which results in an expression pattern difference between class-I and class-II SCR/SP11 alleles.
In heterozygotes with class-I and class-II S haplotypes, the class-II SCR/SP11 is not expressed, indicating that the dominance relationships are regulated at the messenger RNA level of SCR/SP11 alleles15. It was found that class-I SCR/SP11 alleles having promoter defects, which are not transcribed, also caused suppression of recessive class-II SCR/SP11 alleles17. Subsequent studies demonstrated that the expression suppression of recessive class-II SCR/SP11 alleles results from their tissue-specific methylation of promoter sequences in the tapetum18. Further analysis showed that a sequence with high similarity to the target methylated region lies in a region flanked by dominant SLG alleles named SP11-methylation-inducer (Smi). An Smi sequence was used as a template to form a 24-nucleotide small noncoding RNA (sRNA), which induced the methylation of the promoter of a recessive SCR/SP11 allele and repressed its transcription19. Recently, Smi2 has been identified in a class-II S haplotype sequence and has been shown to control the linear dominance hierarchy of the four class-II SCR alleles20.
S-22 in B. rapa has been reported to be recessive to S-24, S-26, and S-43, belonging to class-I S haplotypes, in pollen in the same way class-II S-60 is recessive to S-24, S-26, S-28, and S-4321. In the stigma, S-22 has been revealed to be recessive to S-28 in class-I S haplotypes. The nucleotide sequences of SLG and SRK of S-22 have been determined and deposited in the DDBJ (AB054060 and AB054061, respectively). Comparison of the nucleotide sequences and deduced amino-acid sequences of these alleles with those of other SLG and SRK alleles has revealed that S-22 belongs to the class-I S haplotypes8. Nucleotide sequences of SCR/SP11 of S-22 have not been reported. Since S-22 is ranked the lowest in the dominance hierarchy of S haplotypes among class-I S haplotypes, this haplotype may have some unique characteristics. In the present study, we found that S-22 was recessive to S-44 in class-II haplotypes in pollen. We identified SCR/SP11 of S-22 (SCR-22 hereafter), finding that it has a unique feature in deduced amino-acid sequences. The expression of SCR-22 was suppressed in the S-22/S-44 heterozygote, but the cytosine methylation pattern in the SCR-22 promoter was different from that of recessive class-II SCR alleles. These results may help us to better understand the mechanism controlling dominance relationships among SCR/SP11 alleles.
Results
Dominance relationships between S-22 and other S haplotypes of B. rapa in pollen
We analyzed the dominance relationships in pollen between S-8, S-22, and class-II S haplotypes of B. rapa by pollination tests. The class-I allele S-8 was used as a control to demonstrate the typical behavior of a class-I allele. Our preliminary experiments show that S-8 is codominant with S-22 and S-36 in pollen (data not shown). The pollen of heterozygotes with S-22 or S-8 and one class-II S haplotype was applied to stigmas of S-22, S-8, or class-II S homozygotes. The results showed that the pollen of S-22/S-44 heterozygotes can germinate and penetrate the stigmas of S-22 homozygotes but cannot penetrate the stigmas of S-44 homozygotes (Table 1). This result indicated that S-22 is recessive to S-44, which is different from other class-I S haplotypes generally dominant over S-44. Further pollination tests showed that the pollen grains of S-22/S-60, S-22/S-40, and S-22/S-29 heterozygotes were incompatible with the stigmas of S-22 homozygotes, whereas they were compatible with the stigmas of S-60, S-40, and S-29 homozygotes, respectively, indicating that S-22 is dominant over S-60, S-40, and S-29 in pollen. In addition, the pollen tubes of S-22/S-36 heterozygotes, of which S-36 is a class-I S haplotype, penetrated the stigmas of S-22 homozygotes, whereas these pollen tubes did not penetrate the stigmas of S-36 homozygotes, revealing that S-22 is recessive to S-36. The pollen of heterozygotes with S-8 and one class-II S haplotype was incompatible with the stigmas of S-8 homozygotes, whereas they were compatible with the stigmas of S-44, S-60, S-40, and S-29 homozygotes, indicating that S-8 is dominant over all four class-II alleles in pollen.
Sequence analysis of SCR-22 of B. rapa
We determined the nucleotide sequence of the coding region of SCR-22 and its promoter region sequence in two steps. Since SCR-22 was not amplified by reverse transcription-polymerase chain reaction (RT-PCR) using the primers reported by Watanabe et al.6 and Sato et al.8, we amplified a partial sequence of SCR-22 of B. rapa using many combinations of primers including newly designed primers (Supplementary Table S1). Second, we amplified the flanking sequence of the identified region of SCR-22 by inverse PCR to determine the nucleotide sequence of the whole coding region of SCR-22 and its promoter region. Our results showed that the coding region of SCR-22 is 627 bp in length and contains a 306 bp intron (Supplementary Figure S1).
It was found that three amino acids, i.e., the seventh, tenth, and twelfth amino acids, in the putative signal peptide of SCR-22 are different from those of other class-I SCR sequences (Fig. 1). As with other SCR/SP11 proteins, the eight conserved cysteine residues are present in SCR-22, and a glycine residue between C1 and C2 is also conserved in SCR-22. However, the length of the SCR-22 protein is different from that of other SCR/SP11 proteins. Most SCR/SP11 proteins contain approximately 50 amino acids, whereas the SCR-22 protein contains approximately 70 amino acids. SCR-22 has a longer C-terminal domain (Fig. 1). Linkage analysis showed that this gene was linked to SRK-22 in B. rapa (Supplementary Figure S2). These results confirm that the gene we identified is SCR-22 of B. rapa.
Tapetum isolation
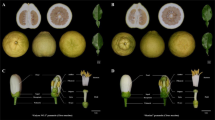
It has been reported that class-II SCR/SP11 alleles are specifically transcribed in the tapetum15 and that DNA methylation of the promoter of recessive SCR/SP11 alleles in heterozygotes also occurs in the tapetum18. In previous studies, DNA was extracted from the nuclei of tapetum cells, but the method was not described in detail18. We therefore developed a method for tapetum isolation. The anther contains endothecium, tapetum, and microspores (Fig. 2a). We cut away both ends of the anthers and released the microspores by vortexing. All microspores were released (Fig. 2b). The anthers were then treated with cellulase RS and pectolyase Y23 for only 7 min to release the tapetum cells (Fig. 2c). We then used this method to isolate the tapetum cells from the anthers of S-60 homozygotes and analyzed the expression of SCR-60 in isolated endothecium, microspores, and tapetum (Fig. 2d). The expression of SCR-60 was detected only in the isolated tapetum cells, not in isolated endothecium or microspores, confirming that the fraction we obtained was the tapetum and that few tapetum cells were present in the isolated endothecium fraction or microspore fraction. The tapetum cells from the anthers of S-22/S-60 heterozygotes were then isolated to assess the methylation rate of the recessive SCR-60 5’ region. Methylated cytosine residues at CpG, CpNpG, and CpNpN sites were widespread, and the methylation frequencies of the two cytosine residues in the region of SCR-60 homologous to the Smi of the S locus19 were 46.9% and 71.9%, respectively (Fig. 2e). The highest methylation frequency of cytosine residues in the 5’ region of class-II SCR/SP11 alleles has been reported to be approximately 80%18,19. Considering that the DNA methylation of SCR/SP11 alleles is detected only in the tapetum and that the highest methylation frequency detected in the present study was close to the highest reported methylation frequency, it can be inferred that the purity of the isolated tapetum fraction prepared using our method is comparable to that obtained by the method of Shiba et al.18 and that the isolated tapetum fraction of our study can be used for the following analyses.
Before the tapetum is isolated, an anther contains endothecium, tapetum, and microspores (a). Microspores were released by vortexing (b). After tapetum isolation, the tapetum and microspores were removed, and the endothecium had integrity (c). The expression of SCR-60 was detected in the isolated tapetum of S-60 homozygotes (d). The DNA methylation state of the SCR-60 promoter in the tapetum of S-22/S-60 heterozygotes was analyzed using the tapetum isolated by our method (e). The thick line under the DNA methylation profile indicates the region homologous to Smi. E endothecium, T tapetum, M microspores, G genomic DNA
Gene expression of SCR/SP11 alleles in S heterozygotes
The relative expression level of each SCR/SP11 allele was investigated in the S heterozygotes. First, the expression level of SCR-22 was measured by real-time quantitative PCR. The results showed that the expression of SCR-22 in the tapetum cells of S-22/S-60, S-22/S-40, and S-22/S-29 heterozygotes was the same as that in S-22 homozygotes, whereas it was suppressed in the tapetum cells of S-22/S-44 and S-22/S-36 heterozygotes (Fig. 3a). SCR-22 was expressed in the microspores of the S-22 homozygotes, whereas the expression of SCR-22 was suppressed in the microspores of the S-22/S-44 and S-22/S-36 heterozygotes (Fig. 3b). SCR-8, also belonging to class I, was expressed in the tapetum cells of all heterozygotes we analyzed (Fig. 3c). Second, the relative expression levels of four class-II SCR/SP11 alleles were investigated. SCR-44 was not expressed in the tapetum cells of S-8/S-44 heterozygotes but was expressed in those of S-22/S-44 heterozygotes (Fig. 3d). SCR-60, SCR-40, and SCR-29 were not expressed in the tapetum cells of any of the S heterozygotes we analyzed (Fig. 3e, f, g). SCR-36 was expressed in the tapetum cells of S-22/S-36 heterozygotes (Fig. 3h). The observed relative expression levels suggested that SCR-22 is dominant to SCR-60, SCR-40, and SCR-29 and recessive to SCR-44 and SCR-36 and that SCR-8 is dominant to all the class-II SCR/SP11 alleles that we analyzed. These results were consistent with the results of the pollination tests.
The relative expression levels of SCR-22 were detected in the tapetum of S-22 homozygotes and S-22/S-36, S-22/S-44, S-22/S-60, S-22/S-40, and S-22/S-29 heterozygotes (a) and in the microspores of S-22 homozygotes and S-22/S-36 and S-22/S-44 heterozygotes (b); and those of SCR-8 were detected in the tapetum cells of S-8 homozygotes and S-8/S-44, S-8/S-60, S-8/S-40, and S-8/S-29 heterozygotes (c). The relative expression levels of SCR-44 were detected in the tapetum cells of S-44 homozygotes and S-22/S-44 and S-8/S-44 heterozygotes (d), SCR-60 levels were detected in S-60 homozygotes and S-22/S-60 and S-8/S-60 heterozygotes (e), SCR-40 levels were detected in S-40 homozygotes and S-22/S-40 and S-8/S-40 heterozygotes (f), SCR-29 levels were detected in S-40 homozygotes and S-22/S-29 and S-8/S-29 heterozygotes (g), and SCR-36 levels were detected in S-36 homozygotes and S-22/S-36 heterozygotes (h). Error bars represent standard errors (SE) of the mean of triplicate samples
The methylation state of recessive SCR alleles in heterozygotes
It has been reported that suppression of the expression of recessive class-II SCR/SP11 alleles results from methylation of the promoter region of recessive SCR/SP11 alleles induced by an sRNA of the class-I or class-II S haplotype in the tapetum19,20. The methylation state of SCR-44 was therefore measured in the present study. Widespread methylated cytosine residues were found in the SCR-44 promoter region in the tapetum of S-8/S-44 heterozygotes (Fig. 4a). All three types of cytosine methylation, i.e., CpG, CpNpG, and CpNpN, were detected in this region. The methylation frequencies of two cytosine residues in the region homologous to Smi in the SCR-44 promoter of S-8/S-44 heterozygotes were 32.1% and 39.3%, respectively. In the tapetum of S-22/S-44 heterozygotes and S-44 homozygotes, where SCR-44 is transcribed, few methylated cytosine residues were detected in the promoter region of SCR-44 (Fig. 4a). In addition, methylated cytosine residues in the region homologous to Smi in recessive SCR-60, SCR-40, or SCR-29 were also observed, and the percentages of methylated cytosine were from 21.7% to 73.7% in the heterozygotes, which are higher than the 2.8% to 11.8% observed in the homozygotes (Supplementary Figure S3). These results indicate that the suppression of class-II SCR/SP11 expression is related to DNA methylation.
Percentages of methylation at all cytosine residues in the SCR-44 promoter (nucleotides –254 to –1) (a) and SCR-22 promoter (nucleotides –600 to –1) (b) are shown in the histograms. c The DNA methylation state of the SCR-22 gene in the tapetum. d The DNA methylation state of the SCR-22 promoter and coding region in microspores. The results are from at least 30 cloned sequences. The thick line under the DNA methylation profile indicates the region homologous to Smi
The methylation states of SCR-22 in S-22 homozygotes and S-22/S-44 and S-22/S-36 heterozygotes were also investigated. In the tapetum of S-22 homozygotes, the methylation rate of the SCR-22 promoter region was very low (Fig. 4b). In the tapetum of S-22/S-44 heterozygotes, where SCR-22 was not transcribed, the cytosine residue located at the –37 position of the promoter was found to be highly methylated, with a methylation rate of 74% (Fig. 4b). No further methylated cytosine was detected in the SCR-22 promoter region, coding region, and intronic region in the tapetum (Fig. 4b, c). At the same time, few methylated cytosines were detected in the SCR-22 promoter region, coding region, and intronic region in the microspores of S-22/S-44 heterozygotes (Fig. 4d), where SCR-22 was also not transcribed. Additionally, in S-22/S-36 heterozygotes, which carry two class-I S haplotypes, a methylated cytosine-rich region was detected at the promoter region (from –350 to –440) of SCR-22 in the tapetum. However, the methylation rates of these cytosine residues were low, with the highest methylation rate being 31.2%, and only two types of cytosine methylation, i.e., CpNpG and CpNpN, were detected in this region. Few methylated cytosine residues were detected in the coding and intronic regions of recessive SCR-22 (Fig. 4b–d). In the microspores of the S-22/S-36 heterozygotes, similar to those of S-22/S-44 heterozygotes, few methylated cytosines were detected in the SCR-22 promoter region, coding region, and intronic region (Fig. 4b–d).
Transcript analysis of Smi trans-acting sRNA
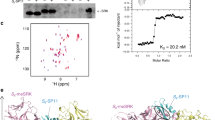
It has been reported that the Smi trans-acting sRNA from the class-I S locus induces the methylation of the promoter of recessive class-II SCR/SP11 alleles19. The primer set SL-F1/SL-R1, designed by Tarutani et al.19, was used to amplify the sequence of the precursors of Smi-8 and Smi-22 from the S locus of S-8 and S-22 haplotypes. The results showed that the precursors can form an imperfect stem-loop structure (Fig. 5a), and the sequences of the Smi-8 and Smi-22 sRNAs are the same as that of the Smi-9 sRNA19.
Stem-loop precursors predicted from Smi-8 and Smi-22 sequences are shown (a). The sequences of the mature sRNA are underlined. Mature Smi sRNA was detected by stem-loop RT-PCR in S-22 homozygotes and heterozygotes with S-22 and one class-II S haplotype (b). The transcription of precursors of Smi-22 was also detected in S-22 homozygotes and heterozygotes with S-22 and one class-II S haplotype (c). Mature Smi sRNA in S-8 homozygotes and heterozygotes with S-8 and one class-II S haplotype (d) and in S-22 homozygotes and class-II S haplotype homozygotes (e) was also detected. Error bars represent standard errors (SE) of the mean of triplicate samples
The mature sRNA expression levels of Smi in the tapetum cells of S-22, S-8, S-44, S-60, S-40, and S-29 homozygotes and S-22/S-44, S-22/S-60, S-22/S-40, S-22/S-29, S-8/S-44, S-8/S-60, S-8/S-40, and S-8/S-29 heterozygotes were analyzed by stem-loop RT-PCR. The results showed that mature sRNA of Smi is expressed in S-22 homozygotes and S-22/S-60, S-22/S-40, and S-22/S-29 heterozygotes but is totally suppressed in S-22/S-44 heterozygotes (Fig. 5b). Furthermore, the expression of precursors of Smi-22 was not detected in S-22/S-44 heterozygotes (Fig. 5c). Mature sRNA of Smi was also detected in all plants having the S-8 haplotype that we analyzed, including S-8/S-44 heterozygotes (Fig. 5d). To confirm that the mature sRNA of Smi is formed from the class-I S locus, not from the class-II S locus, the expression level of mature sRNA of Smi was also analyzed in S-44, S-60, S-40, and S-29 homozygotes. The expression of mature sRNA was hardly detected in the S-29 homozygotes and was not detected in the S-44, S-60, and S-40 homozygotes (Fig. 5e), confirming that the mature sRNA detected in the heterozygotes was mainly from the class-I S haplotypes. These results suggest that SCR-44 expression in S-22/S-44 heterozygotes is due to the suppression of Smi expression.
Discussion
In the present study, we determined the nucleotide sequence of SCR-22 of B. rapa. SCR-22 was found to be 20 amino acids longer than other SCR/SP11 proteins. Such a long SCR/SP11 protein has not previously been reported. In the putative signal peptide of SCR-22, three amino acids, i.e., the seventh, tenth, and twelfth, were not conserved. These changes are not considered to contribute to the difference in the hydrophobicity level of the SCR-22 signal peptide from that of other class-I SCR/SP11 proteins. The grand average of the hydropathicity value of the SCR-22 signal peptide was between those of SCR-8 and SCR-12, indicating that the function of the SCR-22 signal peptide was maintained.
SCR-44 of class-II SCR/SP11 alleles has been thought to be recessive to all class-I SCR/SP11 alleles. In the present study, pollination tests showed that class-I SCR-22 is recessive to SCR-44 but dominant to SCR-60, SCR-40, and SCR-29 (Table 1). The dominance relationships between SCR-22 and class-II SCR/SP11 alleles were further confirmed by gene expression analysis. The expression level of recessive SCR/SP11 alleles is greatly reduced in S heterozygotes15. Our results showed that the expression of SCR-22 is suppressed and that SCR-44 is normally expressed in the tapetum of S-22/S-44 heterozygotes (Fig. 2). Since the suppression of recessive class-II SCR/SP11 transcription is considered to result from methylation of the promoter region induced by Smi, which can be observed in the tapetum19,20, we developed a method for tapetum isolation. Cytosine methylation was detected at a level comparable to that reported previously;18,19 therefore, our tapetum isolation method was found to be usable for analyses of the methylation states of the SCR-22 promoter and the expression level of Smi.
Our investigation of dominance relationships showed that SCR-44 was recessive to class-I SCR-8 but dominant to class-I SCR-22. Methylation state analysis showed that the widespread methylated cytosine residues were present in the SCR-44 promoter in S-8/S-44 heterozygotes (Fig. 4). The methylation profile of the SCR-44 promoter in S-8/S-44 heterozygotes is similar to that of the SCR-60 promoter in S-52/S-60 heterozygotes18,19. The methylated cytosine residues of the SCR-44 promoter at CpG, CpNpG, and CpNpN sites suggest that Smi sRNA triggers monoallelic de novo methylation in the recessive SCR-44 promoter19,22. The methylation percentage of the SCR-44 promoter in S-8/S-44 was clearly lower than that of the SCR-60 promoter in S-52/S-60 heterozygotes and higher than the SCR-60 promoter in S-44/S-60 heterozygotes18,19. The methylation frequencies of two cytosine residues in the region homologous to Smi in the SCR-44 of S-8/S-44 heterozygotes were 32.1% and 39.3%, respectively. Methylated cytosine was also detected in the region homologous to Smi in the recessive SCR-60, SCR-40, or SCR-29 in the heterozygotes (Supplementary Figure S3). In the homozygotes with class-II SCR/SP11 alleles and S-22/S-44 heterozygotes, few methylated cytosines were observed in the promoter of class-II SCR/SP11 alleles. These results confirmed the suppression of recessive class-II SCR/SP11 alleles induced by DNA methylation in their promoter regions.
Because Smi has been revealed to play a key role in inducing the promoter DNA methylation of recessive class-II SCR/SP11 alleles, the precursor sequence of Smi from S-8 and S-22 was identified (Fig. 5a). The sequences of the mature Smi-8 and Smi-22 sRNAs were the same as that of the Smi-9 sRNA19. The mature Smi sRNA was detected in the tapetum cells of S-8 or S-22 homozygotes and S-8/S-44, S-8/S-60, S-8/S-40, S-8/S-29, S-22/S-60, S-22/S-40, and S-22/S-29 heterozygotes. However, mature Smi sRNA could not be detected in S-22/S-44 heterozygotes. At the same time, class-I mature Smi sRNA could not be detected in the tapetum cells of S-44, S-60, or S-40 homozygotes and was hardly detected in S-29 homozygotes. These results confirm that the Smi sRNA detected in the present study was mainly from S-8 or S-22. In addition, the precursors of Smi-22 were not detected in S-22/S-44 heterozygotes (Fig. 5c). Thus, the absence of Smi sRNA in S-22/S-44 heterozygotes is considered to be due to the transcription suppression of Smi-22 rather than a failure to cleave precursors of Smi-22 into mature sRNA. In addition, in S-22/S-44 heterozygotes, few methylated cytosines were detected in the promoter of SCR-44, indicating that the normal transcription of SCR-44 resulted from the absence of Smi sRNA of S-22. Although Smi sRNA is not transcribed in S-22/S-44 heterozygotes, it is transcribed in S-22 homozygotes and in S-22/S-60, S-22/S-40, and S-22/S-29 heterozygotes. The suppression of Smi-22 transcription may be related to the function of the S-44 haplotype. Most conserved microRNA (miRNA) genes are independent transcription units and have their own promoters23. Stress-responsive elements and tissue-specific regulatory elements have been found in the promoters of miRNA genes24,25. Smi is expressed in the tapetum specifically19, indicating that tissue-specific regulatory elements are present in the promoter region of the Smi-22 gene. Transcription of Smi-22 may be suppressed by some factor present in the S-locus sequence of S-44. The sequence of more than 10 kb of the S-44 haplotype has been determined and published26, but it is not available in the sequence database. Therefore, repeated sequencing analysis is required for identification of the factor responsible for the suppression of SCR-22 transcription.
Recently, Smi2 was identified to control the linear dominance hierarchy of the four class-II SCR alleles20. It is possible that SCR-22 expression is suppressed by Smi2 of the S-44 haplotype. However, no region similar to Smi2 was found in the promoter region of SCR-22, indicating that the suppression of SCR-22 expression in S-22/S-44 heterozygotes was not related to the Smi2 of SCR-44.
To examine whether the same suppression mechanism as that for class-II SCR/SP11 participates in the suppression of recessive class-I SCR-22 in S heterozygotes, the methylation state of recessive SCR-22 in the tapetum was analyzed. The cytosine residue located at the –37 position of the SCR-22 5’ region was highly methylated in S-22/S-44 heterozygotes (Fig. 4). In recessive class-II SCR alleles, methylated cytosine residues are widespread in the promoter region, with all three types of cytosine methylation, i.e., CpG, CpNpG, and CpNpN, occurring in the region18,19. However, in recessive class-I SCR-22 in the tapetum of S-22/S-44 heterozygotes, methylated cytosine was restrictedly localized at the –37 cytosine, and only CpNpN methylation was observed. In S-22/S-36 heterozygotes, low levels of CpNpG and CpNpN methylation were detected in the region from –350 to –440 bp of the SCR-22 5’ region. Promoter hypermethylation around cis-regulatory elements could affect transcription repression by interfering with the transcription machinery27,28. It has been reported that the region around –192 bp of SCR-9 contains the elements required for expression in the tapetum12. Alignment revealed that the 5’ region between –1 and –200 bp of SCR-22 and SCR-9 was highly conserved (Supplementary Figure S4), with the two sequences sharing 82.7% identity. Thus, the region around –191 bp of SCR-22 is inferred to contain the elements required for expression in the tapetum. Our results showed that the hypermethylated cytosine located at the –37 position of the SCR-22 5’ region of the S-22/S-44 heterozygotes or the –350 to –440 region of the SCR-22 5’ region with a low methylation rate in S-22/S-36 heterozygotes is far from the core region for expression in the tapetum. Although a putative core binding sequence, CA(A/C)G(T/C)(T/C/A)(T/C/A), for a class of plant-specific NAC transcription factors was suggested to be present within the 5’ region (nucleotides –37 to –31) of SCR-22 by a survey of putative cis-regulatory elements in silico29 (Supplementary Figure S4), this putative cis-regulatory element was not found within the promoter region of SCR-47. Thus, the binding sequence of these NAC transcription factors is not essential for the expression of all class-I SCR/SP11 alleles in the tapetum. In addition, no methylated cytosine was detected in the class-I SCR-22 in microspores of S-22/S-44 or S-22/S-36 heterozygotes, suggesting that the suppression of SCR-22 expression is not induced by DNA methylation. Thus, the recessive SCR-22 is considered to be suppressed through a mechanism without a DNA methylation pathway in general. The hypermethylated cytosine in the SCR-22 promoter in the tapetum of S-22/S-44 heterozygotes or the cytosine with a low methylation rate in the –350 to –440 region of the SCR-22 5’ region in the tapetum of S-22/S-36 heterozygotes may be the result of histone modification22. These findings suggest that the suppression of transcription of SCR-22 is not caused by the DNA methylation-mediated suppression through preventing the transcription factors from binding to their target sequence but possibly by the alteration of chromatin structure22. Therefore, a suppression mechanism different from that for class-II SCR/SP11 may function in the suppression of recessive class-I SCR-22 in S heterozygotes.
Class-I SCR/SP11 alleles have been reported to be transcribed in both the tapetum and microspores7,10,16. The transcription of class-I SCR/SP11 alleles in microspores occurs slightly later than that in the tapetum12. We found that SCR-22 was expressed in microspores of S-22 homozygotes but not in those of S-22/S-44 and S-22/S-36 heterozygotes. Although S-44 and S-36 were not present in the microspores having S-22 in the S-22/S-44 and S-22/S-36 heterozygotes, the expression of SCR-22 was suppressed, indicating that suppression of SCR-22 expression may have been induced to occur before meiosis by a mechanism different from that caused by Smi. Further analyses are required to elucidate the suppression mechanism of recessive class-I SCR/SP11 alleles in S heterozygotes.
Methods
Plant materials
S-8, S-22, S-36, S-44, S-60, S-40, and S-29 homozygotes of B. rapa2,30 were used as the plant materials. Heterozygotes were obtained by cross-pollination of the S homozygotes.
Pollination tests
Pollinated flowers were placed on solid agar for 24 h at 21 °C. Pistils were softened in 1 N NaOH at 55 °C for 1 h. The pistils were then stained with 0.1% aniline blue in 0.1 M K3PO4 and mounted in 60% glycerol. Pollen tubes were observed under a fluorescence microscope. Three flowers were used for each pollination, and the tests were replicated three times on different days.
Amplification of SCR-22 from B. rapa
Total RNA was extracted from anthers of S-22 homozygotes using TRIzol reagent (Invitrogen, Shanghai, China). RNA was reverse-transcribed using a SuperScript™ III First-Strand Synthesis System (Invitrogen, Shanghai, China). A partial sequence of SCR-22 was amplified by nested PCR using the primers SP11-131, SP11-F16, SP11-Fa, and SP11-1F8 as forward primers and Not1-(dT)18 as a reverse primer for the first PCR and SP11-231, SP11-F26, and SP11-2Fa as forward primers and RT1-long9 as a reverse primer for the second PCR. When SP11-1 and SP11-F2 were used for the first and second PCRs, a partial fragment of SCR-22 was amplified. The promoter region and entire DNA sequence of SCR-22 were identified by inverse PCR32. The primer sequences are listed in Supplementary Table S1.
Tapetum isolation
Thirty flowers were collected 3 days before anthesis for collecting anthers. Both ends of the anthers were cut away, and the remaining part of the anthers was cut into two equal sizes. The two pieces were then placed into 1 mL tapetum isolation buffer (50 mM Hepes buffer, 0.5 M sucrose, and KOH to adjust the pH value to 7.5) in 1.5 mL centrifuge tubes. The tube was vortexed for 30 min to release microspores. The solution containing microspores was removed by filtration using nylon net (0.5 mm pore size) and washed three times with tapetum isolation buffer. The solution was centrifuged at 100 × g for 10 min, and the pellet contained microspores. Then, 1 mL isolation buffer (5% cellulase RS and 1% pectolyase Y23 in tapetum cell isolation buffer) was added to the anthers and vortexed for approximately 7 min. The upper solution was transferred to a new tube by filtration using a nylon net (0.5 mm pore size), and the upper solution was centrifuged at 20,000 × g for 10 min. The pellet contained the tapetum. Total RNA and DNA were isolated from the tapetum using TRIzol reagent (Invitrogen, Shanghai, China) for the following experiments.
To examine the adequacy of our method, the anthers collected before and after the isolation process were embedded in 5% agar. The anthers were sliced into 30 μM-thin sections by a DTK-3000 microslicer (Dosaka, Kyoto, Japan), and the sections were observed by a microscope.
SCR/SP11 expression analysis
Total RNA was isolated from the tapetum cells of S-8, S-22, S-36, S-44, S-60, S-40, and S-29 homozygotes and S-22/S-36, S-22/S-44, S-22/S-60, S-22/S-40, S-22/S-29, S-8/S-44, S-8/S-60, S-8/S-40, and S-8/S-29 heterozygotes. RNA was reverse-transcribed by the SuperScript™ III First-Strand Synthesis System (Invitrogen, Shanghai, China). Real-time RT-PCR was performed using SsoAdvancedTM SYBR® Green Supermix (Bio-Rad, Shanghai, China) on a Bio-Rad® CFX96 system, following the manufacturer’s instructions. Each SCR/SP11 region was amplified with specific primers (Supplementary Table S1). The Actin gene was used as an endogenous reference gene. The primers were confirmed to be approximately 90% to 100% efficient for amplification, and the 2−∆∆CT method33 was used for all analyses. All reactions were performed in triplicate, and an average value was calculated for each set of reactions.
DNA methylation state detection
DNA was isolated from the tapeta of S-22 and S-44 homozygotes and S-22/S-36, S-22/S-44, and S-8/S-44 heterozygotes. The DNA was bisulfite treated with a MethylCode™ Bisulfite Conversion Kit (Applied Biosystems, Shanghai, China). The SCR-44 promoter region and the promoter region, coding region, and intronic region of SCR-22 modified by bisulfite were amplified using specific primers (Supplementary Table S1). Amplified PCR products were cloned into pGEM-T Easy vectors (Promega, Beijing, China), and at least 30 clones were sequenced.
Detection of mature and precursor Smi sRNA
Detection of mature Smi sRNA was performed as previously described34. When microspores were in the uninucleate stage, small RNA was isolated from the anthers of S-8, S-22, S-44, S-60, S-40, and S-29 homozygotes and S-22/S-44, S-22/S-60, S-22/S-40, S-22/S-29, S-8/S-44, S-8/S-60, S-8/S-40, and S-8/S-29 heterozygotes using a mirVana™ miRNA Isolation Kit (Ambion, Shanghai, China) and reverse-transcribed using SuperScript™ III RT with RT primers and U6-specific primers (Supplementary Table S1). The transcribed products were quantified using SsoAdvancedTM SYBR® Green Supermix (Bio-Rad, Shanghai, China) with small RNA-specific primers and universal primers (Supplementary Table S1). U6 was used as an endogenous reference gene. The 2−∆∆CT method33 was used for all analyses. All reactions were performed in triplicate, and an average value was calculated for each set of reactions.
For detection of precursor Smi sRNA, total RNA was isolated from anthers when microspores were at the uninucleate stage. RNA was reverse-transcribed by the SuperScript™ III First-Strand Synthesis System (Invitrogen, Shanghai, China) with precursor Smi-22-specific primers and Actin-R (Supplementary Table S1). Real-time RT-PCR was performed using SsoAdvancedTM SYBR® Green Supermix (Bio-Rad, Shanghai, China) on a Bio-Rad® CFX96 system, following the manufacturer’s instructions. The Actin gene was used as an endogenous reference gene. The primers were confirmed to be approximately 90% to 100% efficient for amplification, and the 2−∆∆CT method33 was used for all analyses.
Accession numbers
The sequence data of SCR-22, Smi-8, and Smi-22 were deposited in the GenBank of the National Center for Biotechnology Information (NCBI). The accession numbers are Smi-8, MG708355; Smi-22, MG708356; and SCR-22, MG708357.
References
Stein, J. C., Howlett, B., Boyes, D. C., Nasrallah, M. E. & Nasrallah, J. B. Molecular-cloning of a putative receptor protein-kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl Acad. Sci. USA 88, 8816–8820 (1991).
Takasaki, T. et al. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403, 913–916 (2000).
Schopfer, C. R., Nasrallah, M. E. & Nasrallah, J. B. The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700 (1999).
Suzuki, G. et al. Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S-9 haplotype of Brassica campestris (syn. rapa). Genetics 153, 391–400 (1999).
Nasrallah, J. B., Yu, S. M. & Nasrallah, M. E. Self-incompatibility genes of Brassica oleracea: expression, isolation, and structure. Proc. Natl Acad. Sci. USA 85, 5551–5555 (1988).
Watanabe, M. et al. Highly divergent sequences of the pollen self-incompatibility (S) gene in class-I S haplotypes of Brassica campestris (syn. rapa) L. FEBS Lett. 473, 139–144 (2000).
Schopfer, C. R. & Nasrallah, J. B. Self-incompatibility: prospects for a novel putative peptide-signaling molecule. Plant Physiol. 124, 935–939 (2000).
Sato, K. et al. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B-rapa. Genetics 162, 931–940 (2002).
Okamoto, S., Sato, Y., Sakamoto, K. & Nishio, T. Distribution of similar self-incompatibility (S) haplotypes in different genera, Raphanus and Brassica. Sex. Plant. Reprod. 17, 33–39 (2004).
Takayama, S. et al. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl Acad. Sci. USA 97, 1920–1925 (2000).
Iwano, M. et al. Immunohistochemical studies on translocation of pollen S-haplotype determinant in self-incompatibility of Brassica rapa. Plant Cell Physiol. 44, 428–436 (2003).
Shiba, H. et al. A pollen coat protein, SP11/SCR, determines the pollen S-specificity in the self-incompatibility of Brassica species. Plant Physiol. 125, 2095–2103 (2001).
Nasrallah, J. B., Nishio, T. & Nasrallah, M. E. The self-incompatibility genes of Brassica: expression and use in genetic ablation of floral tissues. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 42, 393–422 (1991).
Kakizaki, T. et al. Linear dominance relationship among four class-II S haplotypes in pollen is determined by the expression of SP11 in Brassica self-incompatibility. Plant Cell Physiol. 44, 70–75 (2003).
Shiba, H. et al. The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell 14, 491–504 (2002).
Kusaba, M., Tung, C. W., Nasrallah, M. E. & Nasrallah, J. B. Monoallelic expression and dominance interactions in anthers of self-incompatible Arabidopsis lyrata. Plant Physiol. 128, 17–20 (2002).
Fujimoto, R., Sugimura, T., Fukai, E. & Nishio, T. Suppression of gene expression of a recessive SP11/SCR allele by an untranscribed SP11/SCR allele in Brassica self-incompatibility. Plant Mol. Biol. 61, 577–587 (2006).
Shiba, H. et al. Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat. Genet. 38, 297–299 (2006).
Tarutani, Y. et al. Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466, 983–U110 (2010).
Yasuda, S. et al. A complex dominance hierarchy is controlled by polymorphism of small RNAs and their targets. Nat. Plants 3, 16206 (2016).
Hatakeyama, K. et al. The S receptor kinase gene determines dominance relationships in stigma expression of self-incompatibility in Brassica. Plant J. 26, 69–76 (2001).
Saze, H., Tsugane, K., Kanno, T. & Nishimura, T. DNA methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol. 53, 766–784 (2012).
Zhang, B. H., Wang, Q. L. & Pan, X. P. MicroRNAs and their regulatory roles in animals and plants. J. Cell Physiol. 210, 279–289 (2007).
Usadel, B. et al. Multilevel genomic analysis of the response of transcripts, enzyme activities and metabolites in Arabidopsis rosettes to a progressive decrease of temperature in the non-freezing range. Plant Cell Environ. 31, 518–547 (2008).
Halford, N. G. in Plant Developmental Biology - Biotechnological Perspectives (eds Pua, E.C & Davey, M. R.) Ch. 4, 67–82 (Springer, Berlin Heidelberg, 2010).
Kakizaki, T. et al. Comparative analysis of the S-intergenic region in class-II S haplotypes of self-incompatible Brassica rapa (syn. campestris). Genes Genet. Syst. 81, 63–67 (2006).
Zhu, W. G. et al. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in thep21(Cip1) promoter. Mol. Cell Biol. 23, 4056–4065 (2003).
Boyes, J. & Bird, A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64, 1123–1134 (1991).
Puranik, S., Sahu, P. P., Srivastava, P. S. & Prasad, M. NAC proteins: regulation and role in stress tolerance. Trends Plant. Sci. 17, 369–381 (2012).
Nou, S., Watanabe, M., Isogai, A. & Hinata, K. Comparison of S-alleles and S-glycoproteins between two wild populations of Brassica campestris in Turkey and Japan. Sex. Plant Reprod. 6, 79–86 (1993).
Kimura, R., Sato, K., Fujimoto, R. & Nishio, T. Recognition specificity of self-incompatibility maintained after the divergence of Brassica oleracea and Brassica rapa. Plant J. 29, 215–223 (2002).
Pavlopoulos, A. in Molecular Methods for Evolutionary Genetics (eds Orgogozo, V. & Rockman, M. V.) 267–275 (Humana Press, New York, USA 2011).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408 (2001).
Varkonyi-Gasic, E., Wu, R., Wood, M., Walton, E. F. & Hellens, R. P. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12 (2007).
Acknowledgements
This work was supported in part by the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Foreign Research to C.-L.W. (P10094), the National Natural Science Foundation of China (No. 31401856 to C.-L.W.), and the Natural Science Foundation of Jiangsu Province (No. BK20140482 to C.-L.W.).
Author information
Authors and Affiliations
Contributions
C. -L.W., H.K., and T.N. designed the experiments and wrote the paper; C.-L.W., Z.-P. Z., and E.O. performed the experiments and analyzed the data.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, CL., Zhang, ZP., Oikawa, E. et al. SCR-22 of pollen-dominant S haplotype class is recessive to SCR-44 of pollen-recessive S haplotype class in Brassica rapa. Hortic Res 6, 25 (2019). https://doi.org/10.1038/s41438-018-0103-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-018-0103-5