Abstract
Our previous studies indicated that tomato miR482b could negatively regulate the resistance of tomato to Phytophthora infestans and the expression of miR482b was decreased after inoculation with P. infestans. However, the mechanism by which the accumulation of miR482b is suppressed remains unclear. In this study, we wrote a program to identify 89 long noncoding RNA (lncRNA)-originated endogenous target mimics (eTMs) for 46 miRNAs from our RNA-Seq data. Three tomato lncRNAs, lncRNA23468, lncRNA01308 and lncRNA13262, contained conserved eTM sites for miR482b. When lncRNA23468 was overexpressed in tomato, miR482b expression was significantly decreased, and the expression of the target genes, NBS-LRRs, was significantly increased, resulting in enhanced resistance to P. infestans. Silencing lncRNA23468 in tomato led to the increased accumulation of miR482b and decreased accumulation of NBS-LRRs, as well as reduced resistance to P. infestans. In addition, the accumulation of both miR482b and NBS-LRRs was not significantly changed in tomato plants that overexpressed lncRNA23468 with a mutated eTM site. Based on the VIGS system, a target gene of miR482b, Solyc02g036270.2, was silenced. The disease symptoms of the VIGS-Solyc02g036270.2 tomato plants were in accordance with those of tomato plants in which lncRNA23468 was silenced after inoculation with P. infestans. More severe disease symptoms were found in the modified plants than in the control plants. Our results demonstrate that lncRNAs functioning as eTMs may modulate the effects of miRNAs in tomato and provide insight into how the lncRNA23468-miR482b-NBS-LRR module regulates tomato resistance to P. infestans.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are noncoding RNAs of 20–24 nucleotides1 that transcriptionally and posttranscriptionally regulate gene expression in various biological processes of plants2. Many miRNAs act in plant responses to various biotic challenges. For example, miR482, belonging to the miR482/2118 family, functions in various plants3,4. After Verticillium dahliae infection, the accumulation of miR482e in potato significantly decreases5. Similarly, after infection with cucumber mosaic virus and V. dahliae, the expression of miR482 is suppressed in tomato and cotton, respectively6,7.
To show the biological function of miRNAs, identification of their target genes is necessary and important8. MiR482 can silence members of the NBS-LRR gene family4. An NBS-LRR protein is a disease resistance (R) protein that is involved in the effector-triggered immunity (ETI) of the plant innate immune system9. An NBS-LRR protein contains a nucleotide-binding site (NBS), a leucine-rich repeat (LRR) and a toll-interleukin receptor-like (TIR) domain or a coiled coil (CC) domain10. An NBS-LRR gene, GbRVd, was cloned and characterized in Gossypium barbadense, and its silencing enhances the susceptibility of G. barbadense to Verticillium wilt11. The resistance of Nicotiana benthamiana to Phytophthora parasitica is effectively enhanced by the overexpression of the grapevine TIR-NB-LRR gene VaRGA112. Plants infected with pathogens show an increased level of NBS-LRR transcripts and reduced level of miR4827,13. Potato miR482 acts by suppressing NBS-LRR genes to regulate the potato resistance against V. dahliae infection5.
Recently, long noncoding RNA (lncRNA), another type of ncRNA, has been identified and analyzed in various biological processes14. LncRNAs are a set of RNA transcripts (>200 nt length) that have no protein-coding ability15. LncRNAs play important roles in flowering time regulation16, fruit development17,18, photomorphogenesis19, gene silencing20,21 and biotic and abiotic stress responses22,23. In plant-pathogen interactions, using an RNA sequencing approach, 125 putative stress-responsive lncRNAs that are induced by powdery mildew infection have been identified in wheat 24. A number of lncRNAs have been identified in Paulownia witches’ broom-infected Paulownia tomentosa by high-throughput sequencing25. In addition, Arabidopsis thaliana lncRNAs have been found to act in Arabidopsis resistance to Fusarium oxysporum22. In tomato, a comprehensive set of lncRNAs was identified26,27,28. These lncRNAs were primarily involved in the regulation of tomato fruit ripening18,28,29,30, the ethylene signal transduction pathway31, chilling injury32, and the tomato-potato spindle tuber viroid (PSTVd)/tomato yellow leaf curl virus (TYLCV) interaction33,34,35.
Tomato is the second most important vegetable crop in the world, constituting a major agricultural industry36. Late blight (LB), caused by Phytophthora infestans, is one of the most serious diseases of tomato. In the early 2000s, LB occurred and hindered tomato production, causing serious economic losses in the USA and China37. In our previous work, we focused on the effects of lncRNAs and miRNAs during tomato resistance to P. infestans. A number of miRNAs were identified by next-generation sequencing38, including miR482b. MiR482b was downregulated after infection with P. infestans and negatively regulated tomato resistance8. Tomato lncRNA16397 was found to induce glutaredoxin expression to enhance resistance to P. infestans39. However, the mechanisms by which lncRNAs, as competing endogenous RNAs (ceRNAs), suppress the accumulation of miRNAs in the tomato-P. infestans interaction are unknown. To determine whether lncRNAs can suppress miR482b accumulation in tomato resistance to pathogen infection, RNA-Seq data were used to identify and characterize a number of lncRNAs, and bioinformatics analysis was used to predict the endogenous target mimics (eTMs) of these lncRNAs. We found that lncRNA23468 modulated the accumulation of NBS-LRRs by suppressing miR482b expression in tomato plants infected with P. infestans. These results will improve our understanding of the regulatory mechanism of miR482b in the response of tomato to P. infestans infection and help future molecular-based breeding approaches of pathogen resistance.
Materials and methods
Bioinformatics pipeline for identifying lncRNAs
To identify the lncRNAs, we used two RNA-Seq datasets obtained from our previous studies. These two datasets were constructed by LC Biotech, Hangzhou, China, using the leaves of miR482b-overexpressing and Zaofen No. 2 tomatoes. The clean reads were de novo assembled using Cufflinks. TopHat was used to align assembled transcripts to the tomato genome iTAGv2.3 (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias = Org_Slycopersicum). All transcripts were required to be more than 200 bp in length. The lncRNAs were identified according to the method of Cui et al.39. According to their genomic locations, the lncRNAs were classified into four categories. The fragments per kilobaseof exon per million fragments mapped (FPKM) value was applied to represent the normalized expression value of the lncRNAs.
Prediction of ceRNAs
All tomato miRNAs were collected from miRBase (http://www.mirbase.org/), and the lncRNAs identified above were used as the ceRNA prediction libraries. CeRNAs for the selected tomato miRNAs were predicted using RNAhybrid software with the following rules: (i) P-value < 0.05 and minimum free energy (mfe) <−25 kcal/mol; (ii) bulges were only permitted at the ninth to 12th positions of the 5’ end of a miRNA sequence; (iii) the bulge in ceRNAs should be composed of 2–4 nucleotides; (iv) G/U pairs were allowed with the ceRNA and miRNA pairing region, and perfect nucleotide pairing was required at the second to eighth positions of the 5’ end of the miRNA sequence; and (v) except for the central bulge, the total mismatches within the ceRNA and miRNA pairing regions should be no more than four, with no more than two consecutive mismatches. Interaction networks among the ceRNAs and miRNAs were constructed using the software Cytoscape. WebLogo software was used to analyze the conserved residues (http://weblogo.berkeley.edu/logo.cgi).
Tomato material and P. infestans inoculation
Tomato Zaofen No. 2, bred by the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China, is an accession susceptible to P. infestans. The tomato was grown in a greenhouse under 16 h light within a temperature range of 22–28 °C. P. infestans strain P12103 was cultured in oat medium in the dark at 20 °C. The tomato plants (4–5-leaf stage) were inoculated with P. infestans spores according to the method of Jiang et al.8. The whole fifth leaves of each sample were collected at the indicated times (0, 1, 2, 3 and 4 dpi). All samples were quickly frozen in liquid nitrogen and stored at −80 °C until RNA isolation.
Cloning of lncRNA23468, mutation of lncRNA23468 and construction of the overexpression plasmid
According to the tomato genome and lncRNA prediction results, a pair of primers (l23468F and l23468R) were designed and used to clone lncRNA23468 from tomato plant (Table S1).
We introduced six point mutations to lncRNA23468 within sequences pairing with miR482b. LncRNA23468 mutation was generated by PCR, which involved amplification and mutagenesis using lncRNA23468 as the backbone. Two more primers were used for this: ml23468-1R and ml23468-2F (Table S1). Three rounds of PCR were performed to amplify the mutated lncRNA23468 (mlncRNA23468). First, the primers l23468F and ml23468-1R were used to amplify a fragment containing mutation points, and then ml23468-2F and l23468R were used to amplify another fragment. Finally, the PCR products of the first and second rounds were used as the template along with l23468F and l23468R.
The PCR fragments of lncRNA23468 or mlncRNA23468 were subcloned into binary vector pBI121, replacing the GUS gene. In plasmids, lncRNA23468 and mlncRNA23468 were controlled by the Cauliflower mosaic virus (CaMV) 35S promoter.
Virus-induced gene silencing (VIGS) constructs
TVR-based vectors (pTRV1 and pTRV2), which were provided by Prof. Liu from Tsinghua University of China, were used for VIGS. The VIGS sequence was designed according to the SGN VIGS Tool (http://vigs.solgenomics.net/) and cloned into the pTRV2 vector by the ligation-independent cloning method40,41.
Agrobacteria infiltration
All the plasmids were transformed into A. tumefaciens strain GV3101 by the freeze-thaw method37. Agrobacteria infiltration was performed according to Jiang’s method8.
A. tumefaciens containing pBI-121-lncRNA23468 or pBI-121-mlncRNA23468 plasmids was introduced into the leaves of Zaofen No. 2 tomato by infiltration. A. tumefaciens with an empty vector was used as a control. The leaves were harvested for the next experiments at 3 dpi.
In the tobacco (N. tabacum) system, we introduced Agrobacterium harboring pBI121-miR482b into tobacco leaf cells. After 3 days, the Agrobacterium harboring pBI121-lncRNA23468 and mlncRNA23468 were introduced into the tobacco leaves that expressed miR482b. The accumulation of miR482b was examined.
Agrobacterium cultures containing pTRV2 derivatives and pTRV1 were mixed at a 1:1 ratio and then infiltrated into a 2-3-leaf-stage tomato. The pTRV2 empty vector was used as the negative control. The plants were maintained for 3 weeks in a greenhouse under 16 h light within 20 °C, and leaflets were harvested from several plants for the isolation of RNA and qRT-PCR analysis to assess the degree of silencing.
P. infestans resistance analysis
For the tomato plants that overexpressed lncRNA23468 or mlncRNA23468, the infiltrated leaf regions were inoculated with 20 μl of P. infestans (106 zoospores/ml) and then were placed at 20 ± 1 °C in a 100%-relative-humidity environment without light. The lesions were observed at the fifth day, and the sizes of the lesions were also calculated.
3 weeks after VIGS, detached leaves from the VIGS tomato plants were inoculated with 20 μl of the P. infestans zoospore suspension (1 × 106 zoospores/ml) according to Jiang’s method8. The whole plants were sprayed to run-off with the same zoospore suspension by the method described above. At 5 dpi, the diameters of the lesions and the abundance of P. infestans were calculated according to Jiang’s method8.
The disease index (DI) was calculated according to disease grade (DG). The DGs were sorted from 0 to 6 on the basis of the lesion area (Table S2). The DI was calculated according to the following formula:
Where DGi is the value of DG, ni is the number of plants in each DG, and n is the total number of plants. Each experiment was carried out at least three times.
RNA isolation, reverse transcription, and qRT-PCR analysis
Total RNA was extracted using RNAiso Plus (TaKaRa, Dalian, China). Reverse transcription and the qRT-PCR reactions of miRNAs were performed with TransScript Green miRNA Two-Step qRT-PCR SuperMix (Transgen Biotech, Beijing, China) according to the manufacturer’s instructions. We used PrimeScriptTM RT Master Mix (TaKaRa, Dalian, China) to synthesize the cDNAs of the mRNAs and lncRNAs. The qRT-PCR reactions were performed by using a SYBR Premix Ex TaqTM II kit (TaKaRa, Dalian, China). The qRT-PCR reactions of the selected genes, miRNAs and lncRNAs were performed on an ABI7500. The tomato actin gene was used as an internal reference gene. All primer sequences are shown in Table S1. The gene sample Ct values were standardized, and the 2–ΔΔCt method was used to analyze the relative changes in expression. Of the nine leaves sampled in each experiment, three leaves were pooled into one biological replicate, resulting in three biological replicates.
Statistical analysis
All statistical analyses of the data were performed with SPSS19.0, and all data were expressed as the means ± SEs from three independent experiments. We used the Tukey method to estimate significance.
Results
Identification of lncRNAs
Two of our group’s RNA-Seq datasets, miR482b-overexpressing (OE482) and Zaofen No. 2 tomatoes (Slz), were used to identify the expressed lncRNAs. After assembly and mapping to the tomato genome, the noncoding transcripts were considered “putative” lncRNAs and filtered according to length, coding potentials and coverage of reads. From these analyses, approximately 9742 unique lncRNAs were obtained in two samples, and 9562 of these lncRNAs were expressed in the OE482 sample (Table S3). Among these lncRNAs, 6,785 were natural antisense transcripts (x); 2,669 were long intergenic noncoding RNAs (u); 72 were generic exonic overlaps with a reference transcript (o); and 36 were potentially novel isoforms (j) in the OE482 sample (Fig. 1a). These lncRNAs were evenly distributed across the 12 chromosomes in tomato (Fig. 1b).
a Composition of different types of lncRNAs. b The expression levels of the lncRNAs (log10 FPKM) along 12 tomato chromosomes. c Cytoscape results of lncRNAs and miRNAs. The 89 lncRNAs that may act as ceRNAs could be bound by 46 miRNAs. The red round and green square nodes represent lncRNAs and miRNAs, respectively
Identification of ceRNAs
LncRNAs may act as ceRNAs via the eTMs of miRNAs. Therefore, a program was written to identify the lncRNAs acting as eTMs of miRNAs in tomato. In total, 89 lncRNAs were found to be bound by 46 miRNAs (Table S4). Interaction networks showed that 38 miRNA-lncRNA duplexes were formed (Fig. 1c). In these duplexes, miR482d-5p was putatively sequestered by 16 lncRNAs, and lncRNA34658 decoyed three miRNAs, including miR168a-5p, miR168b-5p, and miR477-3p. In addition, more than one-third of the regulated relationships had only two nodes, as shown in the networks in Fig. 1c.
The eTMs of miR482b and expression analysis of miR482b ceRNAs after infection with P. infestans
The predicted binding sites of miR482b among these lncRNAs (lncRNA23468, lncRNA13262 and lncRNA01308) were well conserved (Fig. 2a). In our prediction, a two or three-nucleotide bulge on the eTMs located between the ninth and 12th positions at the 5’ end of miR482b was required for the eTMs to decoy miR482b (Fig. 2b). The minimum free energies (mfes) were −33.8 kcal/mol, −35.8 kcal/mol and −33.2 kcal/mol in lncRNA13262-miR482b, lncRNA01308-miR482b and lncRNA23468-miR482b, respectively. The values of the mfes in lncRNA13262-miR482b and lncRNA01308-miR482b were close to that in lncRNA23468-miR482b. This suggests that the abilities of the three lncRNAs to suppress miR482b were not significantly different.
a Conservation analysis of the eTM sites of three lncRNAs that decoy miR482b. The conservation status of the sequence was analyzed by WebLogo. The red letters represent the conserved sequence at the second to eighth positions of the 5’ end of the miRNA sequence. The purple letters represent bulges permitted at the ninth to 12th positions of the 5’ end of a miRNA sequence. b Predicted base-pairing interactions between miR482b and the eTM sites of three lncRNAs. c Expression of the three lncRNAs in tomato inoculated with P. infestans. All data are the means ± SE of three independent experiments. Different letters among the groups indicate a significant difference at the P = 0.05 level
To explore whether the ceRNAs of the miR482b, lncRNA23468, lncRNA13262 and lncRNA01308 had an effect on tomato response to P. infestans, the relative levels of accumulation were examined in tomato leaves infected by P. infestans. These three lncRNAs responded to P. infestans infection. The changes in the expression of both lncRNA23468 and lncRNA01308 followed the same pattern. Both first increased and then decreased, reaching a peak at 2 days after P. infestans infection. LncRNA13262 was downregulated from 0 dpi to 1 dpi and then reached a peak at 2 dpi (Fig. 2c).
Construction of the lncRNA23468 mutant
To examine whether lncRNA23468 indeed functioned via pairing with miR482b, a six-point mutation was introduced into lncRNA23468 by using sequences that paired with miR482b (mlncRNA23468; Fig. 3a). The design of the mlncRNA23468 is shown in Fig. 3b. The eTM of miR482b (5′-AGAUUGGGCGGAAUAGGUAAGA-3′) in lncRNA23468 was replaced with the mutation sequence (5′-AGAUUCGCCCGAAUAGCUAUCA-3′) by oligonucleotide-directed mutagenesis. After three rounds of PCR, the sequence of mlncRNA23468 was amplified (Fig. 3c). As shown in Fig. 3d, the sequencing results showed that six point mutations were introduced into the eTM from lncRNA23468. In addition, except for the eTM region of miR482b, the lncRNA23468 and mlncRNA23468 sequences were conserved after sequence alignment analysis (Fig. 3e). Overall, mlncRNA23468 was successfully constructed by using lncRNA23468 as the backbone.
a Predicted base-pairing interactions between miR482b and the eTM of lncRNA23468 with the designed mutations. b Strategy for introducing mutations by PCR. c PCR products from the different stages of the mutant eTM of lncRNA23468. M, DL2000 DNA Marker; 1, 2, and 3, products from the first, second and third rounds of PCR, respectively. d The sequencing results of the eTM regions of lncRNA23468 (top) and its mutated sequence (bottom). The red box represents the eTM region. Blue triangles represent the six-point mutation. e Sequence alignment of lncRNA23468 and mlncRNA23468 performed with ClustalX. The red box represents the eTM region
LncRNA23468 as a miRNA decoy suppresses the expression of miR482b
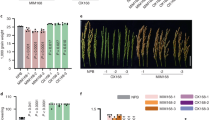
We investigated the possible function of lncRNA23468 using infiltration to upregulate lncRNA23468 expression in Zaofen No. 2 tomato. The plasmids for the overexpression of lncRNA23468 and mlncRNA23468 were carried out on the basis of the pBI121 vector (Fig. 4a). The introduction of A. tumefaciens harboring pBI121-lncRNA23468 and pBI121-mlncRNA23468 into tomato leaf cells resulted in significant upregulation of lncRNA23468 and mlncRNA23468 expression at 3 days: the expression levels of lncRNA23468 and mlncRNA23468 were approximately 6.1-fold and 6.0-fold greater than the tomato leaves that overexpressed empty vector (EV), respectively (Fig. 4b). The expression of miR482b was dramatically suppressed to approximately 40% in the tomato plants that overexpressed lncRNA23468 (OE23468), and the target genes of miR482b, NBS-LRRs, were significantly increased (Fig. 4b). The levels of the transcripts of Solyc02g036270.2, Solyc04g009070.1, Solyc05g008070.2 and Solyc12g016220.2 in the leaves of the OE23468 tomato plants were approximately 2.9-fold, 2.1-fold, 2.4-fold, and 2.1-fold, respectively, to the values in the leaves of the EV tomato plants. The overexpression of mlncRNA23468 in tomato (mOE23468) did not cause expression changes in either miR482b or its target genes (Fig. 4b).
a Schematic diagram of the gene cassette containing lncRNA23468 and mlncRNA23468. b qRT-PCR analysis of lncRNA23468, miR482b and the target genes in the EV, mOE23468 and OE23468 tomato plants. c Disease signs on the detached leaves from the EV, mOE23468 and OE23468 tomato plants at 5 dpi. Scale bars = 0.5 cm. d The diameter of the lesion of the detached leaves from EV, mOE23468 and OE23468 tomato plants at 5 dpi. All data are the means ± SE of three independent experiments. Different letters among the groups indicate a significant difference at the P = 0.05 level
In our previous work, miR172 and miR396 were identified to play important roles in tomato resistance to P. infestans42,43. We examined the accumulation of miR172, miR396 and their target genes in the tomato leaves that overexpressed lncRNA23468 by using qRT-PCR. The overexpression of lncRNA23468 did not cause expression changes in miR172, miR396 and their target genes compared to the control samples (Fig. S1).
Disease resistance tests were performed on the OE23468, mOE23468 and EV plants using the oomycete P. infestans. Five days after infection, the physical appearance of these tomato plants was assessed. Compared with the severe disease symptoms that appeared on the leaves of EV plants, the disease symptoms on the leaves of mOE23468 plants were similar, while fewer disease symptoms were exhibited on the leaves of OE23468 plants (Fig. 4c). The diameter of the lesion was significantly smaller in the leaves of P. infestans-infected OE23468 plants than in those of EV or mOE23468 plants (Fig. 4d).
As overexpression is not enough to fully understand lncRNA functions, lncRNA silencing should be performed to explore these biological functions. LncRNA23468 was silenced in Zaofen No. 2 tomato by VIGS. The empty TRV2 vector and PHYTOENE DESATURASE (PDS)-VIGS were used as the negative control and the positive control, respectively. At 21 days of VIGS treatment, the PDS-VIGS tomato showed visible bleaching, indicating that the lncRNA23468 gene was also silenced (Fig. S2). The expression of lncRNA23468 was suppressed to approximately 40%, and the accumulation of miR482b was upregulated approximately 2-fold, while the four target genes showed decreased abundance (Fig. 5a).
a Relative expression levels of lncRNA23468, miR482b and the target genes of miR482b in the TRV2 and lncRNA23468-VIGS tomato plants. b The silencing efficiency of NBS-LRR in NBS-LRR-VIGS tomato plants. c Phenotypes of the detached leaves from TRV2, lncRNA23468-VIGS and NBS-LRR-VIGS tomato plants at 5 dpi. Scale bars = 0.5 cm. Top: disease symptoms; Bottom: trypan blue staining for detection of dead cells. d The diameter of the lesion of the detached leaves. e Leaf phenotypes at 5 dpi after whole-plant inoculation with P. infestans. Scale bars = 0.5 cm. f Disease index of VIGS-tomato plants at 5 dpi. g Transcript accumulation of the P. infestans actin gene in these inoculated plants at 5 dpi. All data are the means ± SE of three independent experiments. Different letters among the groups indicate a significant difference at the P = 0.05 level
The disease phenotypes for the control (TRV2) and lncRNA23468-VIGS tomato plants were assessed after infection with P. infestans. Compared to the leaves of TRV2 tomato plants, the detached leaves of the lncRNA23468-VIGS tomato plants showed more serious symptoms of the disease. The decreased resistance to P. infestans in the leaves of lncRNA23468-VIGS tomato plants was also revealed by trypan blue staining, as shown by the increased number of necrotic cells relative to those in the leaves of TRV2 tomato plants (Fig. 5c). In addition, Fig. 5d shows that the leaves of the lncRNA23468-VIGS tomato plants had larger lesions than did the leaves of the TRV2 tomato plants.
The leaves of the lncRNA23468-VIGS tomato plants also exhibited more serious symptoms of the disease after whole-plant inoculation with P. infestans (Fig. 5e), with a higher disease index than that observed for the TRV2 tomato plants (Fig. 5f). In addition, there were significant increases in the abundance of P. infestans in the lncRNA23468-VIGS tomato plants compared to the TRV2 tomato plants (Fig. 5g). These results suggest that lncRNA23468 suppressed the accumulation of miR482b, resulting in an increased expression of NBS-LRRs and enhanced tomato resistance to P. infestans.
The regulation between lncRNA23468 and miR482b was also validated using the Nicotiana system. We introduced Agrobacterium harboring pBI121-miR482b into tobacco leaf cells, which led to a significant increase in miR482b expression at 3 days compared to tobacco leaves overexpressing the empty vector (Fig. S3a). Then, the Agrobacterium harboring pBI121-lncRNA23468 and mlncRNA23468 were introduced into the tobacco leaves that overexpressed miR482b. The accumulation of miR482b was significantly decreased after the overexpression of lncRNA23468. The overexpression of mlncRNA23468 did not cause expression changes in miR482b (Fig. S3b). These results suggested that lncRNA23468 as a ceRNA may decoy miR482b.
Silencing of NBS-LRR enhances tomato susceptibility to P. infestans
We also employed the VIGS system to test the role of the target gene of miR482b (Solyc02g036270.2) in tomato resistance to P. infestans. At 21 days after hand-infiltration, a further examination of Solyc02g036270.2 indicated that the abundance of transcripts was significantly suppressed in the tomato plants that silenced Solyc02g036270.2 (NBS-LRR-VIGS) (Fig. 5b).
More serious disease symptoms were shown in the detached leaves of the NBS-LRR-VIGS plants than in the TRV2 plants after infection with P. infestans (Fig. 5c). The darker blue marks from trypan blue staining in the detached leaves indicated that a greater number of dead cells occurred in the NBS-LRR-VIGS tomato plants than in the TRV2 tomato plants (Fig. 5c). Furthermore, these NBS-LRR-VIGS tomato leaves also had larger diameter lesions (approximately 1.91-fold increase compared to TRV2 tomato plants infected with P. infestans) (Fig. 5d). In the whole-plant inoculation assay, the susceptibility of the silenced plants manifested as more serious disease symptoms (Fig. 5e), higher DI (Fig. 5f) and an increased abundance of P. infestans (Fig. 5g).
Discussion
LncRNA as ceRNA regulates the accumulation of miRNA
MicroRNAs (miRNAs), a class of approximately 22 nt endogenous small noncoding RNAs, can direct the posttranscriptional repression of target genes by base-pairing to mRNAs. The mature miRNA is separated from the miRNA/miRNA* duplex, which is processed from a pre-miRNA1.
One focus of miRNA research is the regulation of miRNA accumulation. Various transcription factors can promote the expression of pre-miRNAs at the transcriptional level. For example, in A. thaliana, transcription factor APETALA2 (AP2) positively or negatively regulates the expression of miR156 or miR172 by binding to these miRNA genes44. Another Arabidopsis transcription factor, AtMYB2, has been identified for its roles in the activation of miR399f expression in the context of phosphate homeostasis through sequence-specific interactions with a MYB-binding site in the promoter of the miR399f precursor45. In addition, tomato ripening inhibitor (RIN) binds to a RIN-binding site in the promoter of miR172a to regulate its accumulation46.
Another regulatory mechanism has been proposed to underlie the accumulation of miRNA. During plant-pathogen interactions, pathogen effectors can suppress host defense47. The effectors, RNA-silencing suppressors from the pathogens, affect the resistance of plants by suppressing the accumulation of host miRNAs. P. sojae encoded two RNA-silencing suppressors (PsPSR1 and PsPSR2) that enhance plant susceptibility by impairing the small RNA-mediated defense of the host48. The PsPSR1 virulence target acts in the assembly of sRNA-processing complexes in Arabidopsis and soybean49. PSR2 is involved in modifying plant gene regulation early during Phytophthora infection, and the overexpression of PsPSR2 in Arabidopsis enhances hypersusceptibility to P. capsici50.
Some specific endogenous lncRNAs can act as ceRNAs to interfere with miRNA pathways51. Acting as a ceRNA is an effective posttranscriptional regulatory mechanism by which lncRNAs interfere with target transcripts. The disruption of the equilibrium between ceRNAs and miRNAs could be important to ceRNA activity in diseases such as cancer52. For example, the lncRNA HOTAIR decoys miR-152-3p to promote malignant melanoma progression53. In addition, the lncRNA CRNDE prevents miR-136-5p-mediated downregulation of Bcl-2 and Wnt2 to promote glioma malignancy54. However, studies on ceRNA in plants are limited. In maize, a number of lncRNAs have been identified as miRNA decoys by using computational methods55. Tobacco eTMX27 inhibits the expression of miRX27, resulting in enhanced nicotine biosynthesis56. Arabidopsis IPS1 contains the eTM of ath-miR399 and inhibits the expression of ath-miR39920, and PDIL1 reduces the accumulation of miR399, thus regulating Pi-related pathways in Medicago truncatula57. Wang et al.21 reported that the eTMs of miR160 from lncRNAs can serve as decoys for miR160 and function as the regulator in rice development.
In this study, we wrote a program that systematically identified 89 lncRNAs as 46 miRNA decoys in tomato (Fig. 1c and Table S4). Three of these lncRNAs, lncRNA23468, lncRNA01308 and lncRNA13262, contained an eTM site of miR482b (Fig. 2b). The expression of these three lncRNAs was initially increased but then decreased with P. infestans infection (Fig. 2c). This result was in contrast to the expression trend of miR482b upon infection by P. infestans, which was detected in our previous study8. The expression of miR482b was decreased in the tomato plants that overexpressed lncRNA23468 (Fig. 4a), while silencing lncRNA23468 led to increased miR482b accumulation (Fig. 5a). In addition, the accumulation of miR482b was not significantly changed in the tomato plants that overexpressed mlncRNA23468 with a mutated eTM site (Fig. 4a). These results suggest that the lncRNAs functioned as an eTM that may modulate the effects of miRNAs in tomato.
The lncRNA23468-miR482b-NBS-LRR network in tomato resistance to P. infestans
The miR482b family is involved in various plant interactions with pathogens6,58. The overexpression of miR482e in potato decreased plant resistance to V. dahliae infection5. Similar results were reported in our previous study, showing that miR482b negatively regulates tomato resistance to P. infestans8. To understand the biological functions of miRNAs, the identification of their target genes is an important step. In our previous study, four members of the NBS-LRR family were identified as the target genes of miR482b using the degradome data of tomato infected with P. infestans8. NBS-LRRs are active in the resistance of other plants to pathogens, such as tobacco resistance to P. parasitica12, wheat resistance to powdery mildew59, and rice resistance to blast60. In this study, NBS-LRR accumulation was suppressed by silencing lncRNA23468 and VIGS technology in tomato, resulting in the enhancement of tomato susceptibility to P. infestans (Figs. 4 and 5). Similarly, cotton susceptibility is also increased by silencing a CC-NBS-LRR gene11.
LncRNAs are involved in many biological processes, including biotic stresses. For example, Joshi et al.61 found that 13 lncRNAs were predicted to be the precursors of 96 miRNAs that affect the Brassica napus and Sclerotinia sclerotiorum interaction. The silencing of GhlncNAT-ANX2 and GhlncNAT-RLP7 in cotton seedlings can induce LOX1 and LOX2 expression to increase the resistance to V. dahliae and Botrytis cinerea infection62. In addition, many lncRNAs act in the response of Arabidopsis to F. oxysporum and Pseudomonas syringe pv. tomato DC3000, wheat response to powdery mildew pathogen, tomato response to TYLCV and P. tomentosa response to Paulownia witches’ broom22,24,25,33,35. The overexpression of lncRNA16397 in tomato induced glutaredoxin expression to increase the resistance to P. infestans39. Another tomato lncRNA, lncRNA23468, was also responsive to P. infestans infection (Fig. 2c). In addition, the overexpression of lncRNA23468 in tomato positively modulated the P. infestans defense response (Fig. 4), while the resistance was impaired after lncRNA23468 silencing (Fig. 5).
The overexpression of lncRNA23468 in tomato suppressed the accumulation of miR482b, resulting in the increased abundance of NBS-LRRs, and after lncRNA23468 silencing, the opposite was observed (Figs. 4a and 5a). In other words, lncRNA23468 functions as a ceRNA to modulate NBS-LRR genes by decoying miR482b, which enhances tomato resistance to P. infestans. Similarly, the lncRNA Slylnc0195 might function as a ceRNA to protect miR166 targets, class III HD-Zip transcription factor genes, by binding to miR166 via target mimicry in the tomato response to TYLCV33.
In summary, lncRNA23468 increases resistance to P. infestans infection in tomato. From our results in the lncRNA23468-silencing, lncRNA23468-overexpressing and mlncRNA23468-overexpressing tomato, we propose that lncRNA23468 increases the expression levels of NBS-LRRs by the repression of miR482b. This mechanism would allow for a response to P. infestans stress through lncRNA23468-induced downregulation of miR482b, which increases NBS-LRRs, thus promoting tomato resistance to P. infestans. Our results provide insight into an effective posttranscriptional regulation mechanism of lncRNA and demonstrate that the lncRNA23468-miR482b-NBS-LRR network is an important component of the P. infestans network in tomato.
References
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Janga, S. C. & Vallabhaneni, S. MicroRNAs as post-transcriptional machines and their interplay with cellular networks. Adv. Exp. Med. Biol. 722, 59–74 (2011).
Yin, X., Wang, J., Cheng, H., Wang, X. & Yu, D. Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta 237, 1213–1225 (2013).
de Vries, S., Kloesges, T. & Rose, L. E. Evolutionarily dynamic, but robust, targeting of resistance genes by the miR482/2118 gene family in the Solanaceae. Genome Biol. Evol. 7, 3307–3321 (2015).
Yang, L. et al. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 57, 1078–1088 (2015).
Feng, J., Liu, S., Wang, M., Lang, Q. & Jin, C. Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing. Planta 240, 1335–1352 (2014).
Zhu, Q. H. et al. MiR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE 8, e84390 (2013).
Jiang, N., Meng, J., Cui, J., Sun, G. & Luan, Y. Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans. Hortic. Res. 5, 9 (2018).
Yang, X. & Wang, J. Genome-wide analysis of NBS-LRR genes in Sorghum genome revealed several events contributing to NBS-LRR gene evolution in grass species. Evol. Bioinform. 12, 9–21 (2016).
Zhao, Y. et al. Bioinformatics analysis of NBS-LRR encoding resistance genes in Setaria italica. Biochem. Genet. 54, 232–248 (2016).
Yang, J. et al. Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene 575, 687–694 (2016).
Li, X., Zhang, Y., Yin, L. & Lu, J. Overexpression of pathogen-induced grapevine TIR-NB-LRR gene VaRGA1 enhances disease resistance and drought and salt tolerance in Nicotiana benthamiana. Protoplasma 254, 957–969 (2017).
Ouyang, S. et al. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 10, e1004464 (2014).
Heo, J. B., Lee, Y. S. & Sung, S. Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res. 21, 685–693 (2013).
Mercer, T. R., Dinger, M. E. & Mattick, J. S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159 (2009).
Liu, F., Marquardt, S., Lister, C., Swiezewski, S. & Dean, C. Targeted 3’ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97 (2010).
Wang, X. et al. Expression and diversification analysis reveals transposable elements play important roles in the origin of Lycopersicon-specific lncRNAs in tomato. New Phytol. 209, 1442–1455 (2016).
Zhu, B. et al. RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening. J. Exp. Bot. 66, 4483–4495 (2015).
Wang, H. et al. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 24, 444–453 (2014).
Franco-Zorrilla, J. M. et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037 (2007).
Wang, M., Wu, H. J., Fang, J., Chu, C. C. & Wang, X. J. A long noncoding RNA involved in rice reproductive development by negatively regulating osa-miR160. Sci. Bull. 62, 470–475 (2017).
Zhu, Q. H., Stephen, S., Taylor, J., Helliwell, C. A. & Wang, M. B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 201, 574–584 (2014).
Qin, T., Zhao, H., Cui, P., Albesher, N. & Xiong, L. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 175, 1321–1336 (2017).
Xin, M. et al. Identification and characterization of wheat long non protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 11, 61 (2011).
Wang, Z., Zhai, X., Cao, Y., Dong, Y. & Fan, G. Long non-coding RNAs responsive to witches’ broom disease in Paulownia tomentosa. Forests 8, 348 (2017).
Scarano, D., Rao, R. & Corrado, G. In Silico identification and annotation of non-coding RNAs by RNA-seq and De Novo assembly of the transcriptome of Tomato Fruits. PLoS ONE 12, e0171504 (2017).
Dai, Q. et al. Comparative transcriptome analysis of the different tissues between the cultivated and wild tomato. PLoS ONE 12, e0172411 (2017).
Wang, M., Zhao, W., Gao, L. & Zhao, L. Genome-wide profiling of long non-coding RNAs from tomato and a comparison with mRNAs associated with the regulation of fruit ripening. BMC Plant Biol. 18, 75 (2018).
Sun, Y. & Xiao, H. Identification of alternative splicing events by RNA sequencing in early growth tomato fruits. BMC Genom. 16, 948 (2015).
Li, R., Fu, D., Zhu, B., Luo, Y. & Zhu, H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 94, 513–524 (2018).
Wang, Y. et al. Analysis of long-non-coding RNAs associated with ethylene in tomato. Gene 674, 151–160 (2018).
Wang, Y. et al. Integrative analysis of long non-coding RNA acting as ceRNAs involved in chilling injury in tomato fruit. Gene 667, 25–33 (2018).
Wang, J. et al. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 5, 16946 (2015).
Zheng, Y., Wang, Y., Ding, B. & Fei, Z. Comprehensive transcriptome analyses reveal that Potato Spindle Tuber Viroid triggers genome-wide changes in alternative splicing, inducible trans-acting activity of phased secondary small interfering RNAs, and immune responses. J. Virol. 91, e00247–17 (2017).
Wang, J. et al. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol. 18, 104 (2018).
Bhattarai, K., Louws, F. J., Williamson, J. D. & Panthee, D. R. Differential response of tomato genotypes to Xanthomonas-specific pathogen-associated molecular patterns and correlation with bacterial spot (Xanthomonas perforans) resistance. Hortic. Res. 3, 16035 (2016).
Cui, J. et al. Transcriptome signatures of tomato leaf induced by Phytophthora infestans and functional identification of transcription factor SpWRKY3. Theor. Appl. Genet. 131, 787–800 (2018).
Luan, Y. et al. High-throughput sequencing reveals differential expression of miRNAs in tomato inoculated with Phytophthora infestans. Planta 241, 1405–1416 (2015).
Cui, J., Luan, Y., Jiang, N., Bao, H. & Meng, J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 89, 577–589 (2017).
Liu, Y., Schiff, M. & Dinesh-Kumar, S. P. Virus-induced gene silencing in tomato. Plant J. 31, 777–786 (2002).
Zhao, J. et al. An efficient Potato virus X-based microRNA silencing in Nicotiana benthamiana. Sci. Rep. 6, 20573 (2016).
Luan, Y. et al. Effective enhancement of resistance to Phytophthora infestans by overexpression of miR172a and b in Solanum lycopersicum. Planta 247, 127–138 (2018).
Chen, L., Luan, Y. & Zhai, J. Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 34, 2013–2025 (2015).
Yant, L. et al. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156–2170 (2010).
Baek, D., Park, H. C., Kim, M. C. & Yun, D. J. The role of Arabidopsis MYB2 in miR399f-mediated phosphate-starvation response. Plant Signal. Behav. 8, e23488 (2013).
Gao, C. et al. MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant. Biotechnol. J. 13, 370–382 (2015).
Kong, L. et al. A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Curr. Biol. 27, 981–991 (2017).
Qiao, Y. et al. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45, 330–333 (2013).
Qiao, Y., Shi, J., Zhai, Y., Hou, Y. & Ma, W. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl Acad. Sci. USA 112, 5850–5855 (2015).
Xiong, Q. et al. Phytophthora suppressor of RNA silencing 2 is a conserved RxLR effector that promotes infection in soybean and Arabidopsis thaliana. Mol. Plant Microbe Interact. 27, 1379–1389 (2014).
Liu, D., Mewalal, R., Hu, R., Tuskan, G. A. & Yang, X. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic. Res. 4, 17031 (2017).
Sen, R., Ghosal, S., Das, S., Balti, S. & Chakrabarti, J. Competing endogenousRNA: the key to posttranscriptional regulation. ScientificWorldJ 2014, 896206 (2014).
Luan, W. et al. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote malignant melanoma progression by sponging miR-152-3p. Oncotarget 8, 85401–85414 (2017).
Li, D. X. et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget 8, 88163–88178 (2017).
Zhu, M. et al. Transcriptomic analysis of long Non-coding RNAs and coding genes uncovers a complex regulatory network that is involved in maize seed development. Genes 8, (274 (2017).
Li, F. et al. Regulation of nicotine biosynthesis by an endogenous target mimicry of microRNA in tobacco. Plant Physiol. 169, 1062–1071 (2015).
Wang, T. et al. Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 68, 5937–5948 (2017).
Bao, D. et al. Down-regulation of genes coding for core RNAi components and disease resistance proteins via corresponding microRNAs might be correlated with successful soybean mosaic virus infection in soybean. Mol. Plant Pathol. 19, 948–960 (2017).
Hurni, S. et al. Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76, 957–969 (2013).
Ma, J. et al. Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol. Plant Microbe Interact. 28, 558–568 (2015).
Joshi, R. K., Megha, S., Basu, U., Rahman, M. H. & Kav, N. N. Genome wide identification and functional prediction of long non-coding RNAs responsive to Sclerotinia sclerotiorum infection in Brassica napus. PLoS ONE 11, e0158784 (2016).
Zhang, L. et al. Long non-coding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant. Biotechnol. J. 16, 1172–1185 (2018).
Acknowledgements
This work is supported by grants from the National Natural Science Foundation of China (Nos. 31471880 and 61472061). We thank Prof. Weixing Shan from Northwest A & F, University of China, for providing P. infestans strain P12103 and Yule Liu from Tsinghua University for providing the VIGS system.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jiang, N., Cui, J., Shi, Y. et al. Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic Res 6, 28 (2019). https://doi.org/10.1038/s41438-018-0096-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-018-0096-0
This article is cited by
-
A long non-coding RNA functions as a competitive endogenous RNA to modulate TaNAC018 by acting as a decoy for tae-miR6206
Plant Molecular Biology (2024)
-
The lncRNA20718-miR6022-RLPs module regulates tomato resistance to Phytophthora infestans
Plant Cell Reports (2024)
-
Comprehensive insights into the regulatory mechanisms of lncRNA in alkaline-salt stress tolerance in rice
Molecular Biology Reports (2023)
-
Whole-transcriptome profiles of Chrysanthemum seticuspe improve genome annotation and shed new light on mRNA–miRNA–lncRNA networks in ray florets and disc florets
BMC Plant Biology (2022)
-
Dynamic characteristics and functional analysis provide new insights into long non-coding RNA responsive to Verticillium dahliae infection in Gossypium hirsutum
BMC Plant Biology (2021)