Abstract
Marssonina apple blotch, caused by Diplocarpon mali, is one of the most serious diseases of apple. Autophagy plays a key role in pathogen resistance. We previously showed that MdATG18a has a positive influence on drought tolerance. Herein, we describe how overexpression (OE) of MdATG18a enhances resistance to D. mali infection, probably because less H2O2 but more salicylic acid (SA) is accumulated in the leaves of OE apple plants. Expression of chitinase, β-1,3-glucanase, and SA-related marker genes was induced more strongly by D. mali in OE lines. Transcript levels of other important MdATG genes were also drastically increased by D. mali in OE plants, which indicated increased autophagy activities. Taken together, these results demonstrate that OE of MdATG18a enhances resistance to infection by D. mali and plays positive roles in H2O2-scavenging and SA accumulations. Our findings provide important information for designing strategies which could induce autophagy to minimize the impact of this disease on apple production.
Similar content being viewed by others
Introduction
Marssonina apple blotch is one of the most serious apple diseases, caused by the fungus Diplocarpon mali1. This pathogen infects apple plants using both necrotrophic and biotrophic strategies, suggesting that it behaves in a hemibiotrophic manner2.
Diplocarpon mali ascopores released from overwintered apothecia are responsible for primary infections and conidia produced in acervuli are considered the inoculum for secondary infections2. Upper surface of infected leaves show greyish brown leaf spots and small black acervuli before leaves become yellow and fall off2. Visible symptoms appear in the second half of June and spreads at a faster rate during July–August with average temperature between 23.5 and 25.4 °C and frequent rains of moderate to high intensity1. The development of this disease is positively correlated with relative humidity and rainfall. Diplocarpon mali also produce pseudo-conidia on overwintered disease leaves3, which are dispersed by rain splashes and mainly infect old leaves near the bottom of shoots from the beginning of apple leaf growth3. Ascospores mature from mid May to the end of June, and are discharged in response to rain and dispersed with wind3. Infection leads to premature defoliation and affects fruit quality and quantity4.
Because of long-term interactions with pathogens, plants have developed sophisticated defense systems that can inhibit or alleviate the harm caused by pathogens. When infection occurs, the innate immunity-pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) is generally activated5,6. However, PTI can be overcomed by a secondary immunity-effector triggered immunity5,6. These two phases of plant immunity can be induced by several defense reactions, including oxidative bursts and hormones. Generation of H2O2, an example of a stable reactive oxygen species (ROS), is considered a universal plant response to pathogen attack7. Main ROS-scavenging mechanisms include enzymatic and non-enzymatic antioxidants, both of which regulate redox homeostasis in plant cells. Pathogenesis-related (PR) proteins can be induced by plant basal defense responses. For example, chitinase and β-1,3-glucanase may have direct antimicrobial activities due to their roles in the hydrolyzation of respective chitin and glucans in the fungal cell wall8.
Plant hormones are involved in growth, development, reproduction, and stress reponse. For example, salicylic acid (SA) is a crucial component in disease resistance signaling9. The SA-related pathway is activated by biotrophic pathogens10. SA also activates phenylalanine ammonia-lyase and polyphenol oxidase, which are involved in phenolic compound synthesis and cell wall strengthening11,12. Upregulation of PR genes often indicates the induction of plant defense. In some crops, SA application enhances expression of some PR genes to confer pathogen resistance13,14. Transcripts of PR1 and PR5 are coupled with the accumulation of endogenous SA, which makes both genes molecular markers for the SA-dependent signaling pathway15,16.
Autophagy, a conserved cellular process in eukaryotes, is important for recycling nutrients and cytoplasmic components17,18,19,20. In yeast, WD40 repeat-containing protein AuTophGy-related (Atg) 18 is able to bind phosphatidylinositol 3-phosphate (PtdIns(3)P) and phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2)21. The PtdIns(3)P binding capacity of Atg18 is required for recruitment of Atg8 and Atg16 during phagophore formation22. The access of Atg4 to Atg8-phosphatidylethanolamine at phagophore assembly site is disturbed by the Atg18/21 complex to prevent a premature cleavage. However, the Atg18/21 complex dissociates and allows Atg4 to cleave Atg8-PE and release Atg8 after completion of the autophagosome. Therefore, a key aspect of post-translational regulation of autophagy by Atg4 is closely related with Atg18/2122. Autophagy has both pro-survival and pro-death roles in regulating hypersensitive response programmed cell death (HR-PCD) and plant immunity system under biotic stresses23,24,25,26, depending on factors such as plant age and pathogens. Autophagosome formation in Arabidopsis autophagy-deficient mutant atg18a was disturbed27, resulting in greater susceptibility to necrotrophic fungal pathogens in cooperation with jasmonate-mediated and WRKY33 (containing a highly conserved heptapeptide motif WRKYGQK 33)-mediated signaling pathways26. However, Arabidopsis atg5, atg10, and atg18a-2 plants show enhanced resistance to Pseudomonas syringae pv. tomato28. Liu et al.29 have demonstrated that Nicotiana tabacum with ATG6/BECLIN1 silenced showed increased cell death under viral infection. Some regulatory factors also function in plant immunity by regulating autophagy. RabG3b, a GTP (guanosine triphosphate)-binding protein, facilitates HR-PCD by enhancing autophagosome formation under avirulent bacterial pathogens infection in Arabidopsis, thereby having a positive effect in immunity30. Thus, the molecular mechanisms and functions of autophagy are implicated in the complex life and death decisions under different pathological situations in plants25.
As one of the most serious apple diseases in Asian regions, Marssonina apple blotch causes great losses in fruit production. The function of autophagy in response to the infection by the pathogen is largely unknown. Two transgenic apple lines expressing MdATG18a under the control of the CaMV35S promoter demonstrated that the apple autophagy-related gene MdATG18a enhances plant resistance to drought stress via an improved ROS-scavenging system and activating autophagy31. In addition, these two lines also showed enhanced tolerance to nitrogen depletion32. MdATG18a transcription level was greatly increased by D. mali infection and overexpression (OE) of the gene was associated with improved resistance to D. mali in apple plants, probably due to lower ROS accumulations, higher accumulations of SA, and regulation on some PR genes because of improved autophagy activities. These findings demonstrate that MdATG18a plays a key role in the resistance of apple plants to D. mali.
Results
Pathogen-responsive expression patterns of MdATG18a to D. mali infection
Expression of MdATG18a can be induced by universal stresses33. To examine whether this is also true in response to pathogen attacks, we monitored the transcription level of this gene upon pathogen infection in wild-type (WT) apple leaves and found that the expression was up-regulated in the leaves of the control (mock-inoculated) at the end of the treatment period. The transcripts of the inoculated leaves peaked at 6 days post-inoculation (dpi) (more than 4-fold higher over 0 dpi) before gradually decreased over time (Fig. 1), indicating that MdATG18a was responsive to D. mali infection in apple.
MdATG18a OE enhanced resistance to D. mali infection
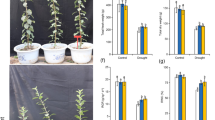
We used two transgenic lines of MdATG18a OE apple which were obtained previously for further investigation of possible functions of MdATG18a in pathogen resistance. Excised, mature leaves from apple plants were inoculated with conidial suspensions of D. mali. As shown in Fig. 2a, the inoculation resulted in serious necrotic spots and chlorosis in WT but failed to cause extensive tissue damage in OE plants. Disease indices on WT leaves were more than 2.5-fold that of OE leaves (Fig. 2b).
a Phenotypes of WT and OE lines at 10 dpi. b Disease indices of WT and OE lines at 10 dpi. Data are means of three replicates with SD. Experimental data were presented as means ± SD. Different letters indicate significant differences between treatments, according to one-way ANOVA Tukey’s multiple range tests (P < 0.05)
MdATG18a OE resulted in reduced H2O2 accumulation and increased antioxidant activity and AsA-GSH cycling upon D. mali infection
To analyze the redox status after leaf inoculation with D. mali, we measured the concentrations of H2O2 and activities of two major H2O2-scavenging antioxidant enzymes. H2O2 increased significantly in all infected leaves, but the level of H2O2 was significantly lower in OE leaves than in WT leaves at 10 dpi (Fig. 3a). Enzyme activities were increased in response to elevated H2O2 accumulations, whereas under control conditions, the levels of catalase (CAT) and peroxidase (POD) did no differ significantly between WT and OE plants. After infection, CAT activity in the OE lines was increased by 1.5 times over that in the WT. A similar pattern was observed for the POD activity.
a H2O2 concentrations and H2O2-scavenging enzymes activity. b Concentrations of antioxidants and transcript levels for genes involved in AsA-GSH cycle. H2O2-scavenging enzymes and concentrations of antioxidants were measured at 10 dpi. Expression levels were calculated relative to the expression of Malus EF-1α mRNA at 6 dpi. Expression in WT under control conditions was set to “1”. Data are means of three replicates with SD. Experimental data were presented as means ± SD. Different letters indicate significant differences between treatments, according to one-way ANOVA Tukey’s multiple range tests (P < 0.05)
As an antioxidant system, the ascorbic acid (AsA)-GSH cycle is involved in scavenging H2O2 under stress. We evaluated the status of ascorbate and glutathione to determine the changes in MdATG18a expression in the AsA-GSH cycle (Fig. 3b). No significant changes in the levels of total ascorbate and total glutathione were found between the OE lines and WT plants under the control conditions. However, the levels of total ascorbates and ratio of AsA to dehydroascorbic acid (DHA) were significantly higher in the transgenic plants than in the WT plants. Similarly, the levels of total glutathione pool and the reduced glutathione/oxidized glutathione (GSH/GSSG) ratio was also determined. We further examined the transcript levels for major genes in the AsA-GSH cycle. Under the normal conditions, expression of Malus × domestica cytoplasm ascorbate peroxidase (MdcAPX), Malus × domestica monodehydroascorbate reductase (MdMDHAR), Malus × domestica dehydroascorbate reductase 1 (MdDHAR1), and Malus × domestica glutathione reductase (MdGR) did not differ significantly among genotypes. However, at 6 dpi, the transcript levels of these four genes were strongly increased, especially in the transgenic lines. For example, expression of MdcAPX was 1.27 times higher in OE samples than in WT (Fig. 3b).
MdATG18a OE enhanced SA accumulation and increased the expression of SA-related PR genes upon D. mali infection
It has been reported that SA signaling is implicated in the autophagy-related immunity response34,35. To examine the relationship between SA and MdATG18a upon D. mali infection, we analyzed SA concentrations and the expression of two SA biosynthesis genes. For the mock control and inoculation, the SA level was significantly higher in the OE lines than in the WT (Fig. 4). For example, in response to inoculation, the level of SA in the OE plants was more than 2-fold that of the WT. Expression of Malus × domestica isochorismate synthase 1 (MdICS1) and Malus × domestica enhanced disease susceptibility 1 (MdEDS1) was up-regulated in parallel with the change in SA concentrations. Furthermore, their expression was up-regulated in the OE plants even under the mock control conditions, and their induction by D. mali infection was much stronger than in the WT. Similarly, the expression level of SA-related PR genes, MdPR1 and MdPR5, was significantly higher in the transgenic lines than in the WT at 6 dpi on the inoculated leaves (Fig. 4).
SA content was measured at 10 dpi. Expression of MdICS1, MdEDS1, MdPR1, and MdPR5 was analyzed at 6 dpi. Expression levels were calculated relative to the expression of Malus EF-1α mRNA. Expression in WT under control conditions was set to “1”. Data are means of three replicates with SD. Experimental data were presented as means ± SD. Different letters indicate significant differences between treatments, according to one-way ANOVA Tukey’s multiple range tests (P < 0.05)
MdATG18a OE promoted the activity and expression of chitinase and β-1,3-glucanase upon D. mali infection
In the mock inoculation control, expression of chitinase and β-1,3-glucanase did not differ significantly among genotypes, but was greatly up-regulated after inoculation; the transcript levels were more than two times higher in the OE plants than in WT at 6 dpi (Fig. 5). Upon inoculation, the chitinase and β-1,3-glucanase activities were significantly greater in the OE lines than in the WT.
Expression was analyzed at 6 dpi. Chitinase and β-1,3-glucanase activity was measured at 10 dpi. Expression levels were calculated relative to the expression of Malus EF-1α mRNA. Expression in WT under control conditions was set to “1”. Data are means of three replicates with SD. Experimental data were presented as means ± SD. Different letters indicate significant differences between treatments, according to one-way ANOVA Tukey’s multiple range tests (P < 0.05)
MdATG18a OE increased the expression of other MdATGs upon D. mali infection
To examine the occurrence of autophagy under pathogen infection, we analyzed expression patterns of several important ATG genes. Under the mock control conditions, expression of MdATG3a, MdATG3b, MdATG5, MdATG7a, MdATG7b, MdATG8f, MdATG8i, MdATG9, and MdATG10 did not differ among genotypes (Fig. 6). However, at 6 dpi, the expression of all studied genes was significantly higher in the OE lines than in WT. These results suggested that the transcripts of other important ATG genes were more responsive to D. mali infection in the OE leaves than in the WT, which might lead to stronger autophagy induction to build defense immunity.
Total RNA was isolated after 6 dpi, and expression levels were calculated relative to the expression of Malus EF-1α mRNA. Expression in WT under control conditions was set to “1”. Data are means of three replicates with SD. Experimental data were presented as means ± SD. Different letters indicate significant differences between treatments, according to one-way ANOVA Tukey’s multiple range tests (P < 0.05)
Discussion
Autophagy is thought to participate in the immune response to infection by various pathogens25,26,34,35,36. We previously demonstrated that MdATG18a is induced by various stress factors in WT apple plants33. The expression of autophagy-related genes and the autophagosome formation are also up-regulated in Arabidopsis in response to Botrytis cinerea infection26. Here, we explored the function of MdATG18a in apple in response to infection by D. mali. Expression of MdATG18a was up-regulated by inoculation with D. mali and its OE led to increased resistance against the pathogen, which might have resulted from increased ROS-scavenging capacity and SA synthesis. In the inoculated tissues, transcripts of chitinase, β-1,3-glucanase, MdPR1, MdPR5, and other important MdATGs were significantly up-regulated in the OE plants than in the WT. These results suggested that OE of MdATG18a enhances resistance to infection by D. mali.
Our previous work showed that MdATG18a transcripts were strongly induced under drought, heat and oxidative stresses, and peaked before a gradual decline33, suggesting the feedback regulation of MdATG18a under stress. The highly expressed MdATG18a appears to be indispensable to the stress response in the late stage of stress. The present work showed that expression of MdATG18a also peaked at 6 dpi and followed by a gradual decline in response to infection by D. mali (Fig. 1). We also found increased expression of MdATG18a even though in the mock-inoculated control (Fig. 1), probably because the mock-inoculated control (detached leaves under dark condition) may also induce MdATG18a expression33.
Pathogens rely upon several toxic factors, including stimulated ROS, to promote host cell death. H2O2 has some possible functions in the host response to pathogen infection and is the most versatile and stable ROS37. It also coordinates a localized HR when attacked by pathogen7. Autophagy is believed to be involved in degrading oxidized proteins and regulating ROS levels under various stresses31,38,39,40. Autophagy induced by pathogens ensures cell survival by preventing excess ROS production, removing excess and damaged organelles, and regulating organelles to a normal level that fulfills cellular requirements25. Antioxidant activity might contribute to infection by D. mali41,42. We previously reported that OE of MdATG18a in apple reduces ROS accumulations by promoting the antioxidant system under the drought stress31. Herein, we found that less H2O2 was accumulated in the OE lines under inoculation. This probably resulted from increased CAT and POD activities as well as improved AsA-GSH cycling.
As PR proteins, chitinase and β-1,3-glucanase can help cells resist pathogen infection by decomposing the pathogen cell walls8, and the products from decomposed cell walls can then activate a series of defense reactions43. Expression of chitinase and β-1,3-glucanase can be up-regulated by pathogen infection42. For example, transgenic lines of Vitis vinifera that contain chitinase (from Triticum aestivum) and β-1,3-glucanase have higher transcripts of chitinase and β-1,3-glucanase and show enhanced resistance to Plasmopara viticola44. After inoculation with D. mali, transcript levels of β-1,3-glucanase in apple are higher in resistant plants of M. sieversii against D. mali than in susceptible plants of M. prunifolia41. Similarly, our results indicated that MdATG18a OE up-regulated the expression of chitinase and β-1,3-glucanase and improved the activities of these two enzymes post-inoculation. This suggests that chitinase and β-1,3-glucanase are involved in limiting the extent of pathogen invasion and colonization in the OE plants.
The ATG18a protein interacts with WRKY33 and plays a key role in resisting to necrotrophic pathogens in Arabidopsis26. A small increase in the bacterial growth was observed in ATG6-silenced Arabidopsis plants over WT only at the early stages of PstDC3000 (Pseudomonas syringae pv. tomato DC3000) infection36. The same bacterium also increased its growth in Arabidopsis atg7 mutants45. All of these findings suggest that autophagy plays a positive role in resisting pathogen. However, Wang et al.35 found both enhanced resistance to powdery mildew and mildew-induced cell death in an Arabidopsis atg2 mutant35. The atg18a-2 mutant has also shown mildew-induced cell death similar to the atg2 mutant and enhanced resistance to the powdery mildew (Golovinomyces cichoracearum)46. These reports demonstrated that autophagy has a negative role in resisting pathogens. These contrasting reports about the role of autophagy in response to pathogen infection are probably related to SA signaling and the age of the leaves that were studied25,34. Young leaves contain less SA and reduced cell death than older leaves, regardless whether they are WT or mutants, suggesting young leaves are more susceptible to Pst-AvrRpm1 than older leaves. In mature leaves, which accumulate higher levels of SA than in young leaves, pathogen-induced cell death is suppressed in WT plants but not in atg mutants25,34. SA is involved in both local defense responses at the infection site and the activation of systemic resistance6,10,47. SA is essential for early senescence and immunity-related PCD in autophagy-deficient mutants34. Our OE lines contained more SA under both the inoculation and control conditions. Two SA synthesis genes, MdICS1 and MdEDS1, were more strongly induced in the OE lines after inoculation. This seems to contradict previous findings that atg mutants accumulate more SA35,46. We previously demonstrated that ectopic expression of apple MdATG8i, MdATG7, and MdATG3 in Arabidopsis slightly promoted leaf senescence and bolting48,49,50. In the present work, the SA level was slightly higher in the OE lines than in the WT under mock control conditions. After inoculation, SA synthesis was up-regulated, especially in the OE lines. This greater accumulation of SA might have contributed to the enhanced resistance to pathogen infection. We speculate that this response resulted from the activation of autophagy via MdATG18a OE, based on our finding that major MdATG genes were significantly up-regulated in transgenic plants. To investigate why OE of MdATG18a also enhanced SA biosynthesis, a knock-out of MdATG18a or interference of MdATG18a in apple would be one viable strategy to pursue. MdATG18a knock-out/interference apple plants may be similar to Arabidopsis atg18a mutant that may have increased SA level compared with WT plants. In the SA signaling defense pathway, the enhanced plant resistance to biotrophic and hemibiotrophic pathogens results from up-regulated PR genes51. Likewise, we noted here that MdPR1 and MdPR5 were much more induced by D. mali in the OE lines than in the WT, demonstrating that regulation of the SA pathway is required for pathogen resistance.
In summary, we have functionally characterized MdATG18a under pathogen infection. Transgenic apple plants had enhanced resistance to D. mali, possibly because of a more-reductive redox state and a higher concentration of SA in the OE plants. Expression levels of chitinase, β-1,3-glucanase, and SA-related genes were greatly up-regulated by D. mali in the OE lines. This might be explained by the improvements in autophagy activity, as supported by increased transcription of important MdATGs. These results provide further insight about the function of autophagy in apple disease resistance. We can design strategies which could induce autophagy to curb the impact of future infections according to our results that increased autophagic activity could increase resistance of apple to pathogen.
Materials and methods
Treatment and plant materials
Tissue-cultured plants of Malus × domestica cv. Royal Gala were cultivated as described previously31. OE transgenic and WT plantlets were transferred to small plastic pots (8.5 × 8.5 × 7.5 cm3) containing a mixture of loam/perlite (1:1, v:v). After 1 month of adaptation in a growth chamber, one plant was moved to each large plastic pot (30 × 26 × 22 cm3) filled with a mixture of clay/sand/organic substrate (5:1:1, v:v:v), and grown in the greenhouse. They were watered regularly and supplied with half-strength Hoagland’s nutrient solution (pH 6.0) once a week. We used theses plants for examining their resistance to D. mali infection after 90 days under such conditions.
For inoculation and disease evaluation, the ninth to twelfth healthy apple leaves from the base of a stem (fully mature leaves) were excised from WT and OE plants. The inoculation assay was performed as previouly described52. A total of 120 WT leaves in two plastic boxes were used for MdATG18a expression following D.mali infection. Three or four entire WT leaves were sampled for per repeat at each 0, 2, 4, 6, 8, and 10 dpi. Leaves were sampled thrice as three repeats with total 10 leaves at each sampling time. Total 60 leaves in two plastic boxes were inoculated from 20 WT/OE-3/OE-11 plants, with four inoculation sites per leaf. Three or four entire leaves were sampled for per repeat at 0, 6, and 10 dpi. Leaves were sampled thrice as three repeats with total 10 leaves at each sampling time. The plastic boxes were sprayed with sterile water twice a day to keep humid conditions for 10 days52.
Inoculum preparation
A monospore culture of D. mali originated from diseased apple leaves showing Marssonina blotch symptoms which were collected from a research orchard at the College of Horticulture, Northwest A&F University, Yangling (34°20′N, 108°24′E), Shaanxi Province, China. Single spores were isolated by a method reported previously53,54. The inoculum suspension was prepared and adjusted to 1 × 106 conidia per milliliter as previously described52,55,56.
Evaluation of infection degree
At 10 dpi, disease severity for each leaf was scored on a scale of 0 to 5, where 0 = no disease symptoms, while 1 = 1 to 10%, 2 = 11 to 30%, 3 = 31 to 50%, 4 = 50 to 80%, or 5 = 80 to 100% of the entire leaf area showed lesions41,52. The following formula was used:
\({\mathrm{Disease}}\,{\mathrm{index}} = \frac{{{\mathrm{Sum}}\,{\mathrm{of(severity}}\,{\mathrm{score \times the}}\,{\mathrm{number}}\,{\mathrm{of}}\,{\mathrm{leaves}}\,{\mathrm{in}}\,{\mathrm{that}}\,{\mathrm{severity}}\,{\mathrm{score) \times 100}}}}{{{\mathrm{Total}}\,{\mathrm{number}}\,{\mathrm{of}}\,{\mathrm{leaves}}\,{\mathrm{ \times 5}}}}.\)
These experiments were conducted three times.
RNA extraction and real-time PCR
Total RNA was extracted according to a CTAB method57. First-strand cDNA synthesis and real-time PCR were performed as previously described31. Malus elongation factor 1α gene (EF-1α; DQ341381) were used as internal control. Primer sequences for real-time PCR are listed in Table S1. Each experiment was repeated three times biologically, based on three separate RNA extracts from three repeats.
Extraction and assay of H2O2 and antioxidant metabolites
Apple leaves (0.1 g) were used for quantifying H2O2 and antioxidant metabolites. Activities of CAT and POD were determined according to established protocols39. Five percent (w/v) of trichloroacetic acid was used for H2O2 extraction and quantification as described previously58. Six percent (v/v) HClO4 was used for AsA and DHA extraction, while 5% (v/v) sulfosalicylic acid was used for GSH and GSSG extraction. Concentrations of AsA, DHA, GSH, and GSSG were quantified as described previously59.
Extraction and assays of chitinase and β-1,3-glucanase activity
Extraction and assays of chitinase and β-1,3-glucanase activity were performed as described previously42,60,61,62,63.
Quantification of SA
SA was extracted and quantified as described previously64,65. Leaf samples (0.1 g) were homogenized in liquid nitrogen and placed in 1 mL of 90% methanol. 3-Hydroxybenzoic acid (Sigma) was added as internal standard. Extracts were analyzed though a fluorescence detector (excitation at 305 nm and emission at 405 nm) on a ZORBAX SB-C18 column (Agilent Technologies, Santa Clara, CA, USA).
Statistical analysis
Experimental data are presented means ± standard deviations of three independent replicates. Data were analyzed via analysis of variance (ANOVA), and mean values were compared by Tukey’s multiple range test (p < 0.05). All the statistical analyses were performed using SPSS18 statistical software package (IBM SPSS Statistics, Chicago, IL, USA).
References
Harada, Y., Sawamura, K. & Konno, K. Diplocarpon mali sp. nov., the perfect state of apple blotch fungus Marssonina coronaria. Jpn. J. Phytopathol. 40, 412–418 (1974).
Zhao, H. et al. Cytology of infection of apple leaves by Diplocarpon mali. Eur. J. Plant Pathol. 136, 41–49 (2013).
Dong, X. et al. Epidemic dynamics of apple marssonina leaf blotch over whole growth season in the central area of shandong peninsula. Sci. Agric. Sin. 48, 479–487 (2015).
Zhao, H., Zhou, T., Cheng, J., Li, X. & Huang, L. Control effect of triazole fungicides in controlling Marsonina coronaria in vitro and in field. Sci. Silvae Sin. 45, 68–73 (2009).
Thomma, B. P. H. J., Nürnberger, T. & Joosten, M. H. A. J. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4 (2011).
Robertseilaniantz, A., Grant, M. & Jones, J. D. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343 (2011).
Levine, A., Tenhaken, R., Dixon, R. & Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593 (1994).
Mauch, F., Mauch-Mani, B. & Boller, T. Antifungal hydrolases in pea tissue: ii. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 88, 936 (1988).
Vlot, A. C., Dempsey, D. A. & Klessig, D. F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206 (2009).
Loake, G. & Grant, M. Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472 (2007).
Tian, S., Wan, Y., Qin, G. & Xu, Y. Induction of defense responses against Alternaria rot by different elicitors in harvested pear fruit. Appl. Microbiol. Biotechnol. 70, 729 (2006).
Mandal, S., Mallick, N. & Mitra, A. Salicylic acid-induced resistance to Fusarium oxysporum f. sp. lycopersici in tomato. Plant Physiol. Biochem. 47, 642–649 (2009).
Niu, J., Liu, R. & Zheng, L. Expression analysis of wheat PR-1, PR-2, PR-5 activated by Bgt and SA, and powdery mildew resistance. J. Triticeae Crops 27, 1132–1137 (2007).
Zhang, Y. et al. Salicylic acid confers enhanced resistance to Glomerella leaf spot in apple. Plant Physiol. Biochem. 106, 64 (2016).
El, O. M. et al. Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23, 2405–2421 (2011).
Eggermont, K., Mauch-Mani, B., Vogelsang, R. & Broekaert, W. F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107 (1998).
Have, M., Marmagne, A., Chardon, F. & Masclaux-Daubresse, C. Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J. Exp. Bot. 68, 2513–2529 (2017).
Masclaux-Daubresse, C., Chen, Q. & Havé, M. Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 39, 8–17 (2017).
Noda, N. N. & Inagaki, F. Mechanisms of autophagy. Ann. Rev. Biophys. 44, 101–122 (2015).
Liu, Y. & Bassham, D. C. Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant. Biol. 63, 215–237 (2012).
Krick, R., Tolstrup, J., Appelles, A., Henke, S. & Thumm, M. The relevance of the phosphatidylinositolphosphate‐binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 580, 4632–4638 (2006).
Nair, U., Cao, Y. & Klionsky, D. J. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J. Biol. Chem. 285, 11476 (2010).
Hayward, A. P. & Dineshkumar, S. P. What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 49, 557 (2011).
Teh, O. K. & Hofius, D. Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J. Exp. Bot. 65, 1297–1312 (2014).
Zhou, J., Yu, J. & Chen, Z. The perplexing role of autophagy in plant innate immune responses. Mol. Plant Pathol. 15, 637 (2014).
Lai, Z., Wang, F., Zheng, Z., Fan, B. & Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66, 953–968 (2011).
Xiong, Y., Contento, A. L. & Bassham, D. C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42, 535–546 (2010).
Lenz, H. D. et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66, 818–830 (2011).
Liu, Y. et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577 (2005).
Kwon, S. I. & Park, O. K. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 161, 1722–1736 (2013).
Sun, X. et al. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 16, 545–557 (2018).
Sun, X. et al. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469–480 (2018).
Wang, P. et al. Isolation and characterization of MdATG18a, a WD40-repeat AuTophaGy-related gene responsive to leaf senescence and abiotic stress in Malus. Sci. Hortic. 165, 51–61 (2014).
Yoshimoto, K. et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914 (2009).
Wang, Y., Nishimura, M. T., Zhao, T. & Tang, D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68, 74–87 (2011).
Patel, S. & Dineshkumar, S. P. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4, 20 (2008).
Shetty, N. P., Jørgensen, H. J. L., Jensen, J. D., Collinge, D. B. & Shetty, H. S. Roles of reactive oxygen species in interactions between plants and pathogens. Eur. J. Plant Pathol. 121, 267–280 (2008).
Xiong, Y., Contento, A. L. & Bassham, D. C. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy 3, 257 (2007).
Yang, X., Srivastava, R., Howell, S. H. & Bassham, D. C. Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 85, 83 (2016).
Zhou, J. et al. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 10, e1004116 (2014).
Yin, L. et al. Resistance of Malus plants to Diplocarpon mali infection is associated with the antioxidant system and defense signaling pathways. Physiol. Mol. Plant Pathol. 84, 146–152 (2013).
Yin, L. et al. Exogenous melatonin improves Malus resistance to Marssonina apple blotch. J. Pineal Res. 54, 426 (2013).
Lawrence, C. B., Singh, N. P., Qiu, J., Gardner, R. G. & Tuzun, S. Constitutive hydrolytic enzymes are associated with polygenic resistance of tomato to Alternaria solani and may function as an elicitor release mechanism. Physiol. Mol. Plant Pathol. 57, 211–220 (2000).
Nookaraju, A. & Agrawal, D. C. Enhanced tolerance of transgenic grapevines expressing chitinase and β-1,3-glucanase genes to downy mildew. Plant Cell Tiss. Org. Cult. 111, 15–28 (2012).
Hofius, D. et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137, 773–783 (2009).
Wang, Y., Wu, Y. & Tang, D. The autophagy gene, ATG18a, plays a negative role in powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant Signal. Behav. 6, 1408 (2011).
Nawrath, C., Heck, S., Parinthawong, N. & Métraux, J. P. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286 (2002).
Wang, P. et al. Characterization of an autophagy-related gene MdATG8i from apple. Front. Plant Sci. 7, 720 (2016).
Wang, P., Sun, X., Jia, X. & Ma, F. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 256, 53–64 (2017).
Wang, P., Sun, X., Wang, N., Jia, X. & Ma, F. Ectopic expression of an autophagy-associated MdATG7b gene from apple alters growth and tolerance to nutrient stress in Arabidopsis thaliana. Plant Cell Tiss. Org. Cult. 128, 9–23 (2016).
Kunkel, B. N. & Brooks, D. M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant. Biol. 5, 325–331 (2002).
Wang, N. et al. Functions of two Malus hupehensis (Pamp.) Rehd. YTPs (MhYTP1 and MhYTP2) in biotic- and abiotic-stress responses. Plant Sci. 261, 18–27 (2017).
Zhao, H., Huang, L. & Kang, Z. S. Culture study of Marssonina coronaria from diseased apple leaves. Mycosystema 28, 490–495 (2009).
Dong-Hyuk, L. et al. Biological characterization of Marssonina coronaria associated with apple blotch disease. Mycobiology 39, 200–205 (2011).
Zhao, H. et al. Influence of culture media and environmental factors on mycelial growth and conidial production of Diplocarpon mali. Lett. Appl. Microbiol. 50, 639 (2010).
Shou, Y., Li, C., Zhao, Y., Chen, D. & Zhang, X. In vitro evaluation of resistance to Marssonina mali in apple. J. Fruit. Sci. 26, 912–914 (2009).
Chang, S., Puryear, J. & Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant. Mol. Biol. Rep. 11, 113–116 (1993).
Patterson, B. D., Macrae, E. A. & Ferguson, I. B. Estimation of hydrogen peroxide in plant extracts using titanium(IV). Anal. Biochem. 139, 487–492 (1984).
Logan, B. A., Grace, S. C., Iii, W. W. A. & Demmig-Adams, B. Seasonal differences in xanthophyll cycle characteristics and antioxidants in Mahonia repens growing in different light environments. Oecologia 116, 9 (1998).
Boller, T., Gehri, A. & Mauch, F. Chitinase in bean leaves: induction by ethylene, purification, properties, and possible function. Planta 157, 22–31 (1983).
Reissig, J. L., Strominger, J. L. & Leloir, L. F. A modified colorimetric method for the estimation of n-acetylamino sugars. J. Biol. Chem. 217, 959 (1955).
Roberts, W. K. & Selitrennikoff, C. P. Plant and bacterial chitinases differ in antifungal activity. Microbiology 134, 169–176 (1988).
Abeles, F. B. & Forrence, L. E. Temporal and hormonal control of beta-1,3-glucanase in Phaseolus vulgaris L. Plant Physiol. 45, 395 (1970).
Aboul-Soud, M. A. M., Cook, K. & Loake, G. J. Measurement of salicylic acid by a high-performance liquid chromatography procedure based on ion-exchange. Chromatographia 59, 129–133 (2004).
Fu, Z. et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232 (2012).
Acknowledgements
This work was supported by the State Key Program of the National Natural Science Foundation of China (31330068), the Young Scientists Fund of the National Natural Science Foundation of China (31601735), and the earmarked fund for the China Agriculture Research System (CARS-27). We are grateful to Dr. Zhihong Zhang for providing the tissue-cultured apple plants.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, X., Huo, L., Jia, X. et al. Overexpression of MdATG18a in apple improves resistance to Diplocarpon mali infection by enhancing antioxidant activity and salicylic acid levels. Hortic Res 5, 57 (2018). https://doi.org/10.1038/s41438-018-0059-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-018-0059-5
This article is cited by
-
VvATG6 contributes to copper stress tolerance by enhancing the antioxidant ability in transgenic grape calli
Physiology and Molecular Biology of Plants (2024)
-
Plant chitinases: Types, structural classification, antifungal potential and transgenic expression in plants for enhanced disease resistance
Plant Cell, Tissue and Organ Culture (PCTOC) (2024)
-
Involvement and Possible Mechanism of Autophagy in Postharvest Tomato Fruit Resistance Against Botrytis cinerea
Journal of Plant Growth Regulation (2024)
-
Transcriptomic and metabolic studies on the role of inorganic and organic iodine compounds in lettuce plants
Scientific Reports (2023)
-
Autophagy promotes jasmonate-mediated defense against nematodes
Nature Communications (2023)