Abstract
The enzyme RNAPII CTD phosphatase-like 1 is known as a transcriptional regulator of the plant response to various abiotic stresses. Here, the isolation of CmCPL1, a chrysanthemum (Chrysanthemum morifolium) gene encoding this enzyme is described. Its predicted 955 residue gene product includes the FCPH catalytic domain, two double-stranded RNA binding motifs, and a nuclear localization signal. A sub-cellular localization assay confirmed that CmCPL1 was expressed in the nucleus. CmCPL1 transcription was shown to be significantly inducible by heat stress. The over-expression and knockdown of CmCPL1, respectively, increased and diminished the tolerance of chrysanthemum to heat stress, which maybe dependent on the regulation of CmCPL1 and on the expression of downstream heat stress-responsive genes.
Similar content being viewed by others
Introduction
RNA polymerase II (RNAP II) is a multi-subunit enzyme complex enabling the transcription of mRNA precursors, and microRNAs1. It is also involved in the regulation of various mRNA maturation processes, including capping, splicing, and polyadenylation2,3. The large RNAP II subunit's carboxyl terminal domain (CTD) includes a conserved heptapeptide (YSPTSPS), which plays important roles in the regulation of gene expression through its own phosphorylation/dephosphorylation4. The RNAPII CTD phosphatase FCP1 is largely responsible for the dephosphorylation of Ser-2 and Ser-5 present in the heptapeptide5,6,7,8. The RNAPII CTD phosphatase-like 1 enzyme (CPL1), a homolog of FCP1, harbors the FCPH domain and displays CTD phosphatase activity9,10. It also features two dsRNA-binding motifs (DSRMs) and a nuclear localization signal (NLS) at its C terminus, which are important for, respectively, the necessary protein–protein and protein–RNA interactions, and for nuclear localization11,12,13.
CPL1 proteins have been implicated, particularly, in Arabidopsis thaliana, in both the plant's development and its stress response5,14,15,16,17. According to Zhang et al.6, both the growth and morphogenesis of the plant are affected by the action of AtCPL1 on Rho GTPase signaling, while Xiong et al.17 have confirmed that the same enzyme contributes to the plant's response to low temperature, drought, and salinity stress by negatively regulating the genes encoding DRE/CRT and CBF/DREB proteins. The over-expression of RCF2 (a CPL1 allele) enhances the heat tolerance of A. thaliana through its control over the phosphorylation status of the transcription factor NAC019 and its influence over the activity of various heat stress transcription factors (HSFs) and heat shock proteins (HSPs)18. The loss-of-function of AtCPL1 results in a transcriptional response similar to that induced by iron deficiency19. In addition, a number of interaction partners of CPL1 involved in the response to abiotic stress have been recently identified20,21,22. As yet, however, nothing is known regarding CPL1 function outside of the model plants.
Chrysanthemum (Chrysanthemum morifolium) is a leading ornamental species. The species is highly sensitive to a number of abiotic stresses, including drought, heat, salinity, heavy metal pollution, and nutrient deficiency23. Heat stress is known to be particularly damaging to both the growth and end-use quality of the chrysanthemum24,25. Supporting the breeding of improved cultivars would therefore benefit from a firmer understanding of the mechanistic basis of the chrysanthemum's adaptation to heat stress. Here, the goal was to characterize the CmCPL1 gene and its product, and to explore the effect of its over-expression and knockdown on the plant's heat tolerance.
Materials and methods
Plant materials, growing conditions, and stress treatments
Cuttings of the chrysanthemum cultivar ‘Jinba’, conserved by the Chrysanthemum Germplasm Resource Preserving Centre (Nanjing Agricultural University, Nanjing, China), were cultivated in a 3:5 mixture of garden soil and vermiculite. The plants were transferred and pre-incubated in an artificial climate chamber delivering a 10-h photoperiod (300 μmol m−2 s−1 light), 70–80% relative humidity, and a day/night temperature regime of 28/22 °C. Plants at the 8–10 leaf stage were subjected to a variable period of exposure to a range of abiotic stresses, namely high (45 °C) and low (4 °C) temperature, salinity (200 mM NaCl), drought stress (20% PEG 6000), iron deficiency, and treatment with 100 μM abscisic acid (ABA). For the purpose of transcriptional analysis, the leaf material was sampled before the onset of stress (0 h), and then after 3, 6, 12, and 24 h, with the exception of the iron deficiency stress, where the first sampling was carried out at 0 h, and subsequent ones after 3, 5, 7, 9, and 11 days. The samples required for RNA extraction were snap-frozen in liquid nitrogen, then stored at −80 °C.
Isolation of CmCPL1
Total RNA was extracted from the frozen leaf samples using the RNAiso Plus reagent (TaKaRa, Japan), following the manufacturer’s protocol. The resulting RNA was reverse-transcribed into the first cDNA strand using SuperScript III reverse transcriptase (Invitrogen, USA), following the manufacturer’s protocol. The primer pair CPL1-F/-R (sequences given in Table S1) was designed to amplify a fragment of CmCPL1 based on the sequence represented in the transcriptome of cultivar 'Yuuka' (SRP029991), and RACE-PCR(rapid amplification of cDNA ends-Polymerase Chain Reaction); was then used to extend the sequence into the full-length cDNA (SMARTer® RACE 5′/3′ Kit, Clontech). The resulting sequence, after its gel purification (Agarose Gel DNA Purification Kit, TaKaRa), was ligated into pMD19-T (TaKaRa) for sequencing. Finally, the CmCPL1-F/-R primer pair (Table S1) was designed to amplify the entire CmCPL1 coding sequence (as confirmed by amplicon sequencing).
Sub-cellular localization of CmCPL1 protein
The CmCPL1 ORF was amplified using a forward primer (CmCPL1-SF) incorporating a SalI restriction site and a reverse primer (CmCPL1-NR) with a NotI site (sequences given in Table S1). The resulting PCR product was transferred into pENTR1A (Invitrogen, USA) and subjected to an LR reaction (a recombination reaction between attL and attR sites); using the binary vector pMDC4326 in order to generate the transgene construct p35S::GFP-CmCPL1. The construct was transiently introduced into onion epidermal cells using a PDS-1000/He helium-driven particle accelerator (Bio-Rad, USA); control cells were transformed in the same way with p35S::GFP. After their transformation, the onion epidermis samples were held for 16 h at 25 °C in the dark, after which GFP (Green Fluorescent Protein) fluorescence was monitored by laser scanning confocal microscopy (Zeiss LSM780, Germany).
Quantitative real-time PCR (qRT-PCR)
The first cDNA strand synthesized from the mRNA extracted from the leaf samples was used as the template for 20 µL qRT-PCRs formulated with SYBR® Premix Ex TaqTM II (TaKaRa) on a Mastercycler® ep realplex Real-Time PCR System (Eppendorf, Germany). Each reaction was composed of 10 µL SYBR Green PCR master mix, 0.4 µL of each primer (10 µM), 4.2 µL H2O, and 5 µL of cDNA. The PCR cycling regime comprised an initial denaturation of 95 °C/30 s, followed by 40 cycles of 95 °C/5 s and 60 °C/30 s. Amplification of the CmCPL1 sequence was driven by the primer pair qCmCPL1-F/R and that of the reference sequence CmEF1α (GenBank: AB679278.1) by the primer pair EF1α-F/R (sequences given in Table S1). Normalized transcript abundances were derived by applying the 2−ΔΔCT method27.
Chrysanthemum transformation
Artificial miRNAs (amiRNAs) designed to repress CmCPL1 were prepared using a protocol slightly modified from the one described by Shida et al.28. The product amplified by the A and B primer pair (sequences given in Table S1) was inserted into pENTR1A via its Sal I and Not I sites, and from there into pMDC32 (Invitrogen, USA) using the LR reaction, finally generating the knockdown construct pMDC32-CmCPL1_amiRNA. The over-expression construct pMDC32-CmCPL1 was obtained following the procedure given above for obtaining p35S::GFP-CmCPL1. The plasmids were introduced separately into Agrobacterium tumefaciens strain EHA105 using the freeze–thaw method, and from there into chrysanthemum following Mao et al.29. The abundance of CmCPL1 transcript in the resulting transgenic and non-transgenic (NT) plants was detected using qRT-PCR analysis.
Evaluation of heat tolerance
Rooted cuttings were grown to the 8–10 leaf stage, then exposed to a 24-h period of 45 °C; the plants were imaged after 0, 1, 3, 6, 12, 15, 20, and 24 h. Measurements were made of the leaves' maximal photochemical efficiency (Fv/Fm) after 0, 12, and 24 h, following the method given by Wang et al.30, and the free proline content and peroxidase (POD) activity of the leaf were assessed after 0, 1, 3, 6, 12, and 24 h, as described by Liu et al.31 and He et al.32, respectively. Each set of measurements was based on 25 plants per genotype (NT and transgenics), and the entire experiment was replicated three times.
Transcription of heat-responsive genes
The abundance of transcript in both NT and transgenic plants generated from a number of heat stress-related genes (HSP70, HSP, sHSP, HSFA2, DREB2A, and WRKY41) was evaluated in triplicate using qRT-PCR. Leaf samples were collected after the plants had been exposed to 45 °C for 0, 1, 4, and 12 h. The CmEF1α sequence was used as the reference for normalization. The relevant primer sequences are given in Table S1.
Statistical analysis
All analyses were performed using routines implemented within the SPSS v20.0 package (SPSS Inc., Chicago, IL, USA). A one-way analysis of variance, based on the Tukey test, was used to identify means differing significantly from one another; the significance thresholds were 0.05 (marked by *) and 0.01 (**).
Results
Isolation of CmCPL1 and the analysis of its nucleotide and peptide sequences
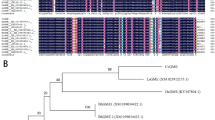
The CmCPL1 sequence isolated from cultivar 'Jinba' was 3159 bp in length, with 2868 bp of an open reading frame. The gene was predicted to encode a 955 residue protein (Fig. S1). A functional domain analysis of the predicted CmCPL1 protein showed that it featured a typical RNAPII CTD phosphatase catalytic domain (FCPH domain), two DSRMs, and one NLS (Fig. S1). A phylogenetic analysis demonstrated that the most similar sequences to CmCPL1 were StCPL1 from potato and SlCPL1 from tomato (Fig. 1).
Sub-cellular localization of CmCPL1
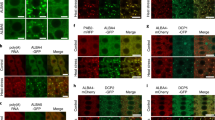
The sub-cellular localization of CmCPL1 was inferred from the transient expression of p35S::GFP-CmCPL1 in onion epidermal cells. The transformed cells expressed GFP strictly in the nucleus (Fig. 2d–f), whereas in control transgenic cells expressing p35S::GFP, GFP signal was detected throughout the cell (Fig. 2a–c).
Transcription pattern of CmCPL1 in NT chrysanthemum
A qRT-PCR analysis showed that CmCPL1 transcription was highest in the leaf, followed by the flower, root, and stem (Fig. S2). The gene was inducible by a range of abiotic stresses (Fig. 3); of particular note was its strong induction following the plants' exposure to 45 °C, which resulted in an approximately four-fold increase in transcript abundance over the untreated control after a 12-h exposure to stress (Fig. 3a).
a Heat stress (40 °C), b low-temperature stress (4 °C), c treatment with 100 μM ABA, d salinity stress (200 mM NaCl), e drought stress (20% PEG 6000), f iron deficiency. Each value represents the mean of three biological replicates, and the whiskers indicate the SE. *, **: Treatment means differed from the control means (CK) at, respectively, P < 0.05 and <0.01
Obtainment of CmCPL1 over-expression and knockdown lines
The over-expression of CmCPL1 in cultivar 'Jinba' was achieved by introducing the transgene pMDC32-CmCPL1 (Fig. S3A), while its knockdown was induced by the introduction of pMDC32-CmCPL1_amiRNA (Fig. S3B). Putative transgenic lines were regenerated on a selective medium (Fig. S4A-D) and validated using a genomic PCR assay (Fig. S4E). When the level of CmCPL1 transcription in these plants was estimated using qRT-PCR, two of the pMDC32-CmCPL1_amiRNA plants (amiR-3 and amiR-8) showed clear evidence of knockdown (Fig. 4a), while the abundance of CmCPL1 transcript was substantially higher in the pMDC32-CmCPL1 lines OE-4 and OE-7 than in NT plants (Fig. 4b). Thus, these four transgenic lines were selected for evaluating the effect of CmCPL1 transcript abundance on the plant's heat tolerance.
The heat tolerance of transgenic chrysanthemum plants
Heat stress induced leaf wilting in NT plants within 15 h, after which the severity of the wilting continued to rise; the two amiR line plants reacted more strongly than NT plants, and were severely wilted at the 24 h time point; however, leaves of the OE-4 and OE-7 line plants only began to wilt after a 20-h exposure (Fig. 5). There was little variation with respect to the photosynthesis parameter Fv/Fm between the four transgenic and the NT plants prior to the onset of stress; but as the stress period was prolonged, both amiR line plants recorded a lower Fv/Fm than NT plants, while both OE line plants recorded a higher one (Fig. 6a, S5). Similarly, while both proline content and POD activity were significantly enhanced by the stress in all genotypes, the levels were lower than NT plants in the amiR plants, and higher in the OE ones (Fig. 6b, c).
a Fv/Fm and Fo images at the end of the stress episode. The pseudocolored bar depicted at the bottom of the panel ranges from 0 (black) to 1.0 (purple). b The leaf content of free proline and c leaf POD activity. Values shown in the form of mean ± SE (n = 3). *, **: Treatment means differed from the control means (NT) at, respectively, P < 0.05 and <0.01
Transcription regulation of several heat-responsive genes by CmCPL1
In both types of transgenic plants, the abundance of HSP70 transcript was less than that in the NT plants during their exposure to heat stress (Fig. 7a). However, compared to NT plants, the level of transcription of HSP, sHSP, and HSF was significantly higher in OE-4 and significantly lower in amiR-3, especially after a 1-h exposure to stress (Fig. 7b–d). Similarly, the abundance of DREB2A transcript was lower in amiR-3 and higher in OE-4 (Fig. 7e). The transcriptional behavior of WRKY41 was very different: the abundance of the transcript was significantly higher in the amiR-3 line than in the NT plants, and lower in OE-4. Overall, the conclusion was that the level of CmCPL1 transcription positively affected that of HSP, sHSP, HSFA2, and DREB2A, and negatively that of WRKY41.
Discussion
The phosphorylation/dephosphorylation of RNAPII CTD mediated by various phosphatases is an important component of eukaryotic transcriptional regulation and mRNA processing3. The CPL1 protein, which acts to dephosphorylate the RNAPII CTD, is known to regulate not just the growth and development of plants, but also their stress responses33. Here, the product of a chrysanthemum gene (CmCPL1) encoding an RNAPII CPL1 protein has been shown to affect the plant's' response to heat stress. The CmCPL1 protein included the characteristic RNAPII CTD phosphatase catalytic domain, along with two DSRMs, mirroring the structure of AtCPL114,17. Previous studies have demonstrated that CPL1 proteins are deposited in the plant cell nucleus, thanks to their C terminal NLS5,18. The CmCPL1 sequence similarly included a putative C terminal NLS, matching that carried by its homologs in both A. thaliana and rice5. Thus, as predicted, when onion epidermal cells were transiently transformed with the p35S::GFP-CmCPL1 construct, GFP accumulated preferentially in the nuclei, confirming CmCPL1 to be a nuclear protein, presumably thanks to its C terminal NLS.
CPL1 is a key player in the plant's response to various abiotic stresses5,14,15,16,17. Since the absence of AtCPL1 results in heightened iron deficiency signaling and enhanced crosstalk with a branch of the osmotic stress/ABA signaling pathway, it has been proposed that this protein acts as a negative regulator of the above process19,22. Similarly, AtCPL1 has been described as a negative regulator of stress-responsive gene transcription in A. thaliana plants subjected to a range of abiotic stresses14. According to Guan et al.18, the allele of AtCPL1 (RCF2) controls the low temperature-responsive genes in both positive and negative manners, and is a major positive regulator of heat stress-responsive gene expression and themotolerance in A. thaliana. Here, the abundance of CmCPL1 transcript was altered by the imposition of a range of abiotic stresses, which indicates that its product is an element of the general stress response of chrysanthemum. Of particular note was the heightened responsiveness of CmCPL1 transcription to heat stress, suggesting the centrality of this protein's involvement in the heat stress response.
In the chrysanthemum plants subjected to heat stress, those which had beeen engineered to over-express CmCPL1 displayed an enhanced level of tolerance, while the CmCPL1 knockdown plants proved to be more sensitive than NT plants. A corresponding experiment in A. thaliana produced a completely similar result18. The expression of CmCPL1 likely contributed to the physiological homeostasis of heat-stressed plants, specifically with respect to their photosynthetic efficiency, osmotic pressure of the cell, and the production of POD. Unfortunately, the contribution of CmCPL1 to stress tolerance cannot exist extensively in other abiotic stress, such as salinity stress, indicating that CmCPl1 may have a specific role in thermotolerance (Fig. 6S). Recently, the genes encoding certain chrysanthemum HSPs and transcription factors have been shown to be inducible by heat stress23,34. In heat-stressed A. thaliana plants, the absence of AtCPL1 suppresses the transcription of a number of HSFs, HSPs, and DREB2, while the stress has the opposite effect on over-expressors of AtCPL118. Here, the observation was that the presence of CmCPL1 was associated in heat-stressed plants with the upregulation of HSP, sHSP, HSFA2, and DREB2A, and the downregulation of WRKY41, in accordance with the outcome of similar experiments performed in other species18,34. Interestingly, the expression of HSP70 was inhibited both in over-expression and knockdown plants under heat stress, implying that CmCPL1 was involved in the regulation of HSP70 transcription, although this process may be complex and multi-factoring, which requires further study. The overall implication was that CmCPL1 acts as a positive regulator of a suite of heat stress-responsive genes, the products of which are required to protect the plant from damage imposed by heat stress. The gene therefore represents an interesting target for devising a genetic engineering-based strategy aimed to develop chrysanthemum cultivars expressing heightened heat tolerance.
References
Sims, R. J., Mandal, S. S. & Reinberg, D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell. Biol. 16, 263–271 (2004).
Hirose, Y. & Manley, J. L. RNA polymerase II and the integration of nuclear events. Gene. Dev. 14, 1415–1429 (2000).
Jin, Y. M. et al. AtCPL5, a novel Ser-2-specific RNA polymerase II C-terminal domain phosphatase, positively regulates ABA and drought responses in Arabidopsis. New Phytol. 190, 57–74 (2011).
Chen, T. et al. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 9, e1003875 (2013).
Koiwa, H. et al. Arabidopsis C-terminal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases. PNAS 101, 14539–14544 (2004).
Zhang, B. et al. C-terminal domain (CTD) phosphatase links Rho GTPase signaling to Pol II CTD phosphorylation in Arabidopsis and yeast. PNAS 113, E8197–E8206 (2016).
Hausmann, S., Koiwa, H., Krishnamurthy, S., Hampsey, M. & Shuman, S. Different strategies for carboxyl-terminal domain (CTD) recognition by serine 5-specific CTD phosphatases. J. Biol. Chem. 280, 37681–37688 (2005).
Hausmann, S., Schwer, B. & Shuman, S. An Encephalitozoon cuniculi ortholog of the RNA polymerase II carboxyl-terminal domain (CTD) serine phosphatase Fcp1. Biochemistry 43, 7111–7120 (2004).
Hausmann, S. & Shuman, S. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. J. Biol. Chem. 277, 21213–21220 (2002).
Hausmann, S. & Shuman, S. Defining the active site of Schizosaccharomyces pombe C-terminal domain phosphatase Fcp1. J. Biol. Chem. 278, 13627–13632 (2003).
Bang, W. Y., Kim, S. W., Jeong, I. S., Koiwa, H. & Bahk, J. D. The C-terminal region (640-967) of Arabidopsis CPL1 interacts with the abiotic stress-and ABA-responsive transcription factors. Biochem. Biophys. Res. Commun. 372, 907–912 (2008).
Fierro-Monti, I. & Mathews, M. B. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci. 25, 241–246 (2000).
Saunders, L. R. & Barber, G. N. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 17, 961–983 (2003).
Koiwa, H. et al. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. PNAS 99, 10893–10898 (2002).
Manavella, P. A. et al. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151, 859–870 (2012).
Matsuda, O., Sakamoto, H., Nakao, Y., Oda, K. & Iba, K. CTD phosphatases in the attenuation of wound-induced transcription of jasmonic acid biosynthetic genes in Arabidopsis. Plant. J. 57, 96–108 (2009).
Xiong, L. et al. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. PNAS 99, 10899–10904 (2002).
Guan, Q., Yue, X., Zeng, H. & Zhu, J. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress–responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26, 438–453 (2014).
Aksoy, E., Jeong, I. S. & Koiwa, H. Loss of function of Arabidopsis C-terminal domain phosphatase-like1 activates iron deficiency responses at the transcriptional level. Plant Physiol. 161, 330–345 (2013).
Jiang, J. et al. TheArabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLoS Genet. 9, e1003625 (2013).
Cui, P. et al. The RNA polymerase II C-terminal domain phosphatase-like protein FIERY2/CPL1 interacts with eIF4AIII and is essential for nonsense-mediated mRNA decay in Arabidopsis. Plant Cell 28, 770–785 (2016).
Jeong, I. S. et al. Regulation of abiotic stress signalling by Arabidopsis C-terminal domain phosphatase-like 1 requires interaction with a K-homology domain-containing protein. PLoS ONE 8, e80509 (2013).
Song, A. et al. A chrysanthemum heat shock protein confers tolerance to abiotic stress. Int. J. Mol. Sci. 15, 5063–5078 (2014).
Janka, E., Körner, O., Rosenqvist, E. & Ottosen, C.-O. High temperature stress monitoring and detection using chlorophyll a fluorescence and infrared thermography in chrysanthemum (Dendranthema grandiflora). Plant Physiol. Biochem. 67, 87–94 (2013).
Willits, D. & Bailey, D. The effect of night temperature on chrysanthemum flowering: heat-tolerant versus heat-sensitive cultivars. Sci. Hortic. 83, 325–330 (2000).
Curtis, M. D. & Grossniklaus, U. Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001).
Shida, T., Fukuda, A., Saito, T., Ito, H. & Kato, A. AtRBP1, which encodes an RNA-binding protein containing RNA-recognition motifs, regulates root growth in Arabidopsis thaliana. Plant Physiol. Biochem. 92, 62–70 (2015).
Mao, Y. et al. Functional analysis of alternative splicing of the FLOWERINGLOCUS T orthologous gene in Chrysanthemum morifolium. Hortic. Res. 3, 16058 (2016).
Wang, W. et al. Overexpression of Camellia sinensis H1 histone gene confers abiotic stress tolerance in transgenic tobacco. Plant Cell Rep. 33, 1829–1841 (2014).
Liu, F., Liu, Q., Liang, X., Huang, H. & Zhang, S. Morphological, anatomical, and physiological assessment of ramie [Boehmeria Nivea (L.) Gaud.] tolerance to soil drought. Genet. Resour. Crop Evol. 52, 497–506 (2005).
He, J. et al. Chrysanthemum leaf epidermal surface morphology and antioxidant and defense enzyme activity in response to aphid infestation. J. Plant. Physiol. 168, 687–693 (2011).
Koiwa, H. Phosphorylation of RNA polymerase II C-terminal domain and plant osmotic-stress responses. In: Rai AK, Takabe T, eds. Abiotic stress tolerance in plants: toward the improvement of global environment and food. Dordrecht, the Netherlands: Springer Publishing, 47–57 (2006).
Sun, J. et al. Identification of differentially expressed genes in Chrysanthemum nankingense (Asteraceae) under heat stress by RNA Seq. Gene 552, 59–66 (2014).
Acknowledgements
This work was supported by funding from the Germplasm Resources Protection (crop) project of Ministry of Agriculture (1120162130135252031), National Natural Science Foundation of China (31572159) and the National Science Fund for Distinguished Young Scholars (31425022).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qi, Y., Liu, Y., Zhang, Z. et al. The over-expression of a chrysanthemum gene encoding an RNA polymerase II CTD phosphatase-like 1 enzyme enhances tolerance to heat stress. Hortic Res 5, 37 (2018). https://doi.org/10.1038/s41438-018-0037-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-018-0037-y
This article is cited by
-
Enhancing crop yield by using Rubisco activase to improve photosynthesis under elevated temperatures
Stress Biology (2021)
-
Current achievements and future prospects in the genetic breeding of chrysanthemum: a review
Horticulture Research (2019)
-
Tools for Chrysanthemum genetic research and breeding: Is genotyping-by-sequencing (GBS) the best approach?
Horticulture, Environment, and Biotechnology (2019)