Abstract
In plants, the vesicle fusion process plays a vital role in pathogen defence. However, the importance of the vesicle fusion process in apple ring rot has not been studied. Here, we isolated and characterised the apple syntaxin gene MdSYP121. Silencing the MdSYP121 gene in transgenic apple calli increased tolerance to Botryosphaeria dothidea infection; this increased tolerance was correlated with salicylic acid (SA) synthesis-related and signalling-related gene transcription. In contrast, overexpressing MdSYP121 in apple calli resulted in the opposite phenotypes. In addition, the results of RNA sequencing (RNA-Seq) and quantitative real-time PCR (qRT-PCR) assays suggested that MdSYP121 plays an important role in responses to oxidation–reduction reactions. Silencing MdSYP121 in apple calli enhanced the expression levels of reactive oxygen species (ROS)-related genes and the activity of ROS-related enzymes. The enhanced defence response status in MdSYP121-RNAi lines suggests that syntaxins are involved in the defence response to B. dothidea. More importantly, we showed that MdSYP121 forms a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex with MdSNAP33, and the complex may participate in regulating resistance to B. dothidea. In conclusion, by regulating the interaction of SA pathway and oxidation–reduction process, MdSYP121 can influence the pathogen infection process in apple.
Similar content being viewed by others
Introduction
During the long-term process of evolution, plants have evolved a secretory pathway, which is critical for biosynthetic and endocytic trafficking to the plasma membrane (PM) and vacuole1. Secretory membrane trafficking mechanisms have recently been shown to be involved in a variety of plant-specific processes, including plant development, tropic responses and pathogen defence2,3. Most endoplasmic reticulum-localised proteins and ER-resident chaperones of the secretory pathway and many antimicrobial compounds are destined for the apoplast or various organelles upon pathogen attack and penetration4,5.
In plants, fusion of the endomembrane system is important and rather difficult. During the evolution of eukaryotes, a specialised class of proteins, the soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), were formed. These receptors function as mediators of fusion between vesicles and target lipid bilayers3,6. Based on the occurrence of either a conserved glutamine or arginine residue in the centre of the SNARE domain, SNAREs can be grouped as target site-localised Q-SNAREs (Qa, Qb, Qc and Q(b+c)) and vesicle-residing R-SNAREs7,8. SNAREs drive membrane merging by forming SNARE complexes via intermolecular interactions among vesicles and target subcellular SNAREs. A typical SNARE complex involves distinct types of SNARE proteins (Qa+Qb+Qc or Qa+Q(b+c)) and one R-SNARE polypeptide that together contribute to a four-helix bundle of intertwined SNARE domains9.
SNARE-domain proteins represent a fundamental role in biotic defence against fungal pathogens. At least two SNARE protein-mediated exocytosis pathways appear to drive the directional or non-directional secretion of antimicrobial compounds, including defence-related proteins and cell wall building blocks, into the apoplastic space to terminate the infection of fungi or bacteria10,11,12. On one hand, plants exhibit an active resistance mechanism to combat extracellular infection. Silencing a specific PM syntaxin, NbSYP132, in Nicotiana benthamiana impairs the accumulation of a subset of pathogenesis-related (PR) proteins in the cell wall. NbSYP132 also contributes to basal and salicylate-associated defence. These results indicate that SYP132-mediated secretion is an important component of resistance against bacterial pathogens in plants11. In addition, a SNAP25 homologue is required for pathogen resistance. Mutants of the barley MLO gene are fully resistant to powdery mildew; silencing the barley HvSNAP34 gene, a SNAP25 homologue, revealed increased fungal entry rates in the mlo genotype10. On the other hand, cell-autonomous immunity is widespread in plant–fungal interactions and terminates fungal infection after pathogen entry12. Plants exhibit resistance by forming SNARE secretory complexes, and pathogen-induced subcellular dynamics enable the execution of immune responses by focal secretion12. The AtSYP122 syntaxin functions in exocytosis during defence responses to strengthen the cell wall and prevent penetration by microbes13,14. In addition, syntaxins can regulate heavy vesicle traffic towards the site of attempted infection when plants are subjected to fungal attack14.
SNARE syntaxin 121 (SYP121) is one of the most important SNARE proteins. SYP121, which is a Qa-SNARE protein, cycles continuously between the PM and endosomes15 and has a fundamental role in plant defence10. Importantly, SYP121-dependent disease resistance acts in vivo mainly by forming SNARE complexes together with the SNAP33 adaptor and a subset of vesicle-associated membrane protein (VAMP) subfamily members, VAMP721/VAMP72212. In Arabidopsis, SYP121 is an important element in resistance to non-host powdery mildew fungi. In addition, the barley ROR2 gene, a functional homologue of AtSYP121, is required for basal penetration resistance against Blumeria graminis f. sp. hordei (Bgh)10. A mechanistic link has been demonstrated between non-host resistance and basal penetration resistance in monocotyledons and dicotyledons10. AtSYP121 is encoded by PEN110. The Arabidopsis mutant pen1-1 caused delayed formation of localised cell wall appositions (so-called papillae), resulting in increased Bgh infection at attack sites16. In addition, using brefeldin A (BFA) showed that interference in endosome recycling to the PM both severely inhibited the SYP121 focal accumulation at fungal entry sites that accompanied delayed callose deposition and compromised defence against powdery mildew fungi17. However, Zhang et al. reported that SYP121 and its homologue SYP122 are negative regulators of salicylic acid (SA) signalling and programmed cell death (PCD)18. The enhanced SA and jasmonic acid (JA) signalling levels and PCD in the syntaxin double mutant syp121-1 syp122-1 contributed to improved disease resistance against the virulent powdery mildew fungus Erysiphe cichoracearum and the bacterial pathogen Pseudomonas DC300 in an SA-independent manner18. These results suggested that SNAREs employs different mechanisms to regulate both the initial penetration resistance process and the subsequent immune signalling process; these processes differentially contribute to disease resistance. The PM-localised syntaxin-related 1 (StSYR1) and StSNAP33 genes in potato are homologues of Arabidopsis AtSYP121 and AtSNAP33, respectively. The resistance of both StSYR1-RNA interference (RNAi) and StSNAP33-RNAi plants increased and was correlated with the constitutive accumulation of SA and PR1 transcripts, and these plants displayed an early senescence-like phenotype showing chlorosis and necrosis. In addition, downregulation of StSYR1 led to enhanced resistance against Phytophthora infestans and a cell death response at the site of infection19.
Apple is one of the most widely cultivated fruits in the world. China has the largest planting area and is the greatest producer of apples worldwide. Apple ring rot is one of the most devastating diseases in China and greatly affects the production of apple;20 this disease occurs mostly in the Circum-Bohai-Sea region (Shandong, Hebei and Liaoning provinces)21. Since the 1980s, in conjunction with the widespread planting of Fuji cultivars, the area of apple ring rot has increased in eastern China20. This disease, which is caused by the fungal pathogen Botryosphaeria dothidea, infects both fruits and branches20. The resistance mechanism of apple ring rot is very complex, and few studies have examined the molecular mechanism of apple resistance to B. dothidea infection. In the present study, based on RNA-Seq results, the MdSYP121 gene was isolated in apple. The results of a series of genomic, genetic and transgenic experiments suggest that MdSYP121 plays an important role in B. dothidea resistance by affecting the oxidation–reduction process in apple. More importantly, we showed that a SNARE complex composed of MdSYP121 and MdSNAP33 may play an important role in pathogen resistance. Together, these results increase the knowledge regarding the biological roles of SNARE proteins in disease resistance and improve our understanding of the pathogenesis of B. dothidea. This study hints at a useful disease management strategy and is helpful for breeding resistance to apple ring rot.
Materials and methods
Cultivation and treatment of plants
Tissue-cultured calli of the apple cultivar ‘Orin’ were subcultured under the basic growth conditions of 24 ± 0.5 °C and 24 h of darkness (at a relative humidity of 60–75%). The ‘Orin’ calli were subcultured in culture dishes (9 cm in diameter) containing 35 mL of Murashige and Skoog (MS) medium (0.4 mg L−1 6-BA, 1.5 mg L−1 2,4-D, 30 g L−1 sucrose and 7.5 g L−1 agar; pH 5.8–6.0; autoclaved at 121 °C for 20 min). Tissue-cultured plants of the apple cultivar ‘Gala’ were incubated under greenhouse conditions of 24 ± 0.5 °C and a 16-h light/8-h dark cycle (at a relative humidity of 60–75%). The ‘Gala’ explants were cultured in culture bottles (5.5 cm in diameter) containing 40 mL of MS subculture medium (0.5 mg L−1 6-BA, 0.2 mg L−1 IAA, 30 g L−1 sucrose and 7.5 g L−1 agar; pH 5.8–6.0; autoclaved at 121 °C for 20 min). Botryosphaeria dothidea was incubated in culture dishes (9 cm in diameter) containing 15 mL of potato dextrose agar medium at 24 ± 0.5 °C in darkness. Nicotiana benthamiana seeds were surface-sterilised and germinated on MS medium under greenhouse conditions of 24 ± 0.5 °C and a 16-h light/8-h dark cycle (at a relative humidity of 60–75%). N. benthamiana seedlings were subsequently transplanted at the two-leaf or three-leaf stage into soil and grown under greenhouse conditions.
Vector construction and genetic transformation
Fragments and full-length coding sequences were cloned from a cDNA library that was reverse-transcribed from RNA isolated from Fuji fruits synthesised using a First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) in accordance with the manufacturer’s instructions. PCR was performed with Pfu/Taq polymerase (Fermentas, USA). The primers used are shown in Supplementary Table S1. All generated amplicons were subcloned into a pLB vector (Tiangen, China). Homologous MdSYP121 protein sequences were retrieved from the National Center Biotechnology Information (NCBI) database and aligned using DNAMAN 5.2.2 software (Lynnon Biosoft, USA). A phylogenetic tree was subsequently generated using the Neighbour Joining method with MEGA 5.0 software. A MdSYP121 fragment from 121 to 613 bp was used for RNAi vector construction. The MdSYP121 RNAi fragments were subsequently transferred to pHANNIBAL and pCB302 vectors by T4 recombination (NEB, USA) to generate RNAi transformation vectors. The full-length products were used to create HA protein fusions by T4 recombination into pCB302-MdSYP121-HA under the control of the cauliflower mosaic virus (CaMV) 35S promoter to generate the overexpression (OE) transformation factor.
The leaves of 4-week-old tissue-cultured ‘Gala’ plants were transformed with Agrobacterium tumefaciens LBA4404 carrying the RNAi transformation vector. The leaves were scratched and pre-differentiated on MS differentiation medium (2.0 mg L−1 TDZ, 0.2 mg L−1 IAA, 30 g L−1 sucrose and 7.5 g L−1 agar; pH 5.8–6.0; autoclaved at 121 °C for 20 min) for 2 days, and then they were incubated with LBA4404 (optical density (OD) = 0.4–0.6) for 20–30 min and co-cultured on MS differentiation medium at 24 °C for 7 days in the dark. The leaves were then transferred to screening differentiation medium containing glufosinate ammonium and carbenicillin under a 16-h light/8-h dark cycle (at a relative humidity of 60–75%). The screened explants were cultured on MS subculture medium.
To generate transgenic apple calli, 10-day-old apple calli were transformed with A. tumefaciens LBA4404 carrying the RNAi or OE transformation vectors, respectively. The apple calli were incubated with LBA4404 (optical density (OD) = 0.4–0.6) for 20–30 min and co-cultured on MS solid media containing no antibiotics at 24 °C for 48 h in the dark. The calli cells were then transferred to screening medium containing glufosinate ammonium and carbenicillin22.
Subcellular localisation of MdSYP121 and MdSNAP33
The open reading frames (ORFs) of MdSYP121 and MdSNAP33 were inserted into a pCB302 green fluorescent protein (GFP) vector, whose N-terminus consists of a GFP under the control of the CaMV 35S promoter. To achieve transient expression, the recombinant plasmids pCB302-MdSYP121-GFP and pCB302-MdSNAP33-GFP were transformed into A. tumefaciens GV3101. After the cell cultures were incubated overnight, A. tumefaciens cells were harvested via centrifugation and resuspended in infiltration medium (100 mL of medium containing 1 mL of 1 M MES-KOH at pH 5.6, 333 μL of 3 M MgCl2 and 100 μL of 150 mM acetosyringone). The leaves of 5-week-old N. benthamiana plants were used for transient expression, and the GFP signals were observed using a LSM 880 META confocal microscope (Carl Zeiss, Germany). A pCB302-GFP construct was used as a control.
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was isolated from apple tissue culture plants and calli using the cetyltrimethylammonium bromide method described by Wang et al.23. First strand cDNA was synthesised using a First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) in accordance with the manufacturer’s instructions. qRT-PCR was used to detect the expression levels of the target genes. The 20 μL PCR mixture comprised 10 μL of Fast Start Universal SYBR® Green Master mix (Roche, USA), 0.6 μL of each primer (10 mM), 2 μL of diluted cDNA and 6.8 μL of PCR-grade H2O and a CFX96TM Real-time Detection System (Bio-Rad, USA) was used to perform PCR. The following PCR programme was used: predenaturation at 98 °C for 10 min; 40 cycles of 98 °C for 15 s and then 60 °C for 30 s; and a final melt cycle from 60 to 98 °C. The PCR process was completed with a melting curve analysis program. The Malus × domestica actin gene was used as a standard control to quantify cDNA abundance. The primers used in the qRT-PCR analyses are shown in Supplementary Table S2.
Pathogen infection assays
For the pathogen infection analysis, transgenic and Vec (serving as an empty vector control) lines were transferred to MS solid medium that lacked glufosinate ammonium and carbenicillin. Ten-day-old Vec, RNAi and OE lines were infected with 0.5-cm-diameter agar discs containing uniform B. dothidea mycelia that were incubated for 5 days. The calli were co-cultured for 4 days in the dark. For expression pattern analyses, treated calli were collected from culture dishes, frozen in liquid nitrogen and stored at −80 °C for RNA extraction. Each experimental treatment was repeated at least three times.
Enzyme activity assays
The activity levels of peroxidase (POD), catalase (CAT), ascorbate peroxidase (APX) and glutathione reductase (GST) were spectrophotometrically measured using hydrogen peroxide test kits (Nanjing Jiancheng Bioengineering Institute, China). Each experimental treatment was repeated at least three times.
Library construction and RNA-Seq analysis
Nine independent calli from RNAi and Vec lines were infected by B. dothidea. Both the total composite RNA from the RNAi lines and the total composite RNA from the Vec lines, respectively, were used for Illumina sequencing at Novogene Technologies (Beijing, China). All procedures during the cDNA library construction were performed in accordance with a standard Illumina sample preparation protocol. The RNA-Seq libraries were sequenced on an Illumina Genome HiSeq platform.
After the RNA was sequenced, the adaptors were trimmed and low-quality sequences were removed from the raw data. The unigenes were annotated using the following databases: the Clusters of Orthologous Groups (COG) (http://www.ncbi.nlm.nih.gov), Gene Ontology (GO) (http://www.geneontol-ogy.org/) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/)24,25,26. To assay the differentially expressed genes (DEGs), trimmed mean of M-values normalisation and DEG seq were used to normalise gene expression levels for differential expression analyses, respectively27. A q-value < 0.005 served as the P-value threshold in multiple tests to determine the significance of differences in gene expression.
Verification of RNA-Seq data using qRT-PCR
RNA was extracted from each RNAi and Vec line to verify the RNA-Seq data. RNA was prepared, and cDNA was synthesised, respectively, as described above. The primers used for the qRT-PCR analyses are shown in Supplementary Table S2. All of the samples were tested at least three times, and the experiments were performed on three biological replicates.
GST pull-down analysis
MdSYP121 with an HA tag was expressed in apple calli. The calli were homogenized in an extraction buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 50 mM EDTA, 1% Triton, 1 mM phenylmethanesulfonyl fluoride and a protease inhibitor cocktail (Roche, USA). For prokaryotic expression assays, a plasmid containing MdSNAP33 with a GST tag was transformed into Escherichia coli Rosetta (Tiangen, China), and the proteins were induced with 1 mM isopropyl 1-thio-β-d-galactopyranoside at 28 °C. The GST-MdSNAP33 protein was purified using a Pierce® GST Spin Purification Kit in accordance with the manufacturer’s protocol (Thermo Fisher Scientific, USA). The mixture of MdSYP121-HA protein extracted from the apple calli and the purified GST-MdSNAP33 protein with 2 μL of GST antibody were incubated together with gentle shaking for 2 h at 4 °C. The protein mixture was then immunoprecipitated with G-agarose beads (Sigma-Aldrich, USA) by gentle shaking for 2 h at 4 °C. The beads were collected and washed three times with washing buffer (100 mM NaCl, 10 mM HEPES, pH 7.5; 1 mM EDTA; a protease inhibitor cocktail; and 10% glycerol) and once with 50 mM Tris-HCl (pH 7.5). The immunoprecipitated proteins were analysed with an HA antibody. Equal amounts of total protein were electrophoresed on 10% SDS-PAGE.
Bimolecular fluorescence complementation assays
Full-length MdSYP121 and MdSNAP33 were fused to yellow fluorescent protein (YFP) vectors to pUC-SPYCE-35S and pUC-SPYNE-35S, respectively, by T4 recombination. The primers used are listed in Supplementary Table S1. These two recombinant plasmids were transiently expressed in tobacco leaves by A. tumefaciens (GV3101)-mediated infiltration28. The YFP fluorescence of tobacco leaves was detected after infiltration for 4 days using a LSM 880 META confocal microscope (Carl Zeiss, Germany).
SDS resistance assays
For in vitro-binding studies, MdSYP121-HA homogenized in extraction buffer (as mentioned above) and GST-MdSNAP33 purified protein (as mentioned above) were incubated together for 12 h at 4 °C (head-over-head rotation). The bound material was eluted by incubation (30 min at 37 °C) together with 4× SDS loading buffer, because it is SDS-resistant but heat-sensitive; the controls were incubated for 10 min at 100 °C29. Protein samples were then separated on 10% SDS-PAGE. Western blot was used to detect the expression of proteins.
Statistical analysis
All experiments were performed at least three times. The error bars in each graph indicate the mean values ± SEs of three repetitions. Statistical significance between different measurements was determined using Tukey’s honestly significant difference (HSD) test via IBM Statistical Product and Service Solutions (SPSS) statistics software version 19 (IBM, USA).
Results
Cloning and characterisation of MdSYP121
To study the resistance mechanism of apple to apple ring rot, we analysed the RNA-Seq data of Fuji cultivars inoculated with B. dothidea. MDP0000709455 was chosen for detailed characterisation because it was induced in ‘Fuji’ apple with B. dothidea inoculation and served as an important node by analysing the interaction network using Cytoscape (version 3.6.0, USA) (data not shown), and the orthologous genes of MDP0000709455 played critical roles in controlling disease resistance towards powdery mildew in Arabidopsis and barley10. A fragment was isolated from the cDNA library of Fuji fruit using the primers listed in supplementary Table S1. The full-length cDNA sequence consisted of a 1068-bp ORF. The ORF encoded a protein that was 356 amino acids in length and had a calculated molecular mass of 43.83 kDa and an isoelectric point of 7.15. This clone exhibited a high level of sequence similarity to AtSYP121 of Arabidopsis. Based on multiple sequence alignments with other plant SYP121s, MDP0000709455 contains Ha-conserved, Hb-conserved and Hc-conserved subdomains, a Qa-SNARE subdomain and a transmembrane domain (Supplementary Fig. S1a). In addition, MDP0000709455 exhibits high homology to AtSYP121 (AEE75103), PpSYP121 (XP_007202232), CsSYP121 (XP_00648162) and OsSYP121 (ABB22872) (Supplementary Fig. S1a); so, this gene was designated MdSYP121.
To investigate the evolutionary relationships among SYP121s from different species, a phylogenetic analysis based on their amino acid sequences was performed by the Neighbour Joining method using the software MEGA version 5.0. As shown in Supplementary Fig. S1b, MdSYP121 exhibited high similarity to the members of the Qa-SNARE group, including AtSYP121, PpSYP121, CsSYP121 and QsSYP121. These results suggest that MdSYP121 is a member of the Qa-SNARE.
Subcellular localisation of MdSYP121
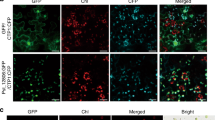
To determine the actual localisation of the MdSYP121 protein within cells, MdSYP121 was fused to an HA tag in the OE vector 35S::MdSYP121-HA; the recombinant plasmid was transferred into apple calli, and OE transgenic lines were obtained (Supplementary Fig. S2). The membrane proteins of the MdSYP121-OE lines were separated using a membrane protein extraction kit (BestBio, China). MdSYP121 was detected among both total proteins and membrane proteins by using western blot with anti-HA antibody (Fig. 1a).
a Expression of 35S::MdSYP121-HA fusion constructs in apple calli. Total proteins, non-membrane proteins and membrane proteins (indicated in the figure as T, NM and M, respectively) were detected using anti-HA. b Schematic diagram of the 35S::MdSYP121-GFP fusion construct and the 35S::GFP construct. c Transient expression of the 35S::MdSYP121-GFP fusion construct and the 35S::GFP construct in N. benthamiana. Green fluorescence was observed after transient infection for 4 days using a confocal microscope. Bar = 10 μm
In addition, MdSYP121 was fused to a GFP tag in the OE vector 35S::MdSYP121-GFP (Fig. 1b). The fusion protein was transiently expressed in N. benthamiana leaves using agroinfiltration. A 35S::GFP construct served as a control. Fluorescence was detected in the PM of N. benthamiana leaves (Fig. 1c). These results suggested that MdSYP121 is a membrane-localised protein.
Silencing MdSYP121 increased resistance to Botryosphaeria dothidea
To investigate the function of MdSYP121, we silenced MdSYP121 in apple ‘Gala’ using the RNAi method. However, the transgenic plants showed the dwarfism and necrosis phenotypes, and only two transgenic plants were ultimately obtained (Supplementary Fig. S3). The transgenic plants were tiny and difficult to differentiate, so the functional analysis was difficult to perform.
To further analyse the function of this gene, we produced calli tissue whose expression of MdSYP121 was inhibited by the RNAi method. Ten independent transgenic lines were selected using glufosinate ammonium resistance selection and qRT-PCR (Supplementary Fig. S4). Three typical MdSYP121-RNAi lines (RNAi3, RNAi5 and RNAi7) were randomly selected for further functional analysis. As a control, Vec lines were subcultured at the same time.
To identify the function of MdSYP121 during B. dothidea infection, MdSYP121-RNAi lines growing on MS agar medium were inoculated with B. dothidea. Although the untreated Vec and MdSYP121-RNAi lines have slight difference, the spot extension areas of the MdSYP121-RNAi lines were clearly fewer than those of the Vec lines after inoculation with B. dothidea for 4 days (Fig. 2a and Supplementary Fig. S5b). The RNAi lines showed a nearly twofold decrease in fungal growth (Fig. 2b).
a Representative phenotypes of Vec and MdSYP121-RNAi calli both untreated and infected with B. dothidea for 4 days, respectively. b Pathogen disease indexes in Vec and MdSYP121-RNAi calli lines after B. dothidea infection for 4 days. c qRT-PCR analysis for the expression of SA signalling-related genes in Vec, RNAi3, RNAi5 and RNAi7 calli lines after B. dothidea infection for 4 days. d qRT-PCR analysis for the expression of SA-related genes in Vec, RNAi3, RNAi5 and RNAi7 calli lines after B. dothidea infection for 4 days. The error bars in b, c and d indicate the mean values ± SEs of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey’s HSD test. Vec served as the empty vector control
The phytohormone SA serves as an endogenous messenger during biotic stress in plants and is required for the activation of systemic acquired resistance (SAR)30. The expression of SA signalling-related and synthesis-related genes was analysed in the MdSYP121-RNAi and Vec lines after inoculation with B. dothidea for 4 days. As shown in Fig. 2c,d, the expression levels of both the SA signalling-related genes PR1, PR5 and NPR1 and the SA synthesis-related genes EDS1, PAD4 and PAL were higher in the MdSYP121-RNAi lines than those in the Vec line. These results show that silencing MdSYP121 increases tolerance to B. dothidea.
Overexpression of MdSYP121 decreased resistance to Botryosphaeria dothidea
To investigate the biological role of MdSYP121, we produced calli tissue that overexpressed MdSYP121. Six independent transgenic lines were selected using glufosinate ammonium resistance selection and western blot analysis (Supplementary Fig. S2). Three typical lines (OE3, OE4 and OE5) were randomly selected and confirmed, and these lines were used for further functional analysis. As a control, Vec lines were subcultured at the same time.
As shown in Fig. 3a and Supplementary Fig. S5a, 4 days after B. dothidea inoculation, fungal extension was significantly greater in the OE lines than that in the Vec lines. Compared with that of the Vec lines, the spot extension areas of the OE lines exhibited a nearly twofold increase in fungal growth (Fig. 3b).
a Representative phenotypes of Vec and MdSYP121-OE calli lines both untreated and infected with B. dothidea for 4 days, respectively. b Pathogen disease indexes in Vec and MdSYP121-OE calli lines after B. dothidea infection for 4 days. c qRT-PCR analysis for the expression of SA signalling-related genes in Vec, OE3, OE4 and OE5 calli lines after B. dothidea infection for 4 days. d qRT-PCR analysis for the expression of SA-related genes in Vec, OE3, OE4 and OE5 calli lines after B. dothidea infection for 4 days. The error bars in b, c and d indicate the mean values ± SEs of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey’s HSD test. Vec served as the empty vector control
The SA signalling-related and synthesis-related genes were also analysed in the MdSYP121-OE and Vec lines after B. dothidea infection for 4 days. As shown in Fig. 3c,d, the expression of the SA-related genes was lower in the MdSYP121-OE lines than that in the Vec lines. These results show that overexpressing MdSYP121 decreased tolerance to B. dothidea.
Silencing MdSYP121 influenced the oxidative activity of plants
To further analyse the molecular mechanism of MdSYP121 involved in B. dothidea resistance, we performed an RNA-Seq analysis of Vec and MdSYP121-RNAi calli lines under mock conditions or 4 days after treatment with B. dothidea. After trimming the adaptor sequences and removing low-quality reads, we generated 29.65 Gb of clean reads. Among these unigenes, many DEGs were identified between the Vec and RNAi samples: in the absence of B. dothidea inoculation, 637 genes were upregulated in the RNAi samples, and 720 genes were downregulated; after B. dothidea inoculation, 364 genes were upregulated and 553 genes were downregulated (>twofold changes) (Fig. 4a,b).
a The number of DEGs (twofold change cut-off) between Vec and MdSYP121-RNAi calli lines under mock conditions or in response to B. dothidea infection. b Venn diagram showing the numbers of genes whose expression was independent or dependent of B. dothidea infection. Left: the expression of genes regulated with or without B. dothidea inoculation. Right: the expression of genes upregulated with or without B. dothidea inoculation. Mock indicates the RNAi mock compared with the Vec mock; inoculation indicates RNAi with B. dothidea inoculation compared with Vec with B. dothidea inoculation. c GO analysis of DEGs independent or dependent of B. dothidea infection. Histograms of the values were generated to highlight the GO enrichment of representative GO classes. P-values for each enriched class are indicated (p–v)
To analyse functional differences between the Vec and MdSYP121-RNAi samples, the identified DEGs were characterised using GO enrichment analysis to explore their relevant biological functions. Based on the GO annotation analysis of these genes, we found that the oxidation–reduction process (GO:0055114) and oxidoreductase activity (GO:0016491) were the major enriched GO terms among the upregulated genes (Fig. 4c and Table 1). Oxidation–reduction enzymes have important functions; they can function as part of protective mechanisms involved in plant disease resistance. To test whether the MdSYP121 is related to oxidative regulation, four genes from GO:0055114 or GO:0016491 were randomly selected and verified using qRT-PCR. We confirmed that the expression of the selected genes in the MdSYP121-RNAi lines (RNAi3, RNAi5 and RNAi7) showed the same increasing tendency and that the expression was higher in those lines than in the Vec lines (Fig. 5a). We then analysed the related oxidation–reduction enzyme activity in the Vec and MdSYP121-RNAi lines. As shown in Fig. 5b, the activity of oxidation–reduction enzymes was higher in the MdSYP121-RNAi transgenic lines than that in the Vec lines both in the presence and absence of B. dothidea infection. In conclusion, these results show that MdSYP121 plays a vital role in plant disease resistance together with a set of reactive oxygen biotic stress genes.
a qRT-PCR analysis of four genes randomly selected from transcriptomic experiments following inoculation with B. dothidea for 4 days and untreated with B. dothidea in the Vec and MdSYP121-RNAi calli lines. b The activities of the antioxidant enzymes in Vec and RNAi calli lines after inoculation with B. dothidea for 4 days and in untreated controls. The error bars in a and b indicate the mean values ± SEs of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey’s HSD test
MdSYP121 interacted with MdSNAP33
Binary target membrane (t)-SNARE complexes are typically formed by SYP121 and SNAP25 at the PM during exocytosis. The Arabidopsis SNAP25 homologue, SNAP33, has been detected in the target tissue of powdery mildew fungi in leaves12. In the present study, the full length of MdSNAP33 (MDP0000242070) was cloned and analysed in apple. In Arabidopsis, AtSNAP33 is located in the PM3,31,32. To determine the actual localisation of the protein within cells, MdSNAP33 was fused to a GFP tag in the OE vector 35S::MdSNAP33-GFP. The fusion protein was transiently expressed in N. benthamiana leaves using agroinfiltration. The result showed that fluorescence was detected in the PM in N. benthamiana leaves (Fig. 6a).
a Transient expression of a 35S::MdSNAP33-GFP fusion construct in N. benthamiana. Green fluorescence was observed after transient infection for 4 days using a confocal microscope. Bars = 10 μm. In vivo interactions between MdSYP121 and MdSNAP33 were determined using BiFC. C-terminal and N-terminal fragments of YFP (indicated in the figure as YFPC and YFPN, respectively) were fused to the C-terminus of MdSYP121 and MdSNAP33, respectively. Combinations of YFPN and YFPC with the corresponding MdSYP121 and MdSNAP33 constructs were used as negative controls. Fluorescence of YFP represents protein-protein interactions. Yellow fluorescence was observed using a confocal microscope after transient infection for 4 days. Bars = 10 μm. b GST pull-down assays were performed using HA antibody. Western blot analysis results showing protein expression levels. c MdSYP121 together with MdSNAP33 forms SDS-resistant SNARE complexes. SDS-resistant MdSYP121-containing complexes are detectable in Vec and MdSYP121-OE calli lines. Proteins were extracted from Vec and MdSYP121-OE lines and incubated together with GST-MdSNAP33 protein. The proteins were loaded on a 10% polyacrylamide gel and subjected to immunoblot analysis in conjunction with anti-GST. The lower panel shows the same immunoblot protein samples that were detected with anti-HA
Interaction between MdSYP121 and MdSNAP33 was verified by bimolecular fluorescence complementation (BiFC) assays using tobacco leaves. Co-expression of MdSYP121-YFPC and MdSNAP33-YFPN produced strong signals in the PM (Fig. 6a). As negative combinations, YFPN/MdSYP121-YFPC, MdSNAP33-YFPN/YFPC and YFPN/YFPC produced no detectable fluorescence signals.
The physical interaction between MdSYP121 and MdSNAP33 was also examined by pull-down analysis. We performed MdSYP121 pull-down assays using an antibody against the HA epitope, and the presence of MdSNAP33 was assayed using an antibody against the GST epitope (Fig. 6b). Immunoblot analysis with an anti-HA antibody indicated that MdSYP121 co-immunoprecipitated by MdSNAP33. These findings indicated that MdSYP121 specifically interacts with MdSNAP33.
SYP121 forms SNARE complexes with SNAP33 adaptors and two functionally redundant VAMP72 subfamily members, VAMP721/VAMP722; these complexes are important for defence resistance in Arabidopsis12. SYP121-dependent disease resistance acts in vivo mainly via SNAP33 and VAMP721/VAMP72212. The typical features of exocytic ternary SNARE complexes are SDS resistance and heat sensitivity in animals and yeast29. We detected SDS-resistant SNARE complexes by incubating MdSYP121-HA protein extracted from apple calli together with GST-MdSNAP33 protein bound to glutathione sepharose. Sedimented bead-bound GST-MdSNAP33 and MdSYP121-HA were released and detected by immunoblot analysis using GST and HA antibodies, respectively (Fig. 6c). The GST-MdSNAP33 protein and MdSYP121-HA apple calli proteins were able to form SDS-resistant and heat-sensitive complexes. These results showed that the MdSYP121 can form a SNARE complex together with MdSNAP33.
Discussion
Many studies have reported that SYP121 is an important element in host and/or non-host resistance, and SYP121 may play a role in penetration resistance to host powdery mildew fungi (E. cichoracearum) and non-host powdery mildew fungi (Bgh)12. In the present study, we isolated and identified a Qa-SNARE group gene, MdSYP121, from Fuji apple (Supplementary Fig. S1). The results of a series of genomic, genetic and transgenic experiments suggested that MdSYP121 plays an important role in B. dothidea resistance in apple. An SNARE complex was also identified and may play an important role in pathogen resistance by affecting oxidation–reduction processes in apple following B. dothidea infection.
SNAREs proteins function as mediators of fusion between vesicular and target membranes3, and SYP121 syntaxin participates in vesicle fusion processes33. In Arabidopsis, SYP121 is a PM-localised protein10. Our analysis of the subcellular location revealed that MdSYP121 is located in the cell membrane (Fig. 1), indicating that the gene may function in membrane fusion and vesicular transport.
In plants, SA is an essential signalling molecule that induces SAR and is implicated in resistance to pathogens34. Many studies have shown that EDS1 (enhanced disease susceptibility 1), PAD4 (phytoalexin-deficient 4) and PAL (Phenylalanine ammonia-lyase) play important roles in SA biosynthesis35,36. EDS1 and PAD4 specifically promote the expression of principal SA biosynthetic enzyme gene ICS1 (Isochorismate synthase 1)35,37. PR genes and NPR1 are the marker genes of the SA signalling pathway38. The expression level of PR genes indicates the activity of SA signalling39. NPR1 is considered as a positive regulator of SA-mediated plant immune responses, and AtNPR1 is considered as a key regulator of SAR40. In the Arabidopsis syntaxin double mutant syp121-1 syp122-1, the SA level is dramatically elevated, resulting in necrosis and dwarfism. Interference of the SA signalling pathway in syp121-1 syp122-1 mutants partially rescues the necrotic and dwarfed phenotype18. In the present study, silencing MdSYP121 in ‘Gala’ apple plants lead to a necrotic and dwarfed phenotype (Supplementary Fig. S3), which is similar to the Arabidopsis syntaxin double mutant. We speculated that MdSYP121 may play a negative role in SA signalling pathway. Due to physiological and biochemical changes in cultured cells and tissues infected by pathogens41, plant tissue culture systems can be used as model systems to study the plant defence responses to pathogenic bacteria42,43. In recent years, apple calli have been used as model experimental materials to study gene function. An et al. used Agrobacterium-mediated transformation to obtain transgenic apple calli overexpressing MdHY5 to further investigate the functions of MdHY5 in apple anthocyanin accumulation and nitrate assimilation44. Wang et al. reported that the roles of MYB12 and MYB22 in apple flavonoid biosynthesis were identified by OE of the genes in apple callus22. In this study, apple calli were used to further study the function of MdSYP121 in response to B. dothidea. Based on our results, we noticed that silencing MdSYP121 could increase the disease resistance to B. dothidea, while OE of the gene decreases resistance to B. dothidea. The expression of SA synthesis-related genes (EDS1, PAD4 and PAL) and SA signalling pathway genes (PR1, PR5 and NPR1) were highly elevated in the MdSYP121-RNAi lines after B. dothidea inoculation compared with the Vec line, and they were lower in the MdSYP121-OE lines than in the Vec line (Figs. 2 and 3). The results indicated that, by regulating the biosynthesis of SA and/or the activity of SA signalling, MdSYP121 plays important roles in apple resistance to B. dothidea.
To better investigate the molecular mechanism of MdSYP121 in regulating resistance to B. dothidea, RNA-Seq was used to study the expression levels of the genes before and after inoculation. Based on the results of RNA-Seq and the qRT-PCR assays, many genes associated with biotic stress were upregulated in the RNAi lines. The oxidation–reduction process (GO:0055114) and oxidoreductase activity (GO:0016491) constituted the major enriched GO terms in the upregulated group of genes (Fig. 4c). Most of the upregulated genes, including APX (MDP0000169497), CAT (MDP0000147628), POD (MDP0000611163) and GST (MDP0000300208), encoded proteins related to the synthesis of oxidoreductase. Enzymatic antioxidants such as POD and CAT are involved in scavenging H2O2 in living cells45. PODs are α-helical heme-containing proteins and play important roles in scavenging late massive reactive oxygen species (ROS) during plant–pathogen interactions. POD and CAT have prevailing functions in basal resistance and lignification against Alternaria tenuissima and are key resistance markers in potato46. Generation of ROS is among the earliest plant defence responses to various biotic stresses. ROS can enhance the hypersensitive response (HR) or act as secondary messengers in resistance mechanisms, leading to the upregulation of defence-related genes and interactions with other signalling molecules47. Previous studies have shown that SA pathway interacts with ROS in stressed plants48. EDS1 plays an important role during oxidative stress caused by the release of singlet oxygen49. And PAD4 also involves in the integrated regulation of ROS homoeostasis50. The pathogen B. dothidea causes cankers characterised by the collapse of cells and discoloured areas; these cankers alter cellular regulatory processes and increase the production of ROS. In this study, the results of B. dothidea inoculation assays suggested that the MdSYP121-RNAi lines displayed better tolerance than the Vec lines (Fig. 2). Higher expression levels of SA pathway-related genes and higher activities of APX, CAT, POD and GST were observed in the MdSYP121-RNAi lines than those in the Vec lines (Figs. 2 and 5). Based on our results, we speculated that MdSYP121 influence disease resistance to B. dothidea by regulating the interaction of SA pathway and oxidation–reduction process.
The Arabidopsis thaliana SYP121 syntaxin resides in the PM and was previously shown to act together with its partner SNAREs, the adaptor protein SNAP33 and endomembrane-anchored VAMP721/722 to form a ternary SYP121-SNAP33-VAMP721/VAMP722 SNARE complex, which is required for in the execution of secretory immune responses against powdery mildew fungi12. The results of the analysis of the subcellular localisation of MdSYP121 and MdSNAP33 revealed that green fluorescence was detected in the cell membrane, indicating that MdSYP121 and MdSNAP33 are located in the PM and may function together in vesicular transport and membrane fusion (Figs. 1 and 6). In apple, MdSYP121 and MdSNAP33 may also function in disease resistance by forming a complex. The ternary SNARE complexes are SDS-resistant but heat sensitive12,29. The abundance of SYP121-containing SNARE complexes were examined in Vec and MdSYP121-OE lines by comparing band disappearances in response to boiling in an immunoblot solution containing anti-GST antibody (Fig. 6c). However, we detected the presence of the complex in both Vec and MdSYP121-OE lines, which is consistent with the higher levels of MdSYP121 monomers in the MdSYP121-OE lines. Previous studies have shown that the formation of SYP121-dependent ternary SNARE complexes is critical for plant pre-invasive resistance to powdery mildew fungi12. The MdSYP121 complex identified in our research may participate in resistance to B. dothidea.
In conclusion, MdSYP121 is involved in balancing penetration resistance and regulating SA-mediated defences and oxidation–reduction processes; both of these functions are important in apple ring rot resistance. The results show that MdSYP121 plays a central regulatory role in resistance against B. dothidea penetration in apple. Our results imply a potential function of MdSYP121 in apple resistance to B. dothidea. This knowledge concerning defence mechanisms involved in B. dothidea resistance could be useful in breeding programmes aiming to introduce apple genotypes that exhibit high levels of immunity against this destructive fungus. This knowledge could also promote the breeding of new cultivars that display enhanced disease resistance capabilities against this devastating disease.
References
Nebenführ, A. Vesicle traffic in the endomembrane system: a tale of COPs, Rabs and SNAREs. Curr. Opin. Plant Biol. 5, 507–512 (2002).
Carter, C. J., Bednarek, S. Y. & Raikhel, N. V. Membrane trafficking in plants: new discoveries and approaches. Curr. Opin. Plant Biol. 7, 701–707 (2004).
Lipka, V., Kwon, C. & Panstruga, R. SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 23, 147–174 (2007).
Wang, D., Weaver, N. D., Kesarwani, M. & Dong, X. Induction of protein secretory pathway is required for systemic acquired resistance. Science 308, 1036–1040 (2005).
Carr, J. P., Dixon, D. C., Nikolau, B. J., Voelkerding, K. V. & Klessig, D. F. Synthesis and localization of pathogenesis-related proteins in tobacco. Mol. Cell Biol. 7, 1580–1583 (1987).
Sanderfoot, A. Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 144, 6–17 (2007).
Fasshauer, D., Sutton, R. B., Brunger, A. T. & Jahn, R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl Acad. Sci. USA 95, 15781–15786 (1998).
Bock, J. B., Matern, H. T., Peden, A. A. & Scheller, R. H. A genomic perspective on membrane compartment organization. Nature 409, 839–841 (2001).
Jahn, R. & Scheller, R. H. SNAREs–engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 (2006).
Collins, N. C. et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977 (2003).
Kalde, M., Nühse, T. S., Findlay, K. & Peck, S. C. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl Acad. Sci. USA 104, 11850–11855 (2007).
Kwon, C. et al. Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840 (2008).
Nühse, T. S., Boller, T. & Peck, S. C. A plasma membrane syntaxin is phosphorylated in response to the bacterial elicitor flagellin. J. Biol. Chem. 278, 45248–45254 (2003).
Schmelzer, E. Cell polarization, a crucial process in fungal defence. Trends Plant Sci. 7, 411–415 (2002).
Reichardt, I. et al. Mechanisms of functional specificity among plasma-membrane syntaxins in Arabidopsis. Traffic 12, 1269–1280 (2011).
Assaad, F. F. et al. The PEN1 Syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15, 5118–5129 (2004).
Nielsen, M. E., Feechan, A., Böhlenius, H., Ueda, T. & Thordal-Christensen, H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl Acad. Sci. USA 109, 11443–11448 (2012).
Zhang, Z. et al. A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49, 302–312 (2007).
Eschen-Lippold, L. et al. Activation of defense against phytophthora infestans in potato by down-regulation of syntaxin gene expression. New Phytol. 193, 985–996 (2012).
Tang, W., Ding, Z., Zhou, Z. Q., Wang, Y. Z. & Guo, L. Y. Phylogenetic and pathogenic analyses show that the causal agent of apple ring rot in China is Botryosphaeria dothidea. Plant Dis. 96, 486–496 (2012).
Guo, L., Li, J. & Li, B. Investigations on the occurrence and chemical control of Botryosphaeria canker of apple in China. Plant Prot. 35, 120–123 (2009).
Wang, N. et al. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292 (2017).
Wang, X. et al. Isolation of high-quality RNA from Reaumuria soongorica, a desert plant rich in secondary metabolites. Mol. Biotechnol. 48, 165–172 (2011).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Tatusov, R. L. et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 4, 41 (2003).
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y. & Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280 (2004).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Li, S. J. et al. The citrus transcription factor, CitERF13, regulates citric acid accumulation via a protein-protein interaction with the vacuolar proton pump, CitVHA-c4. Sci. Rep. 6, 20151 (2016).
Hayashi, T. et al. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 13, 5051–5061 (1994).
Durrant, W. E. & Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209 (2004).
Söllner, T. et al. SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324 (1993).
Heese, M. et al. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J. Cell Biol. 155, 239–249 (2001).
Ungar, D. & Hughson, F. M. SNARE protein structure and function. Annu. Rev. Cell Dev. Biol. 19, 493–517 (2003).
Spoel, S. H. & Dong, X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3, 348–351 (2008).
Rietz, S. et al. Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from phytoalexin deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119 (2011).
Diaz-Vivancos, P., Bernal-Vicente, A., Cantabella, D., Petri, C. & Hernández, J. A. Metabolomics and biochemical approaches link salicylic acid biosynthesis to cyanogenesis in peach plants. Plant Cell Physiol. 58, 2057–2066 (2017).
Cui, H. et al. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 213, 1802–1817 (2017).
Zhang, L. et al. Cotton GhMKK5 affects disease resistance, induces HR-like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana. J. Exp. Bot. 63, 3935–3951 (2012).
Herrera-Vásquez, A., Salinas, P. & Holuigue, L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front. Plant Sci. 6, 171 (2015).
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S. & Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63 (1997).
Sircar, D. & Mitra, A. Evidence for p-hydroxybenzoate formation involving enzymatic phenylpropanoid side-chain cleavage in hairy roots of Daucus carota. J. Plant Physiol. 165, 407–414 (2008).
Kneer, R., Poulev, A. A., Olesinski, A. & Raskin, I. Characterization of the elicitor-induced biosynthesis and secretion of genistein from roots of Lupinus luteus L. J. Exp. Bot. 50, 1553–1559 (1999).
Stratmann, J., Scheer, J. & Ryan, C. A. Suramin inhibits initiation of defense signaling by systemin, chitosan, and a beta-glucan elicitor in suspension-cultured Lycopersicon peruvianum cells. Proc. Natl Acad. Sci. USA 97, 8862–8867 (2000).
An, J. P. et al. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hort. Res. 4, 17023 (2017).
Barna, B., Fodor, J., Harrach, B. D., Pogány, M. & Király, Z. The Janus face of reactive oxygen species in resistance and susceptibility of plants to necrotrophic and biotrophic pathogens. Plant Physiol. Biochem. 59, 37–43 (2012).
Taheri, P. & Kakooee, T. Reactive oxygen species accumulation and homeostasis are involved in plant immunity to an opportunistic fungal pathogen. J. Plant Physiol. 216, 152–163 (2017).
Chen, Y.-J., Lyngkjær, M. F. & Collinge, D. B. Future prospects for genetically engineering disease resistance plants. In: Molecular Plant Immunity (ed Sessa, G.) 251–275 (Wiley-Blackwell Press, 2013).
Chen, Z., Silva, H. & Klessig, D. F. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262, 1883–1886 (1993).
Ochsenbein, C. et al. The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. Plant J. 47, 445–456 (2006).
Ślesak, I. et al. Phytoalexin Deficient 4 affects reactive oxygen species metabolism, cell wall and wood properties in hybrid aspen (Populus tremula L. × tremuloides). Plant Cell Environ. 38, 1275–1284 (2015).
Acknowledgements
We thank Professors Libo Shan and Ping He at Texas A&M University for initiating this project. This work was financially supported by grants from the National Natural Science Foundation of China (No. 31272132), the Taishan Scholar Assistance Program (No. tshw20120712) from Shandong Provincial Government, China and the fund from State Key Laboratory of Crop Biology in Shandong Agricultural University (No. dxkt201713).
Authors' contributions
X.W.H., S.J.W and X.S.C. conceived and designed the experiments. X.W.H. performed most of the experiments. Y.H.H. cloned the genes. X.X.L. and Q.Q.Z. constructed the plasmids used in this article. B.H.L. provided the Botryosphaeria dothidea strain. X.W.H. analysed the data and wrote the paper. S.Q.F., X.S., S.J.W. and X.S.C. corrected the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, X., Huo, Y., Liu, X. et al. Activation of disease resistance against Botryosphaeria dothidea by downregulating the expression of MdSYP121 in apple. Hortic Res 5, 24 (2018). https://doi.org/10.1038/s41438-018-0030-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-018-0030-5
This article is cited by
-
MdWRKY74 is involved in resistance response to apple replant disease
Plant Growth Regulation (2022)
-
Application of CRISPR/Cas9 technology in wild apple (Malus sieverii) for paired sites gene editing
Plant Methods (2021)
-
Development of potent promoters that drive the efficient expression of genes in apple protoplasts
Horticulture Research (2021)
-
SNARE proteins and their role in plant ion channel regulation
Plant Growth Regulation (2020)
-
Time-series expression profiling of sugarcane leaves infected with Puccinia kuehnii reveals an ineffective defense system leading to susceptibility
Plant Cell Reports (2020)