Abstract
Sugar content is an important component of fruit quality. Although sugar transporters are known to be crucial for sugar accumulation, the role of genes encoding SWEET sugar transporters in fruit sugar accumulation remains elusive. Here we report the effect of the SWEET genes on fruit sugar accumulation in apple. A total of 25 MdSWEET genes were identified in the apple genome, and 9 were highly expressed throughout fruit development. Molecular markers of these 9 MdSWEET genes were developed and used for genotyping of 188 apple cultivars. The association of polymorphic MdSWEET genes with soluble sugar content in mature fruit was analyzed. Three genes, MdSWEET2e, MdSWEET9b, and MdSWEET15a, were significantly associated with fruit sugar content, with MdSWEET15a and MdSWEET9b accounting for a relatively large proportion of phenotypic variation in sugar content. Moreover, both MdSWEET9b and MdSWEET15a are located on chromosomal regions harboring QTLs for sugar content. Hence, MdSWEET9b and MdSWEET15a are likely candidates regulating fruit sugar accumulation in apple. Our study not only presents an efficient way of implementing gene functional study but also provides molecular tools for genetic improvement of fruit quality in apple-breeding programs.
Similar content being viewed by others
INTRODUCTION
Sugar is the main carbon source and energy-supplying substance in organisms, and it plays an important role in plant growth and development. Sugar is the main product of photosynthesis and carbon dioxide assimilation occurs mainly in stromal cells of chloroplast. The main assimilation product synthesized in chloroplasts is triose phosphate, most of which will be converted to either sucrose in the cytosol or starch in the chloroplast1,2. Sucrose is commonly translocated to other carbon-demanding organs through the long-distance transport occurring in phloem. Hence, sugar transportation is critical for maintaining the source-sink balance3.
Over the past two decades, various sugar transporters have been identified in living organisms, including plants, animals, humans, and fungi4,5,6. These transporters can be categorized into four families: sodium solute symporter transporters; major facilitator superfamily transporters; phosphotransferase system transporters; and sugar will eventually be exported transporters (SWEETs). Among these transporters, SWEETs emerge as a unique and novel class of sugar transporters. SWEETs are conserved evolutionarily and exist widely in eukaryotes, and prokaryotes such as arechaea and eubacteria. The first member of the SWEET gene family, MtN3, was identified in Medicago truncatula, and its expression was induced during root nodule development7. Later, a homolog of the MtN3 gene, designated Saliva, was identified in Drosophila and displayed salivary gland-specific expression during embryonic development8. Recently, MtN3/Saliva-type genes were functionally characterized as sugar transporters in both animals and plants, and thus gave the name “SWEET”9. SWEET proteins are characterized by the MtN3/Saliva motif (also known as the PQ-loop repeat), which comprises three alpha-helical transmembrane domains (3-TMs). Eukaryotic SWEETs consists of a tandem repeat of the basic 3-TM unit separated by a single transmembrane domain, which constitutes a 3-1-3 TM structure. In contrast, prokaryotic SWEETs, also called SemiSWEETs, contain only a single 3-TM unit, suggesting that eukaryotic SWEETs evolved through a duplication of the 3-TM unit10.
In plants, SWEETs function as bidirectional uniporters that mediate influx and efflux of sugars across cell membranes. SWEETs can be divided into four clades11. Clades I, II, and IV SWEETs transport predominantly hexoses, whilst clade III SWEETs appear to be sucrose transporters (SUTs)12,13,14,15. For example, the clade I SWEET AtSWEET1 and the clade II SWEET OsSWEET5 mediate the uptake and efflux of glucose or galactose, respectively, across the cell membrane9,16. Two clade III SWEETs, AtSWEET11 and AtSWEET12, are involved in efflux of photosynthesized sucrose from phloem parenchyma cells into intercellular space for phloem loading and long-distance translocation of sucrose17. A newly identified clade III SWEET, AtSWEET9, transports sucrose to the apoplast and plays an essential role in nectar secretion18. The clade IV SWEET, AtSWEET17, functions as a fructose-specific uniporter, with a key role in facilitating bidirectional transport of fructose across the tonoplast in leaf and root12,13. In addition, SWEETs have been shown to affect various physiological processes, such as pollen development19,20, seed filling21,22, stress and senescence14,16,23,24, modulating gibberellins response25, and host–pathogen interaction26. Usually, abiotic stress such as cold, high salinity, and drought, and biotic stress caused by fungi or bacteria result in an induction of specific SWEET genes27,28.
The accumulation of carbohydrates in storage organs such as seeds and fruits mainly depends on the supply of photoassimilates from photosynthetic tissues, especially source leaves. In most plants, sucrose is the major carbohydrate transported over a long distance in the veins to support the growth and development of storage organs6. Sugar transporter genes exhibit divergent evolutionary patterns and play important roles in sugar accumulation in plants29. For example, genes encoding SUT proteins are involved in sugar translocation toward storage organs30,31,32. Recently, the SWEET4 gene is also found to play an important role in seed filling22. In maize and rice, SWEET4 shows high expression during seed development and contributes to seed filling by enhancing the importation of hexoses into the endosperm. This study sheds light on our understanding on the mechanism by which sucrose is released from maternal tissues such as seed coat to support filial tissues such as embryonic tissue. Since sugar content is an important component of fruit quality, increasing attention has also been paid to investigate the SWEET gene family in fruit crops such as apple33, grapevine34, and banana35. However, the effect of SWEETs on fruit sugar accumulation remains elusive.
The domesticated apple, Malus x domestica Borkh., is an economically important fruit crop worldwide. Apple belongs to the family Rosaceae, and the cultivated apple is a diploidized autopolyploid species with a basic chromosome number of x = 17. The draft genome sequence of the domesticated apple has been released, and accounts for an approximately 750 Mb per haploid36. In this study, we report the identification of the MdSWEET genes with high expression during apple fruit development. DNA markers for these MdSWEETs were developed to investigate their association with fruit sugar content. Our study not only aids our better understanding of the effect of SWEETs on fruit sugar accumulation but it will also be helpful for genetic improvement of fruit sugar accumulation in apple-breeding programs.
MATERIALS AND METHODS
Plant material
All 188 apple cultivars (Table S1) used in this study that show a great variation in fruit sweetness are maintained at Xingcheng Institute of Pomology of the Chinese Academy of Agricultural Sciences, Xingcheng, Liaoning, China. Young leaves used for genomic DNA extraction were collected in the spring of 2015. Leaf samples were immediately frozen in liquid nitrogen, and then stored at −80 °C until use. Fruits at mature stage were randomly collected in 2015 and fruit maturity was comprehensively estimated based on skin background color and blush development, seed color turning into brown, and the previous records of maturity date. Each cultivar had three replicates, consisting of nine fruits. Fruit samples were cut into small pieces, immediately frozen in liquid nitrogen, and then stored either at −40 °C for sugar measurement or at −80 °C for real-time PCR (RT-PCR) analysis.
Measurement of soluble sugar content
The content of soluble sugar components was measured using high-performance liquid chromatography (HPLC) according to our previous report37. Briefly, fruit samples were ground into fine powder in liquid nitrogen using an A11 basic Analytical mill (IKA, Germany). One gram of powder was dissolved in 6 ml sterilized deionized water, the mixture was extracted in an ultrasonic bath for 30 min, and then centrifuged at 6000 r/s for 15 min. The supernatant was collected, purified using a SEPC18 syringe (Supelclean ENVI C18 SPE), and subsequently filtered through a 0.22 μm Sep-Pak filter. The filtered supernatant was used for sugar content measurement using a Dionex P680 HPLC system (Dionex Corporation, CA, USA) equipped with a refractive index detector (Shodex RI-101; Shodex Munich, Germany). The separation was performed on a Transgenomic COREGET-87C column (7.8 mm × 300 mm, 10 μm) together with a guard column Transgenomic CARB Sep Coregel 87C. The column temperature was maintained at 85 °C by using a Dionex TCC-100 thermostated column compartment. The mobile phase was set at a flow rate of 0.6 ml/min with degassed, distilled, deionized water. Peak areas were integrated with the Chromeleon chromatography data system according to external standard solution calibrations (reagents from Sigma Chemical Co., Castle Hill, NSW, Australia). Sugar concentrations were expressed on a fresh weight (FW) basis, and total sugar content was indicated by the amount of four sugars found in apple fruit, i.e., glucose, sucrose, fructose, and sorbitol. In addition, the measurement of soluble solid content (SSC) was conducted using a pocket refractometer (Atago, Tokyo, Japan).
Identification of the SWEET genes in apple and their phylogenetic analysis
Coding DNA sequences of the SWEET gene family in Arabidopsis thaliana were retrieved from the Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/). These coding sequences were used as query sequences to compare against the apple genome sequence database (GDDH13 V1.1, https://www.rosaceae.org/blast/) by BlastX with a cutoff E-value of 1.00E-10. The homologs of Arabidopsis SWEET genes in the apple genome were named according to their phylogenetic relationships to the founding members of the family in Arabidopsis9. Chromosome lengths and gene locations were presented according to the draft genome sequence of the doubled haploid (GDDH13 V1.1)38.
Multiple alignment of amino-acid sequences of SWEET genes in Arabidopsis and apple was conducted using the integrated MUSCLE alignment program in MEGA5 (Molecular Evolutionary GenetiMd Analysis) with default parameters39. The resulting data matrix was analyzed using the Neighbor-Joining method. The bootstrap consensus tree was inferred from 1000 replicates and the bootstrap values <50% were collapsed.
RNA isolation and quantitative RT-PCR
Two apple cultivars, K9 and Shizishan 2, were randomly selected for quantitative RT-PCR (qRT-PCR) analysis. Fruit samples were collected at 30, 60, and 90 days after full bloom. Each cultivar had three biological replicates, containing of nine fruits. Fruits of each replicate were cut into small pieces, mixed, and used for total RNA extraction. RNA extraction was conducted using RNA prep Pure Plant Kit (TianGen, Beijing, China) according to the manufacturer’s instructions. RNA concentration and quantity were detected and assessed with NanoDrop2000 (Thermo Scientific).
Approximately 1 µg of total RNA was used to synthesize the first strand of cDNA using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TRANS, Beijing, China) following the manufacturer’s instructions. qRT-PCR was performed in 20 µL reaction containing 1× SYBR Green II Master Mix (Takara, Dalian, China), 0.2 µM of each primer, and 0.5 µL of template cDNA. The qRT-PCR amplifications were performed using the Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, USA), and the reaction program was set as follows: 95 °C for 1 min, one cycle; and 95 °C for 5 s, 60 °C for 34 s, 40 cycles. Melting curve analysis was performed at the end of 40 cycles by heating from 55 to 95 °C at a rate of 0.5 °C/s. An apple polymer ubiquitin enzyme gene (UBQ) was selected as a constitute control40. The relative expression level of all detected genes was calculated according to the cycle threshold 2−ΔΔCT method. All analyses were performed in triplicates. The primer sequences are listed in Table S2.
Development of gene-tagged simple sequence repeat and cleaved amplified polymorphism sequence markers
Simple sequence repeat (SSR) or cleaved amplified polymorphism sequence (CAPS) markers were development for SWEETs that showed high expression in fruit of apple. SSR Hunter software and dCAPS Finder 2.0 (http://helix.wustl.edu/dcaps/dcaps.html) were used to screen SSRs with ≥7 repeats or CAPS markers, respectively, in genomic DNA sequences of each SWEET gene, including 2 kb upstream of the start codon, the entire coding region, and 2 kb downstream of the translation stop codon. Primers flanking the SSR and CAPS loci were designed using the Primer 5 program, and polymorphism of the SSR and CAPS markers was evaluated using four apple cultivars with high sugar content and four cultivars with low sugar content.
PCR amplification was performed using the GeneAmp PCR System 9700 (ABI, USA) with the following condition: 3 min at 94 °C, followed by 35 cycles consisting of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and with a final extension of 72 °C for 7 min. For the SSR markers, 3 μL of amplification products was mixed with an equal volume of formamide loading buffer (98 % formamide, 10 mM EDTA, pH 8.0, 0.025% bromophenol blue, and xylene cyanol). The mixture was denatured at 95 °C for 5 min, and then immediately put on ice for 5 min. An aliquot of 2 μL mixture was on an 8% polyacrylamide gel and electrophoresed for 1–1.5 h at 1000 V. DNA bands were visualized after silver staining, and their sizes were estimated on the basis of a standard 25 bp DNA ladder. For the CAPS markers, amplification products were digested with restriction enzymes and then separated on 2% agarose gel. DNA bands in agrose gel were visualized under ultraviolet light after staining with ethidium bromide.
Statistical analysis
A total of 188 apple cultivars were subjected to statistical analysis. Each cultivar was genotyped for the SWEET loci following analysis of molecular marker profiles. The detection of association between molecular markers and fruit sugar accumulation was performed with the software package TASSEL version 3.0 according to our previous report41. The criterion for marker–trait association was set at P ≤ 0.01. Fisher’s least significant difference at P < 0.01 was used to compare mean soluble sugar contents between cultivars.
RESULTS
The SWEETs gene family in the apple genome
A total of 25 MdSWEET genes were identified in the apple genome. Of the 25 MdSWEET genes, 24 were located on five homologous pairs of chromosomes (3–11, 5–10, 4–12, 6–14, 13–16), and 1 on chromosome 17 (Fig. 1a). Genomic structural analysis showed that the majority of MdSWEET genes consisted of six exons, while 4 MdSWEET genes, MdSWEET5b, MdSWEET7a, MdSWEET7b, and MdSWEET11, contained five exons (Fig. 1b and Table S3). The open reading frames of the MdSWEET genes ranged from 645 to 1020 bp in length and their deduced proteins ranging from 215 to 340 amino acids in length (Table S3). The conserved domain prediction indicated that 21 MdSWEET genes had seven alpha-helical TMs. By contrast, 3 MdSWEET genes, MdSWEET5a, MdSWEET9a, and MdSWEET5b, had six TMs, with absence of the TM7 domain (Table S3). Interestingly, the remaining MdSWEET11 gene had eight TMs. In addition, it is worth noting that an additional SWEET gene (GDR accession no. MD01G1215700) was also found in the apple genome. This SWEET gene was located on chromosome 1, with four exons (Fig. 1b), contained only one MtN3/Saliva motif, and showed extremely low expression throughout fruit development. Hence, this SWEET gene was deemed a pseudogene and was not included in the later analysis.
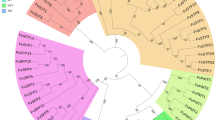
Phylogenetic analysis revealed that all the MdSWEET genes were divided into four clades as previously defined by Chen et al.9, and each MdSWEET gene was named following its homologous genes in Arabidopsis (Fig. 2). Clade I, II, III, and IV contained 10, 5, 9, and 1 MdSWEET genes, respectively. The MdSWEET2 genes had nine homologs, MdSWEET2a, MdSWEET2b, MdSWEET2c, MdSWEET2d, MdSWEET2e, MdSWEET2f, MdSWEET2g, MdSWEET2h, and MdSWEET2i, while only one copy was observed for MdSWEET1, MdSWEET8, MdSWEET11, and MdSWEET17. The remaining MdSWEET genes, MdSWEET2, MdSWEET5, MdSWEET7, MdSWEET9, MdSWEET10, MdSWEET12, and MdSWEET15 contained two homologs.
Expression profiling of MdSWEET genes in fruit at different developmental stages
To identify SWEET genes that are potentially involved in fruit sugar accumulation, we investigated the expression profiling of the MdSWEET genes in fruits of two cultivars, K9 and Shishan 2, at different developmental stages, including juvenile, expanding, and mature stages (Fig. 3). Of the 25 MdSWEET genes, 16 showed extremely low expression throughout fruit development, with relative expression levels ranging from 0 to 0.1. In contrast, 9 MdSWEET genes, MdSWEET2a, MdSWEET2b, MdSWEET2d, MdSWEET2e, MdSWEET7a, MdSWEET7b, MdSWEET9b, MdSWEET12a, and MdSWEET15a, were highly expressed in fruits at all the three stages tested. Hence, these 9 MdSWEET genes were assumed to be potential candidates related to fruit sugar accumulation, and were further subjected to develop gene-tagged markers for evaluation of their genetic association with fruit sugar content.
The heat map was constructed based on relative expression levels of the MdSWEETs. S1, S2, and S3 represent young fruitlets, expanding fruits, and mature fruits, which were collected 30, 60, and 90 days after full bloom, respectively. The MdSWEET genes showing high expression throughout fruit development are highlighted in red color. The different colors represent different levels of gene expression, with the red color representing the highest values of gene expression, followed by yellow, gray, light green, and dark green
Development of gene-tagged markers for SWEETs with high expression in fruit and their polymorphisms in a collection of apple cultivars
Two types of molecular markers, SSR and CAPS, were developed for the nine MdSWEET genes mentioned above (Table 1). The SSR markers for two SWEET genes, MdSWEET7a and MdSWEET9b, were developed based on a (CT) n microsatellite located in the second intron, whereas dinucleotide microsatellites, including (AG) n , (GA) n , (AT) n , (CT) n , and (TA) n , located upstream of the start codon were used to develop SSR markers for six SWEET genes, MdSWEET2a, MdSWEET2b, MdSWEET2d, MdSWEET2e, MdSWEET7b, and MdSWEET12a (Table 1). A T/C single-nucleotide polymorphism (SNP) in the first intron of MdSWEET15a was successfully used to develop a CAPS marker. PCR products harboring a “C” nucleotide at the polymorphic site could be digested with the NdeI enzyme, producing two fragments of 438 and 203 bp in size, while no digestion for PCR products harboring a “T” nucleotide.
These nine gene-tagged markers were subsequently used to screen a collection of 188 cultivars (Fig. 4). As a result, two alleles at each polymorphic locus were detected for three MdSWEET genes, MdSWEET2a, MdSWEET12a, and MdSWEET7a, while three alleles at each polymorphic locus were observed for five MdSWEET genes, MdSWEET2b, MdSWEET2d, MdSWEET2e, MdSWEET7b, and MdSWEET9b. Six genotypes derived from the random combination of three alleles were identified at each of the four gene loci, including MdSWEET2b, MdSWEET2d, MdSWEET2e, and MdSWEET7b, while only four genotypes were detected at the MdSWEET9b locus (Table S4). Three genotypes at each polymorphic locus were identified for three MdSWEET genes, MdSWEET2a, MdSWEET7a, and MdSWEET12a. By contrast, the CAPS marker revealed only two genotypes, T/T and T/C, and the homologous genotype C/C was not identified in all the cultivars tested.
a–h Polymorphic bands of SSR markers detected at the MdSWEET2a, MdSWEET2b, MdSWEET2d, MdSWEET2e, MdSWEET7a, MdSWEET7b, MdSWEET9b, and MdSWEET12a loci, respectively. i An agarose gel electrophoresis of PCR products derived from the first intron of MdSWEET15a, which are digested with the NdeI enzyme. Polymerase chain reactions products containing CATATG sequences can be digested, whereas PCR products with TATATG sequences cannot be digested. 1: Laidi; 2: Fulaibao; 3: Zaoshengchi; 4: Qingxiangjiao; 5: Beinaoni; 6: Fujin; 7: Qingguan; 8: Xingcheng 2310; 9: Bankeluofute; 10: Senmalan; 11: Yeweilin; 12: Kangbingjinguan 51; 13: Zhanxuan 14; 14: Xingcheng 1518; 15: Zhongxin; 16: Zhanxuan 4; 17: Hebeikangbingjinguan; 18: Jinguang; 19: Lisijin; 20: Tianhongyu
Association between MdSWEET genes and fruit sugar accumulation in apple
SSC and soluble sugar content in mature fruit for all the tested cultivars are listed in Table S1 and their distributions are shown in Fig. 5. SSC and the contents of sucrose, fructose, glucose, and total sugar components showed a normal distribution, whereas the distribution of sorbitol content was skewed toward low sorbitol contents. A wide range was observed for the concentration of various sugar components, including sucrose (3.8–41.75 mg/g FW), glucose (3.86–31.80 mg/g FW), fructose (30.48–83.82 mg/g FW), sorbitol (0.14–22.62 mg/g FW), and total sugar components (47.93–142.40 mg/g FW), with an average of 21.65, 14.98, 49.07, 3.90, and 89.60 mg/g FW, respectively. SSC ranged from 7.96 to 20.02, with an average of 12.76. Overall, the apple cultivar collection has a great variation in fruit sugar content, which suggests that they are suitable for investigating genetic association of MdSWEET genes with fruit sugar accumulation.
Candidate gene-based association mapping was further performed to investigate association between the polymorphic loci of MdSWEET genes and fruit sugar accumulation in apple. As a result, three genes, MdSWEET2e, MdSWEET9b, and MdSWEET15a, showed a significant association with fruit sugar accumulation, whereas no significant association was observed for the remaining six genes, MdSWEET2a, MdSWEET2b, MdSWEET2d, MdSWEET7a, MdSWEET7b, and MdSWEET12a (Table 2). Based on the presence or absence of the (AT)13 allele on the MdSWEET2e locus, all the tested cultivars were grouped into three genotypes, (AT)13/13, (AT)13/7 or 17, and (AT)7 or 17/7 or 17. The (AT)13/13 genotype had significantly higher than both (AT)13/7 or 17 and (AT)7 or 17/7 or 17 genotypes for the sucrose, fructose, or total sugar content (Fig. 6). Similarly, all the cultivars were divided into three genotypes, (CT)19/19, (CT)19/23, and (CT)23/23 or 26, based on the presence or absence of the (CT)19 allele on the MdSWEET9b locus. Cultivars with two (CT)19 alleles had significantly higher than cultivars with one or no (CT)19 allele for the fructose or total sugar content. Based on the polymorphic locus of MdSWEET15a, all the cultivars were assigned to two genotypes, T/T and T/C. The T/T genotype had significantly higher than the T/C genotype for SSC and the glucose, sorbitol, or total sugar content. However, the (AT)13 allele of MdSWEET2e and the (CT)19 allele of MdSWEET9b had no effect on the soluble solids, glucose, or sorbitol content and the soluble solids, sucrose, glucose, or sorbitol content, respectively (data not shown). Similarly, the polymorphic locus of MdSWEET15a had no effect on accumulation of sucrose and fructose.
Of the above three genes associated with fruit sugar accumulation, MdSWEET15a had relatively higher contributions to the observed phenotypic variation, and accounted for 6.4%, 6.8%, 5.7%, and 8.4% of the observed phenotypic variation in the soluble solids, glucose, sorbitol and total sugar content, respectively. The MdSWEET9b gene accounted for 6.6% and 2.5% of the observed phenotypic variation in the fructose and total sugar content, respectively. By contrast, MdSWEET2e had lower contribution, accounting for 0.7%, 2.7%, and 3.6% of the observed phenotypic variation in the sucrose, fructose, and total sugar content, respectively.
Taken together, all the above results suggest that one clade II SWEET gene, MdSWEET2e, and two clade III SWEET genes, MdSWEET9b and MdSWEET15a, are genetically associated with sugar content in mature apple fruit, with MdSWEET15a and MdSWEET9b showing relatively higher contribution.
DISCUSSION
Sugar transporters play a crucial role in plant growth and development as they mediate sugar uptake or release from cells or subcellular compartments21. SWEETs are newly identified sugar transporters and they are critical for sugar transportation and sink tissue development17,22,42. Although preliminary analyses of the SWEET gene family have been reported in several fruit crops33,34,35, little is known about the role of SWEETs in fruit sugar accumulation. In this study, we report for the first time genetic association of the SWEET genes with fruit sugar content in apple. Our study indicates that developing gene-tagged markers is an efficient way to investigate gene functionality, and the gene-tagged markers can be directly used in breeding programs if they are associated with horticultural traits of interest.
Duplication of SWEETs in the apple genome
Gene duplication is a major driving force for recruitment of genes in plants. Apple is diploid, with an autopolyploidization origin36,43. Here our study reveals 25 SWEETs in the apple genome, which is inconsistent with a previous report that identifies a total of 29 apple SWEETs33. This inconsistency is likely due to the fact that our analysis is based on of the draft genome sequence of “Golden delicious” doubled-haploid tree (GDDH13 V1.1)38, while the genome sequence of apple cv. Golden Delicious36 was used in the study reported by Wei et al33. All the MdSWEET genes except MdSWEET8 are located on homologous pairs of chromosomes. For example, chromosomes 6 and 14 are homologous pairs and both contain one copy of each of the four SWEET genes, MdSWEET7, MdSWEET12, MdSWEET10, and MdSWEET5. The MdSWEET9 gene has two homologs, which are separately located on the bottom of two homologously paired chromosomes 4 and 12. Chromosomes 5 and 10 are homologous pairs and both contain four MdSWEET genes. These results unambiguously demonstrate that duplication of SWEET genes is related to whole-genome duplication (WGD).
Chromosome 3 is homologous to chromosome 11. Chromosome 3 contains a single SWEET gene MdSWEET2a on the bottom chromosome, whilst a cluster of two SWEET2 genes, MdSWEET2b and MdSWEET11, are found on the bottom of chromosome 11. Similarly, chromosomes 13 and 16 are homologous pairs. A cluster of two SWEET2 genes, MdSWEET15b and MdSWEET17, are found on the top of chromosome 13, whilst only single SWEET gene MdSWEET15a located on the homologous region of chromosome 16. These results also suggest that tandem duplication of SWEET genes has probably occurred on chromosomes 11 and 13. In addition, it is worth noting that one SWEET gene, MdSWEET8, is located on the bottom of chromosome 17, but no SWEET gene was found on chromosome 9 that is homologous to chromosome 17. Since duplicated gene copies following WGD are prone to be rapidly lost44, it is reasonable to speculate that a SWEET gene on chromosome 9 may have been lost in the ancestor of apple.
Most WGD-derived duplicated genes are prone to diverge in expression45. This case is also detected for the apple SWEET genes. For example, two SWEET homologs, MdSWEET15a and MdSWEET15b, which are located on homologous pair of chromosomes 16 and 13, respectively, have undergone divergence in expression. MdSWEET15a is highly expressed throughout fruit development, whilst MdSWEET15b shows no expression in fruit. Similarly, MdSWEET9a and MdSWEET9b are located on homologous pair of chromosomes 12 and 4, respectively. MdSWEET9b shows high expression throughout fruit development, but MdSWEET9a with no expression in fruit. In addition, expression divergence was also observed for tandem duplicated SWEET genes. For example, MdSWEET2b and MdSWEET11 are clustered on the bottom of chromosome 11. MdSWEET2b is highly expressed throughout fruit development, but MdSWEET11 shows no expression in fruit. However, it is unclear whether or not the remaining 16 MdSWEET genes with extremely low expression in fruit have also diverged in expression.
Taken together, all the results above suggest that the SWEET genes in apple have undergone polyploidization and/or segmental duplication during the process of speciation, and some duplicated SWEET genes have diverged in expression.
Candidate MdSWEETs involved in the regulation of fruit sugar accumulation in apple
Measurement of soluble sugar content in mature fruits of 188 apple cultivars reveals that the average concentration of fructose is over twofold higher than those of sucrose, glucose, and sorbitol, which confirms our previous report of fructose being the major sugar component in mature apple fruit37. Genotyping of the apple cultivar collection using nine gene-tagged molecular markers further indicates that three MdSWEET genes, MdSWEET2e, MdSWEET15a, and MdSWEET9b, are associated with fruit sugar accumulation. Six genotypes produced by three alleles at single polymorphic locus were detected for four SWEET genes, MdSWEET7b, MdSWEET2d, MdSWEET2b, and MdSWEET2e, and three genotypes arisen from two alleles at single polymorphic locus were observed for three SWEET genes, MdSWEET12a, MdSWEET2a, and MdSWEET7a. In contrast, the (CT)26/26 and (CT)19/26 genotypes and the C/C genotype were not found at the MdSWEET9b or MdSWEET15a loci, respectively, in the apple cultivar collection. Since selection is well-known to be a directional process that results in changes in the frequency of various genotypes in the population, the MdSWEET15a and MdSWEET9b loci have probably undergone selection during the process of apple domestication.
Linkage mapping of quantitative trait loci (QTLs) for sugar content have revealed many QTLs with minor effects on all linkage groups (LGs) except LG7 and LG1746,47,48,49. MdSWEET15a is located on the region of LG16, which contains several QTLs for Brix and the amount of sorbitol and fructose48,50. Similarly, MdSWEET9b is located on the region of LG4 harboring a QTL for individual sugar content49. By contrast, MdSWEET2e is located far away (approximately 11.2 Mb) from the region of LG10 harboring a QTL for Brix and sucrose content. In addition, MdSWEET2e has a relative smaller contribution to phenotypic variation in sugar content compared with both MdSWEET15a and MdSWEET9b.
Taken together, all these results above suggest that MdSWEET15a and MdSWEET9b are likely candidates involved in sugar accumulation in apple fruit. In peach, two major QTLs for fruit sugar content have been reported on the top of LG4 and the bottom of LG5, respectively51. Interestingly, these two QTL regions both contain SWEET genes, based on the draft genome sequence of peach cv. Lovell50. The two QTL regions on peach LG4 and LG5 correspond to syntenic blocks on apple LG3 and LG6, respectively, which harbor QTLs for sugar content52. Thus, it seems that SWEET genes may also function as candidates for sugar accumulation in peach and other Rosaceae fruit crops.
Both MdSWEET9b and MdSWEET15a belong to the clade III SWEETs that are proved to be efficient SUTs11. In this study, the cultivars with the T/C genotype at the MdSWEET15a locus have higher average sucrose content in mature fruit compared with those with the T/T genotype. Similarly, the cultivars with two (CT)19 alleles have higher average sucrose content in mature fruit than those with one or no (CT)19 allele. Thus, the clade III SWEET genes in apple, similar to clade III SWEETs in Arabidopsis9, may be involved in sucrose transportation. However, the difference in average sucrose content between various genotypes in the MdSWEET9b or MdSWEET15a loci does not reach a significance level (P < 0.01). This might be partially due to the reason that sucrose in the vacuolar has a trend to conversion into hexoses, resulting in fructose being the major sugar component in mature apple fruit37.
Besides MdSWEET15a and MdSWEET9b, two additional SWEET genes, MdSWEET2a and MdSWEET2b, are also located on the regions of LGs 3 and 11, respectively, which contain QTLs for fruit sorbitol content in apple53. However, these two MdSWEET genes both have no significant association with sorbitol content. This could be attributed to certain process of sugar metabolism in fruit, where sorbitol unloaded from leaves into the fruit is converted to fructose or glucose54. It is worth noting that gene-tagged markers developed in this study may not correspond to functional variants that account for associations with sugar content. Thus, further studies are still needed to ascertain whether MdSWEET2a and MdSWEET2b have an influence on fruit sugar accumulation in apple.
Marker-assisted selection (MAS) is a valuable tool in breeding programs of plants, particularly fruit crops. In this study, a T/C SNP in the first intron of MdSWEET15a accounts for 6–8% of phenotypic variation for SSC (6.4%) and total sugar content (8.4%) among apple germplasm, while a (CT) n SSR locus in the second intron of MdSWEET9b explains approximately 7% of phenotypic variation for the concentration of fructose, the major sugar component in apple fruit. Since these two gene-tagged makers account for considerable phenotypic variation, they can serve as efficient tools for genetic improvement of fruit sweetness in apple-breeding programs with MAS.
In summary, our study suggests that both MdSWEET9b and MdSWEET15a are likely candidates regulating fruit sugar accumulation in apple. It is worthy of study in the future to clarify the functions of MdSWEET9b and MdSWEET15a.
References
Kuhn, C., Franceschi, V. R., Schulz, A., Lemoine, R. & Frommer, W. B. Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275, 1298–1300 (1997).
Doerflinger, F. C., Miller, W. B., Nock, J. F. & Watkins, C. B. Variations in zonal fruit starch concentrations of apples - a developmental phenomenon or an indication of ripening? Hortic. Res-Engl. 2, 47 (2015).
Gottwald, J. R., Krysan, P. J., Young, J. C., Evert, R. F. & Sussman, M. R. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl Acad. Sci. USA 97, 13979–13984 (2000).
Walgren, R. A., Lin, J. T., Kinne, R. K. H. & Walle, T. Cellular uptake of dietary flavonoid quercetin 4 ‘-beta-glucoside by sodium-dependent glucose transporter SGLT1. J. Pharmacol. Exp. Ther. 294, 837–843 (2000).
Severi, E., Hosie, A. H. F., Hawkhead, J. A. & Thomas, G. H. Characterization of a novel sialic acid transporter of the sodium solute symporter (SSS) family and in vivo comparison with known bacterial sialic acid transporters. FEMS Microbiol. Lett. 304, 47–54 (2010).
Baker, R. F., Leach, K. A. & Braun, D. M. SWEET as sugar: new sucrose effluxers in plants. Mol. Plant 5, 766–768 (2012).
Gamas, P., de Carvalho-Niebel, F., Lescure, N. & Cullimore, J. V. Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol. Plant Microbe Interact. 9, 233–242 (1996).
Artero, R. D. et al. Saliva, a new drosophila gene expressed in the embryonic salivary glands with homologues in plants and vertebrates. Mech. Dev. 75, 159–162 (1998).
Chen, L. Q. et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–U199 (2010).
Xuan, Y. H. et al. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl Acad. Sci. USA 110, E3685–E3694 (2013).
Eom, J. S. et al. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 25, 53–62 (2015).
Chardon, F. et al. Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr. Biol. 23, 697–702 (2013).
Guo, W. J. et al. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis Roots and Leaves. Plant Physiol. 164, 777–789 (2014).
Klemens, P. A. W. et al. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 163, 1338–1352 (2013).
Chen, L. Q., Cheung, L. S., Feng, L., Tanner, W. & Frommer, W. B. Transport of sugars. Annu. Rev. Biochem. 84, 865–894 (2015).
Zhou, Y. et al. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS ONE 9, e94210 (2014).
Chen, L. Q. et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211 (2012).
Lin, I. W. et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508, 546–549 (2014).
Guan, Y. F. et al. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 147, 852–863 (2008).
Sun, M. X., Huang, X. Y., Yang, J., Guan, Y. F. & Yang, Z. N. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 26, 83–91 (2013).
Chen, L. Q. et al. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27, 607–619 (2015).
Sosso, D. et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 47, 1489–1493 (2015).
Seo, P. J., Park, J. M., Kang, S. K., Kim, S. G. & Park, C. M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 233, 189–200 (2011).
Le Hir, R. et al. Disruption of the sugar transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Mol. Plant 8, 1687–1690 (2015).
Kanno, Y. et al. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 7, 13245 (2016).
Chen, L. Q. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 201, 1150–1155 (2014).
Verdier, V. et al. Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol. 196, 1197–1207 (2012).
Chong, J. et al. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 65, 6589–6601 (2014).
Wang, W. et al. Divergent evolutionary pattern of sugar transporter genes is associated with the difference in sugar accumulation between grasses and eudicots. Sci. Rep. 6, 29153 (2016).
Baud, S. et al. The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J. 43, 824–836 (2005).
Hackel, A. et al. Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J. 45, 180–192 (2006).
Zhang, W. H. et al. Nutrient loading of developing seeds. Funct. Plant. Biol. 34, 314–331 (2007).
Wei, X. Y., Liu, F. L., Chen, C., Ma, F. W. & Li, M. J. The Malus domestica sugar transporter gene family: identifications based on genome and expression prof ling related to the accumulation of fruit sugars. Front. Plant Sci. 5, 569 (2014).
Lecourieux, F. et al. An update on sugar transport and signalling in grapevine. J. Exp. Bot. 65, 821–832 (2014).
Miao, H. X. et al. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci. Rep. 7, 3536 (2017).
Velasco, R. et al. The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42, 833–839 (2010).
Ma, B. Q. et al. Comparative assessment of sugar and malic acid composition in cultivated and wild apples. Food Chem. 172, 86–91 (2015).
Daccord, N. et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49, 3886 (2017).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Fang, T. et al. Variation of ascorbic acid concentration in fruits of cultivated and wild apples. Food Chem. 225, 132–137 (2017).
Ma, B. Q. et al. Genes encoding aluminum-activated malate transporter II and their association with fruit acidity in apple. Plant Genome 8, 3 (2015).
Patil, G. et al. Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics 16, 520 (2015).
Han, Y. P. et al. Integration of physical and genetic maps in apple confirms whole-genome and segmental duplications in the apple genome. J. Exp. Bot. 62, 5117–5130 (2011).
Maere, S. et al. Modeling gene and genome duplications in eukaryotes. Proc. Natl Acad. Sci. USA 102, 5454–5459 (2005).
Blanc, G. & Wolfe, K. H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691 (2004).
Liebhard, R., Kellerhals, M., Pfammatter, W., Jertmini, M. & Gessler, C. Mapping quantitative physiological traits in apple (Malus x domestica Borkh.). Plant Mol. Biol. 52, 511–526 (2003).
Kenis, K., Keulemans, J. & Davey, M. W. Identification and stability of QTLs for fruit quality traits in apple. Tree Genet. Genomes 4, 647–661 (2008).
Kunihisa, M. et al. Identification of QTLs for fruit quality traits in Japanese apples: QTLs for early ripening are tightly related to preharvest fruit drop. Breed. Sci. 64, 240–251 (2014).
Ma, B. Q. et al. Construction of a high density linkage map and its application in the identification of QTLs for soluble sugar and organic acid components in apple. Tree Genet. Genomes 12, 1 (2016).
Verde, I. et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45, 487–U447 (2013).
Zeballos, J. L. et al. Mapping QTLs associated with fruit quality traits in peach [Prunus persica (L.) Batsch] using SNP maps. Tree Genet. Genomes 12, 37 (2016).
Illa, E. et al. Comparative analysis of rosaceous genomes and the reconstruction of a putative ancestral genome for the family. BMC Evol. Biol. 11, 9 (2011).
Guan, Y. Z., Peace, C., Rudell, D., Verma, S. & Evans, K. QTLs detected for individual sugars and soluble solids content in apple. Mol. Breed. 35, 135 (2015).
Yamaki, S. Metabolism and accumulation of sugars translocated to fruit and their regulation. J. Jpn Soc. Hortic. Sci. 79, 1–15 (2010).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31420103914) and the Overseas Construction Plan for Science and Education Base, China-Africa Center for Research and Education, Chinese Academy of Sciences (Grant No. SAJC201327).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhen, Q., Fang, T., Peng, Q. et al. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic Res 5, 14 (2018). https://doi.org/10.1038/s41438-018-0024-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-018-0024-3
This article is cited by
-
Genomic survey and expression analysis of LcARFs reveal multiple functions to somatic embryogenesis in Liriodendron
BMC Plant Biology (2024)
-
The SWEET gene family in watermelon: genome-wide identification, phylogeny, duplication and expression analyses in male sterile buds and under sucrose, fructose and glucose treatments
Biologia (2023)
-
Decoding Sugar Regulation and Homeostasis in Plants: Cracking Functional Roles Under Stresses
Journal of Plant Growth Regulation (2023)
-
Identification and expression analysis of the SWEET genes in radish reveal their potential functions in reproductive organ development
Molecular Biology Reports (2023)
-
Exploring genetic diversity and ascertaining genetic loci associated with important fruit quality traits in apple (Malus × domestica Borkh.)
Physiology and Molecular Biology of Plants (2023)