Abstract
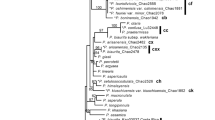
Repeated homoploid hybrid speciation (HHS) events with the same parental species have rarely been reported. In this study, we used population transcriptome data to test paraphyly and HHS events in the conifer Picea brachytyla. Our analyses revealed non-sister relationships for two lineages of P. brachytyla, with the southern lineage being placed within the re-circumscribed P. likiangensis species complex (PLSC) and P. brachytyla sensu stricto (s.s.) consisted solely of the northern lineage, forming a distinct clade that is paratactic to both the PLSC and P. wilsonii. Our phylogenetic and coalescent analyses suggested that P. brachytyla s.s. arose from HHS between the ancestor of the PLSC before its diversification and P. wilsonii through an intermediate hybrid lineage at an early stage and backcrossing to the ancestral PLSC. Additionally, P. purpurea shares the same parents and an extinct lineage with P. brachytyla s.s. but backcrossing to the other parent, P. wilsonii at a later stage. We reveal the first case that backcrossing to different parents of the same extinct hybrid lineage produced two different hybrid species. Our results highlight the existence of more reticulate evolution during species diversification in the spruce genus and more complex homoploid hybrid events than previously identified.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The sequencing data have been deposited in National Genomics Data Center under the BioProject ID: PRJCA003239.
References
Alexander DH, Lange K (2011) Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinforma 12:246. https://doi.org/10.1186/1471-2105-12-246
Anderegg WRL, Flint A, Huang C-Y, Flint L, Berry JA, Davis FW et al. (2015) Tree mortality predicted from drought-induced vascular damage. Nat Geosci 8:367–371
Battipaglia G, DeMicco V, Brand WA, Saurer M, Aronne G, Linke P et al. (2013) Drought impact on water use efficiency and intra-annual density fluctuations in Erica arborea on Elba (Italy). Plant Cell Environ 37:382–391
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bouille M, Senneville S, Bousquet J (2011) Discordant mtDNA and cpDNA phylogenies indicate geographic speciation and reticulation as driving factors for the diversification of the genus Picea. Tree Genet Genomes 7:469–484
Brennan AC, Hiscock SJ, Abbott RJ (2019) Completing the hybridization triangle, the inheritance of genetic incompatibilities during homoploid hybrid speciation in ragworts (Senecio). AoB Plants 11:ply078. https://doi.org/10.1093/aobpla/ply078
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009) BLAST+: architecture and applications. BMC Bioinforma 10:421. https://doi.org/10.1186/1471-2105-10-421
Capella-Gutiérrez S, Silla-Martinez JM, Gabaldon T (2009) TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Chapman MA, Hiscock SJ, Filatov DA (2013) Genomic divergence during speciation driven by adaptation to altitude. Mol Biol Evol 30:2553–2567. https://doi.org/10.1093/molbev/mst168
Choat B, Brodribb TJ, Brodersen CR, Duursma RA, López R, Medlyn BE (2018) Triggers of tree mortality under drought. Nature 558:531–539
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
De La Torre AR, Li Z, Van D, Peer Y, Ingvarsson PK (2017) Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Mol Biol Evol 34:1363–1377. https://doi.org/10.1093/molbev/msx069
Deng T, Ding L (2015) Paleoaltimetry reconstructions of the Tibetan Plateau, progress and contradictions. Natl Sci Rev 2:417–437. https://doi.org/10.1093/nsr/nwv062
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45:e18. https://doi.org/10.1093/nar/gkw955
Du F, Hou M, Wang W, Mao K, Hampe A (2017) Phylogeography of Quercus aquifolioides provides novel insights into the Neogene history of a major global hotspot of plant diversity in south-west China. J Biogeogr 44:294–307. https://doi.org/10.1111/jbi.12836
Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M (2013) Robust demographic inference from genomic and SNP data. PLoS Genet 9:e1003905. https://doi.org/10.1371/journal.pgen.1003905
Feng S, Ru D, Sun Y, Mao K, Milne R, Liu J (2019) Trans‐lineage polymorphism and nonbifurcating diversification of the genus Picea. N Phytologist 222:576–587. https://doi.org/10.1111/nph.15590
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL et al. (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–285. https://doi.org/10.1093/nar/gkv1344
Foote AD, Vijay N, Avila-Arcos MC, Baird RW, Durban JW, Fumagalli M et al. (2016) Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat Commun 7:11693. https://doi.org/10.1038/ncomms11693
Fu L, Li N, Mills R (1999) Pinaceae, flora of China. China, Science Press, Beijing
Fumagalli M, Vieira FG, Linderoth T, Nielsen R (2014) ngsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics 30:1486–1487. https://doi.org/10.1093/bioinformatics/btu041
Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evolut 11:725–736. https://doi.org/10.1093/oxfordjournals.molbev.a040153
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Gross BL, Rieseberg LH (2005) The ecological genetics of homoploid hybrid speciation. J Heredity 96:241–252. https://doi.org/10.1093/jhered/esi026
Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD (2009) Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5:e1000695. https://doi.org/10.1371/journal.pgen.1000695
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. https://doi.org/10.1038/nprot.2013.084
Hahn C, Bachmann L, Chevreux B (2013) Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res 41:e129. https://doi.org/10.1093/nar/gkt371
Hember RA, Kurz WA, Coops NC (2017) Relationships between the individual-tree mortality and water-balance variables indicate positive trends in water stress-induced tree mortality across North America. Glob Change Biol 23:1691–1710
Hermansen JS, Saether SA, Elgvin TO, Borge T, Hjelle E, Saetre GP (2011) Hybrid speciation in sparrows I: phenotypic intermediacy, genetic admixture and barriers to gene flow. Mol Ecol 20:3812–3822. https://doi.org/10.1111/j.1365-294X.2011.05183.x
Huang Y, Niu B, Gao Y, Fu L, Li W (2010) CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680. https://doi.org/10.1093/bioinformatics/btq003
Huson DH (1998) Splitstree, analyzing and visualizing evolutionary data. Bioinformatics 14:68–73. https://doi.org/10.1093/bioinformatics/14.1.68
Huson DH, Bryant D (2005) Estimating phylogenetic trees and networks using SplitsTree 4. Manuscript in preparation, software retrieved from www.splitstree.org
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evolut 30:772–780. https://doi.org/10.1093/molbev/mst010
Lamichhaney S, Han F, Webster MT, Andersson L, Grant BR, Grant PR (2018) Rapid hybrid speciation in Darwin’s finches. Science 359:224–228. https://doi.org/10.1126/science.aao4593
Li H, Durbin R (2019) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Li L, Stoeckert JC, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. https://doi.org/10.1101/gr.1224503
Li M, Tian S, Yeung C, Meng X, Tang Q, Niu L et al. (2014) Whole-genome sequencing of Berkshire (European native pig) provides insights into its origin and domestication. Sci Rep. 4:4678. https://doi.org/10.1038/srep04678
Li Y, Stocks M, Hemmila S, Kallman T, Zhu H, Zhou Y et al. (2010) Demographic histories of four spruce (Picea) species of the Qinghai-Tibetan Plateau and neighboring areas inferred from multiple nuclear loci. Mol Biol Evolut 27:1001–1014. https://doi.org/10.1093/molbev/msp301
Liu J, Moller M, Provan J, Gao L, Poudel RC, Li D (2013) Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. N Phytologist 199:1093–1108. https://doi.org/10.1111/nph.12336
Lockwood JD, Aleksic JM, Zou J, Wang J, Liu J, Renner SS (2013) A new phylogeny for the genus Picea from plastid, mitochondrial, and nuclear sequences. Mol Phylogenetics Evol 69:717–727. https://doi.org/10.1016/j.ympev.2013.07.004
Lyu L, Wang D, Li L, Zhu Y, Jiang D, Liu J et al. (2020) Polyphyly and species delimitation of Picea brachytyla (Pinaceae) based on population genetic data. J Syst Evol 59:515–523. https://doi.org/10.1111/jse.12584
Ma Y, Wang J, Hu Q, Li J, Sun Y, Zhang L et al. (2019) Ancient introgression drives adaptation to cooler and drier mountain habitats in a cypress species complex. Commun Biol 2:213. https://doi.org/10.1038/s42003-019-0445-z
Mavárez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, Linares M (2006) Speciation by hybridization in Heliconius butterflies. Nature 441:868–871. https://doi.org/10.1038/nature04738
Meinzer FC, Brooks JR, Gartner BL, Warren JM, Wodruff DR, Bible K et al. (2006) Dynamics of water transport and storage in conifers studied with deuterium and heat tracing techniques. Plant, Cell Environ 29:105–114
Mulch A, Chamberlain CP (2006) Earth science, the rise and growth of Tibet. Nature 439:670–671. https://doi.org/10.1038/439670a
Nieto Feliner G, Álvarez I, Fuertes-Aguilar J, Heuertz M, Marques I, Moharrek F et al. (2017) Is homoploid hybrid speciation that rare? An empiricist’s view. Heredity 118:513–516. https://doi.org/10.1038/hdy.2017.7
Nolte AW, Tautz D (2010) Understanding the onset of hybrid speciation. Trends Genet 26:54–58. https://doi.org/10.1016/j.tig.2009.12.001
Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. https://doi.org/10.1038/nature12211
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909. https://doi.org/10.1038/ng1847
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Ran J, Shen T, Liu W, Wang P, Wang X (2015) Mitochondrial introgression and complex biogeographic history of the genus Picea. Mol Phylogenet Evol 93:63–76. https://doi.org/10.1016/j.ympev.2015.07.020
Ran J, Wei X, Wang X (2006) Molecular phylogeny and biogeography of Picea (Pinaceae): implications for phylogeographical studies using cytoplasmic haplotypes. Mol Phylogenet Evol 41:405–419
Rieseberg LH (1997) Hybrid origins of plant species. Annu Rev Ecol Syst 28:359–389. https://doi.org/10.1146/annurev.ecolsys.28.1.359
Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T et al. (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216. https://doi.org/10.1126/science.1086949
Ru D, Sun Y, Wang D, Chen Y, Wang T, Hu Q et al. (2018) Population genomic analysis reveals that homoploid hybrid speciation can be a lengthy process. Mol Ecol 27:4875–4887. https://doi.org/10.1111/mec.14909
Ru D, Ma K, Zhang L, Wang X, Lu Z, Sun Y (2016) Genomic evidence for polyphyletic origins and interlineage gene flow within complex taxa: a case study of Picea brachytyla in the Qinghai-Tibet Plateau. Mol Ecol 25:2373–2386. https://doi.org/10.1111/mec.13656
Schumer M, Cui R, Rosenthal GG, Andolfatto P (2015) Reproductive isolation of hybrid populations driven by genetic incompatibilities. PLoS Genet 11:e1005041. https://doi.org/10.1371/journal.pgen.1005041
Schumer M, Rosenthal GG, Andolfatto P (2014) How common is homoploid hybrid speciation? Evolution 68:1553–1560. https://doi.org/10.1111/evo.12399
Shao C, Shen T, Jin W, Mao H, Ran J, Wang X (2019) Phylotranscriptomics resolves interspecific relationships and indicates multiple historical out-of-North America dispersals through the Bering Land Bridge for the genus Picea (Pinaceae). Mol Phylogenet Evol 141:106610. https://doi.org/10.1016/j.ympev.2019.106610
Shen T, Ran J, Wang X (2019) Phylogenomics disentangles the evolutionary history of spruces (Picea) in the Qinghai-Tibetan Plateau: implications for the design of population genetic studies and species delimitation of conifers. Mol Phylogenet Evol 141:106612. https://doi.org/10.1016/j.ympev.2019.106612
Simão FA, Waterhouse RM, loannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatic 31:3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Stamatakis A (2014) RAxML version 8, a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Sun Y, Abbott RJ, Li L, Li L, Zou J, Liu J (2014) Evolutionary history of purple cone spruce (Picea purpurea) in the Qinghai-Tibet Plateau: homoploid hybrid origin and Pleistocene expansion. Mol Ecol 23:343–359. https://doi.org/10.1111/mec.12599
Sun Y, Abbott RJ, Lu Z, Mao K, Zhang L, Wang X et al. (2018) Reticulate evolution within a spruce (Picea) species complex revealed by population genomic analysis. Evolution 72:2669–2681. https://doi.org/10.1111/evo.13624
Sun Y, Lu Z, Zhu X, Ma H (2020) Genomic basis of homoploid hybrid speciation within chestnut trees. Nat Commun 11:3375. https://doi.org/10.1038/s41467-020-17111-w
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–612. https://doi.org/10.1093/nar/gkl315
Than C, Ruths D, Nakhleh L (2008) PhyloNet: a software package for analyzing and reconstructing reticulate evolutionary relationships. BMC Bioinforma 9:322. https://doi.org/10.1186/1471-2105-9-322
Todesco M, Owens GL, Bercovich N, Légaré J-S, Soudi S, Burge DO et al. (2020) Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature 584:602–607
Wang G, Li H, Zhao H, Zhang W (2017) Detecting climatically driven phylogenetic and morphological divergence among spruce (Picea) species worldwide. Biogeosciences 14:2307–2319. https://doi.org/10.5194/bg-14-2307-2017
Wang Z, Jiang Y, Bi H, Lu Z, Ma Y, Yang X et al. (2021) Hybrid speciation via inheritance of alternate alleles of parental isolating genes. Mol Plant 14:208–222. https://doi.org/10.1016/j.molp.2020.11.008
Weir BS, Cockerham CC (1984) Estimating f-statistics for the analysis of population-structure. Evolution 38:1358–1370. https://doi.org/10.1111/j.1558-5646.1984.tb05657.x
Wright JW (1955) Species crossability in spruce in relation to distribution and taxonomy. For Sci 1:319–349. https://doi.org/10.1093/forestscience/1.4.319
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556. https://doi.org/10.1093/bioinformatics/13.5.555
Yu Y, Dong J, Liu KJ, Nakhleh L (2014) Maximum likelihood inference of reticulate evolutionary histories. Proc Natl Acad Sci USA 111:16448–16453. https://doi.org/10.1073/pnas.1407950111
Zou J, Yue W, Li L, Wang X, Lu J, Duan B et al. (2016) DNA barcoding of recently diversified tree species: a case study on spruces based on 20 DNA fragments from three different genomes. Trees 30:959–969. https://doi.org/10.1007/s00468-015-1337-6
Acknowledgements
We would like to thank the HighEdit company for assistance with English language editing of this manuscript. This work was supported by grants from the National Natural Science Foundation of China (grant numbers 32001085, 31971392, 31590821, 31670665, 91731301, 31960319), the National Key Research and Development Program (2017YFC0505203), the Fundamental Research Funds for the Central Universities (Grant No. lzujbky-2020-34, lzujbky-2020-ct02), the Strategic Priority Research Program of Chinese Academy of Sciences (Grant No. XDB01000000), and the Project of Qinghai Science & Technology Department (2020-ZJ-944Q).
Author information
Authors and Affiliations
Contributions
J.L. and D.R. planned and designed the research. D.W., Y.S., W.L. and D.R. conducted fieldwork, performed experiments and analysed data etc. D. R., J.L., Y.S. and D.W. wrote the manuscript; and all authors revised and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Dario Grattapaglia.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, D., Sun, Y., Lei, W. et al. Backcrossing to different parents produced two distinct hybrid species. Heredity 131, 145–155 (2023). https://doi.org/10.1038/s41437-023-00630-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-023-00630-9