Abstract
Seed size affects individual fitness in wild plant populations, but its ability to evolve may be limited by low narrow-sense heritability (h2). h2 is estimated as the proportion of total phenotypic variance (σ2P) attributable to additive genetic variance (σ2A), so low values of h2 may be due to low σ2A (potentially eroded by natural selection) or to high values of the other factors that contribute to σ2P, such as extranuclear maternal effects (m2) and environmental variance effects (e2). Here, we reviewed the published literature and performed a meta-analysis to determine whether h2 of seed size is routinely low in wild populations and, if so, which components of σ2P contribute most strongly to total phenotypic variance. We analyzed available estimates of narrow-sense heritability (h2) of seed size, as well as the variance components contributing to these parameters. Maternal and environmental components of σ2P were significantly greater than σ2A, dominance, paternal, and epistatic components. These results suggest that low h2 of seed size in wild populations (the mean value observed in this study was 0.13) is due to both high values of maternally derived and environmental (residual) σ2, and low values of σ2A in seed size. The type of breeding design used to estimate h2 and m2 also influenced their values, with studies using diallel designs generating lower variance ratios than nested and other designs. e2 was not influenced by breeding design. For some breeding designs, the number of genotypes included in a study also influenced the resulting h2 and e2 estimates, but not m2. Our data support the view that a diallel design is better suited than the alternatives for the accurate estimation of σ2A in seed size due to its factorial design and the inclusion of reciprocal crosses, which allows the independent estimation of both additive and non-additive components of variance.
Similar content being viewed by others
Introduction
Seed mass or size (hereafter, “seed size”) is a functional trait that has important fitness consequences in wild plant populations of many species and is therefore potentially under the influence of natural selection. Seed size influences individual plant performance and population dynamics throughout the life cycle through its effects on the timing and probability of germination, on juvenile survivorship, and on adult size and fecundity. Within populations, plants originated from relatively large seeds have higher probabilities of germination (Cideciyan and Malloch 1982), seedling survival and establishment (Bonfil 1998; Dalling and Hubbell 2002; Moles and Westoby 2004; Seiwa and Kikuzawa 1996; Winn 1988; Wulff 1986) and tend to be more fecund than plants derived from small seeds (Halpern 2005; Larios and Venable 2018; Mojonnier 1998; Stanton 1984). The evolutionary potential of seed size however, is thought to be limited by its low levels of heritable, additive genetic variation within populations (Gustafsson 1986; Merilä and Sheldon 2000; Mousseau and Roff 1987). Furthermore, because the narrow-sense heritability (h2) of a given trait is estimated as the proportion of total phenotypic variance that is attributed to the additive effects of nuclear genes, the magnitudes of non-additive genetic and environmental sources of variation (both of which contribute to total phenotypic variance) are likely to be important factors in determining the magnitude of h2 of a trait at a given time. In particular, maternal genetic and maternal environmental sources of variation may either reduce h2 estimates by inflating total phenotypic variance, or inflate heritability estimates if they are not successfully isolated from additive genetic sources of variation.
With respect to individual seed size, two classes of maternal effects (genetic and environmental) are well known to influence phenotype (Roach and Wulff 1987). Maternal genetic effects on seed size have three distinct potential sources: (1) maternal nuclear genes, transmitted to female gametophytes within ovules, that influence their ability, once fertilized, to garner and to store maternal resources; (2) maternal cytoplasmic genes that are similarly expressed by fertilized ovules; and (3) maternal genes expressed by the sporophyte (typically diploid) that affect the allocation of resources per offspring. Maternal environmental sources of variance in seed size result from the conditions in which a maternal plant develops before and during seed maturation (Roach and Wulff 1987). The competitive environment in which plants provision their offspring may influence the expression of both maternal genetic and maternal environmental variance in seed size. For example, the magnitude of maternal genetic effects on seed size in wild radish (Raphanus sativus (L.) was negatively affected by planting density in an experimental garden in California (Mazer and Wolfe 1992). When pedigreed seeds were sown and raised at low vs. high density and the seeds that they produced were harvested and weighed, maternal genetic effects on mean individual seed mass were much higher among plants sown at low density. In the winter annual Nemophila menziesii (Hook & Arn.), plants grown in competition with the grass Bromus diandrus (Roth) produced smaller seeds than those grown without competition, but this effect was largely environmental (Platenkamp and Shaw 1993). No significant maternal genetic effects on seed size were detected by Platenkamp and Shaw (1993), but a strong interaction between maternal genetic and maternal environmental effects was reported. However, another study on the same species, using a diallel crossing scheme and employing a three-generation design conducted under greenhouse conditions, detected maternal genetic effects on seed size (Byers et al. 1997). In the desert annual, Dithyrea californica (Harv.), plants growing under relatively strong competitive conditions in wild populations were smaller and produced smaller seeds than those growing under relatively weak competition, a pattern that was attributed to maternal environmental effects (Larios and Venable 2015).
Because both maternal genetic and environmental effects on seed size can inflate total phenotypic variance in this trait, and because they are components of the denominator of the fraction that defines h2, they can lower estimates of h2 and lead to the misguided inference that additive genetic variance in seed size is low. We hypothesize that these sources of variance may mask the evolutionary potential of seed size in wild plant populations, and the meta-analysis conducted here was motivated in part to explore this possibility.
Quantitative geneticists estimate the genetic structure of traits within populations by estimating their phenotypic means and variances among individuals of known relatedness. Genetic and environmental variance components that influence individual phenotype can be estimated by performing controlled pollinations—usually under greenhouse conditions—guided by the use of specific breeding designs, and then phenotyping the resulting progeny under one or more environmental conditions (Hallauer et al. 2010; Lynch and Walsh 1998). Estimates of the sources of genetic and environmental variance in mean individual seed size in wild plant species have been used to address questions related to its capacity to evolve in response to natural selection in wild populations (Andersson 1992; Cain et al. 1995; Castellanos et al. 2008). More recently, these estimates have been used to assess the likely evolutionary response of seed size to climate change (Caignard et al. 2018). Variance components of seed size have also been used to investigate the pervasiveness of maternal genetic effects on seed size (Antonovics and Schmitt 1986; Byers et al. 1997; Galloway et al. 2009; Mazer 1987), and less frequently, the adaptive nature of phenotypic plasticity of seed size (Zas and Sampedro 2015).
The most common breeding designs used in quantitative genetic studies of seed size in wild populations comprise controlled crosses between pollen-donating individual plants (males, or “sires”) and pollen-recipients (females, or “dams”). These include nested designs comprised of maternal sibships, in which individual plants are each pollinated by a different group of genetically distinct pollen donors sampled at random from the focal population, as well as nested designs comprised of paternal sibships, in which individual pollen donors are each used to pollinate a different group of genetically distinct plants (similarly randomly sampled). Frequently used designs also include a variety of diallel designs, a common one of which is characterized by a small number of individuals (e.g., 4–10 cosexual plants) used as both pollen donors (sires) and pollen recipients (dams), with all potential pairs being crossed multiple times in both directions. Breeding designs that include reciprocal crosses and the pollination of multiple sires per dam and multiple dams per sire generate pedigreed seeds that allow the most complete estimation of genetic and environmental sources of phenotypic variance in functional traits (Cockerham and Weir 1977). Accordingly, these designs generally provide the most accurate estimates of h2, which may in turn be used to predict the response to selection of the focal traits. Breeding designs that lack reciprocal crosses—including clonal, selfing, and nested designs—tend to overestimate additive genetic variance due to their inability to fully isolate maternal environmental effects and non-additive genetic effects (e.g., epistasis and dominance) from additive genetic effects. The diallel breeding design is therefore considered to be the most complete design in terms of its ability to allow the partitioning of total phenotypic variance in a given trait into its distinct variance components (Hallauer et al. 2010; Hill et al. 2012). We hypothesize that values of additive genetic variance in seed size that are estimated with a diallel design will be lower than those estimated using any other breeding design.
In the current study, we performed a review of the literature and a meta-analysis to seek and to evaluate studies designed to estimate either the heritability of seed size or its variance components in wild plant species; our primary objectives are to answer the following questions:
-
1.
Do studies of wild plant species collectively support the view that the h2 of individual seed size is low, due to either low levels of additive genetic variance, or high values of total phenotypic variance?
-
2.
Across species that have been investigated to date, are low values of h2 in seed mass due to low additive genetic variance expressed by the nuclear genes of seeds, to the contributions of maternal genetic or environmental sources of variation to total phenotypic variance, or to both? We predicted that because seeds are directly provisioned by the maternal plant and (in angiosperms) the endosperm in most species is represented by two or more copies of maternally-derived genes (but only one copy of paternally derived genes), maternal genetic variance component estimates will be high and tend to inflate total phenotypic variance in seed size, contributing to low values of h2.
-
3.
Does the type of breeding design used to estimate variance components influence the magnitude of h2 or the variance components used to estimate it? We expected that, due to the capacity for analyses of diallel breeding designs to isolate the additive effects of nuclear genes on offspring phenotype from the effects of non-additive, extra-nuclear, and maternal genetic effects, diallel designs would generate lower estimates of h2 than alternative breeding designs that do not permit such clear partitioning of sources of variation.
Materials and methods
Data search
We performed a literature search to investigate published estimates of variance components for seed size in wild plant species. We used a query containing the following keywords: (“heritab*” or “variance component*”) and (“seed size” or “seed mass” or “seed weight”). We included only studies that examined the quantitative genetics of seed size of wild plant populations and excluded those that investigated agricultural or commercial species. We also excluded studies that pooled genotypes from multiple populations to estimate species-level heritabilities. Studies included for analysis reported narrow-sense heritability from individual populations, the variance components (as raw values) used to estimate these parameters, or both. We performed our search using the Web of Science search engine.
Data extraction
From all selected studies, we extracted the following estimated parameters: 1) raw values of variance components of seed size (σ2A, σ2M, σ2P, σ2E, σ2D, σ2K; Table 1); and 2) narrow-sense heritability (h2 = σ2A: σ2P) when available. Whenever a reported value of a raw variance component was measured for the same population but in different year, we averaged them and reported a single value. When parameters were reported for more than one population, we included each estimate in our data set. In cases where seed size was measured in multiple ways (e.g., seed weight, seed length, or seed area), we used the value of seed weight and discarded the others.
Additionally, for each published parameter we recorded the following: publication identity, breeding design, and the number of maternal and paternal genotypes used in the breeding design that generated the parameter estimate. Breeding design refers to the pattern by which controlled pollinations were performed or by which natural pollinations occurred in each study. Categories examined here include: diallel and nested designs, clonal replication (hereafter “clones”), and autogamously self-fertilizing genotypes (hereafter “selfing”). Studies that reported heritability estimates but no breeding design (e.g., naturally pollinated maternal lines; Andersson 1992; Gottlieb 1977; Weber and Kolb 2014) were also excluded because such estimates were derived from open-pollinated genotypes and were likely to be confounded by environmentally induced maternal effects.
Data analysis
Standardization of variance components
In order to compare the relative contributions to seed size of each type of variance component, in each published study used here, we standardized the raw variance components by calculating variance ratios, which were computed by dividing a given raw variance component by the total phenotypic variance in seed size (i.e., the sum of all reported variance components, including the focal raw variance component). h2, for instance, is equivalent to additive genetic variance divided by total phenotypic variance (σ2A:σ2P). In the same manner, we defined m2 as the maternal variance component divided by total phenotypic variance (σ2M:σ2P); and e2 as the environmental (residual) variance component divided by total phenotypic variance (σ2E:σ2P). The same procedure was applied to the remaining components of seed size: paternal variance (Pat2 or σ2Pat:σ2P), dominance variance (d2 orσ2D:σ2P), and epistasis variance (k2 or σ2K:σ2P). Such standardized values are well-suited for comparisons among species, among independently conducted studies, and among different units of measurement (e.g., mass vs. linear measures) because standardized components are unitless.
Model construction
Because the variance ratios analyzed here originate from publications that, in some cases, reported multiple estimates of the same variance component per publication, such estimates were not fully independent, thereby violating an assumption of ordinary least-squares methods (Madden et al. 2016). To account for variation among publications in the variance ratios of seed size, we analyzed the data with generalized linear mixed-effects models (GLMMs) that included publication identity as a random effect. We chose to include only publication identity as a random effect because most studies estimated variance components for a single species, making publication and species identity contain nearly the same information, such that their simultaneous inclusion in the model as random effects would be problematic.
The number of maternal and paternal genotypes used to estimate each variance component varied greatly among published studies (range = 2–170 genotypes; Table S1), and we reasoned that the number of genotypes contributing to a genetically determined parameter estimate might influence its estimated value. Accordingly, we controlled for variation among studies in the number of genotypes sampled by including a weighting factor in the GLMMs. Weights were estimated as the number of genotypes—maternal or paternal genotypes, depending on the variance component—used in each study. Specifically, we used the number of paternal genotypes as weightings for models using estimates of h2 or Pat2 because these parameters are usually estimated using trait variation derived from the paternal lines (Falconer and MacKay 1996; Lynch and Walsh 1998). We used the number of maternal genotypes as weightings for models using estimates of m2 and e2 because maternal and environmental sources of variance in seed size are more likely to influenced by maternal than by paternal genotypes (Roach and Wulff 1987). We used the number of maternal genotypes as weightings for models using estimates of d2 and k2 because these parameters may be influenced by the number of either maternal or paternal genotypes. Among the studies analyzed here, the number of maternal genotypes equaled or exceeded the number of paternal genotypes (Table S1), so using the higher number would take this into account.
GLMMs are robust tools for the analysis of variables that vary over multiple levels and can be used with alternative distributional assumptions of the residuals (Schielzeth et al. 2020). Because the variance ratios that we used as response variables have values in the closed interval from zero to one (with many zero values present), distributions commonly employed to model proportion data, such as the beta distribution, were inappropriate for our response variables. Because of this, we used a pseudo-likelihood approach where the variance structure between the mean and the variance of the observations, and the range of the response (but not its precise distribution), are assumed (Wedderburn 1974). Specifically, we used a quasi-binomial GLMM with a logit link function for all models, which uses the variance structure of a binomial distribution while allowing for continuous values in the [0, 1] range. We fitted all GLMMs with the ‘glmmPQL’ function of the R package ‘MASS’ version 7.3–58.1 (Ripley et al. 2013), which uses penalized quasi-likelihood (PQL) for parameter estimation (Mammen and van de Geer 1997).
Comparison of variance components of seed size
To compare the means of the distinct proportional variance component types (h2, d2, pat2, m2, e2, and k2), we used a quasi-binomial GLMM with the observed value of each variance ratio estimate as a response, and the type of variance ratio for each observation as a categorical predictor. Publication identity was included as a random effect, and each observation was weighted based on the number of paternal or maternal genotypes used in estimating the reported ratio. As described above, whether we used the number of maternal or paternal genotypes depended on the identity of the variance ratio to which each observation corresponded. Marginal means of each variance ratio type and pairwise statistical comparisons between ratio types were conducted using the ‘emmeans’ function of the ‘emmeans’ R package version 1.7.5 (Searle et al. 1980), with Tukey contrasts used to assess the significance of pairwise differences, and a Tukey correction of p-values to account for multiple hypothesis testing.
Heritability and variance components of seed mass in relation to breeding design and number of genotypes
To determine whether breeding design and number of genotypes influenced h2, m2, and e2 of seed size, we fitted quasi-binomial GLMMs using each of these variance ratio types as a separate response variable. These models included breeding design as the main explanatory variable, and the number of genotypes (maternal or paternal as assigned above) and the interaction between breeding design and number of genotypes as control variables. The model also included publication identity as a random effect. Number of genotypes were also included as a weight in order to assign more information value to estimates obtained from a greater number of genotypes. In each model, we used the ‘emmeans’ package to estimate the marginal means for each breeding design, using Tukey multiple comparisons tests to assess the significance of differences in mean ratios between breeding designs. In each model, p-values obtained from pairwise comparison of breeding designs were adjusted for multiple hypothesis testing using the Tukey method. Significance testing for the effects of breeding design and genotype number were obtained using Type III ANOVA as implemented in the ‘car’ package version 3.1–0 in R (Fox and Weisberg 2019).
Results
Our search yielded papers from the following journals: Annals of Forest Research, American Journal of Botany, American Naturalist, Biological Journal of the Linnaean Society, Botany, Conservation Genetics, Ecology, Evolution, Evolutionary Ecology, Heredity, Journal of Ecology, Journal of Evolutionary Biology, New Forests, New Phytologist, Oecologia, Plant Systematics and Evolution, Romanian Biotechnological Letters, Silvae Genetica, Systematic Botany, Theoretical Applied Genetics, and Tree Genetics & Genomes. We found 39 studies that reported quantitative genetics parameters for seed size. Altogether, studies included 41 species for which variance components and heritabilities of seed size were provided (see Supplementary Table S1).
Differences among sources of variance (as a proportion of σ2 P)
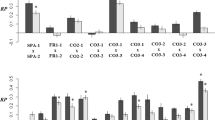
Mean values of all of the alternative variance ratios contributing to σ2P in seed size differed significantly from zero as no values of the lower confidence intervals were estimated to be zero or lower (Table 2, Fig. 1). Marginal means were highest for e2 and m2 (Table 2, Fig. 1). e2 was 0.52 (95% confidence interval: 0.46–0.58) and m2 was 0.39 (95% CI: 0.34–0.44), respectively. h2, d2, Pat2, and k2 were all very low (≤ 0.13; Table 2). Mean h2 was 0.13 (95% CI: 0.08–0.20; Table 2). These results suggest that the mean value of h2 is low but significantly different from zero across taxa, and lower than the mean values of m2 (Pairwise comparison, P < 0.0001; Supplementary Table S2) and e2 (Pairwise comparison, P < 0.0001; Supplementary Table S2).
Variance ratios are: Va:Vp = additive genetic variance ratio, Ve:Vp = environmental (residual) variance ratio, Vpat:Vp = paternal variance ratio, Vm:Vp = maternal variance ratio, Vd:Vp = dominance variance ratio, Vk:Vp = epistatic variance ratio. Black dots represent a marginal mean and bars represent their standard errors.
Effects of breeding design, and the number of genotypes sampled on h2, m2, and e2
The effect of breeding design on h2 was statistically significant (Likelihood-ratio test: P = 0.049 for breeding design, Table 3A). Specifically, controlling for the effects of the number of paternal genotypes on the estimated variance components, the mean h2 estimated from diallel breeding designs was lower than that estimated from nested designs (Fig. 2A); a Tukey pairwise comparison indicated this difference approached significance (based on a threshold of 0.05) (Pairwise comparison: P = 0.074, Supplementary Table S3, Fig. 2A). The interaction between breeding design and the number of paternal genotypes had a significant effect on h2 estimates, indicating that the number of genotypes affected estimates differently depending on breeding design (Likelihood-ratio test: P = 0.034, Table 3A), with non-significant negative effects for nested designs (odds ratio = −0.82 for an increase of ~11 genotypes, or 1 SD, P = 0.58), and a positive effect that approached significance (based on a threshold of 0.05) for diallel crosses (odds ratio= 1.56 for an increase of 1 SD, P = 0.062).
The effects of the type of breeding design on mean m2 approached significance (based on a threshold of 0.05), with non-significant effects for the number of maternal genotypes and their interaction with breeding design (Likelihood-ratio test: P = 0.099 for breeding design, P = 0.77 for maternal genotypes, and P = 0.47 for their interaction; Table 3B, Supplementary Table S3, Fig. 2B). Diallel crosses showed lower estimates of m2 than other breeding design, but such differences were not statistically significant after adjusting for multiple hypothesis testing (Fig. 2B, Table S3). Finally, breeding designs did not differ significantly with respect to mean e2 (Likelihood-ratio test: P = 0.27, Table 3C, Fig. 2C).
Discussion
This study provides the first cross-species comparison of the magnitudes of alternative sources of variance in seed mass, revealing that maternal genetic sources of variance in seed mass are about twice the magnitude of additive genetic variance; that dominance, epistatic, paternal, and additive sources of genetic variance are all similar in magnitude; and that environmental sources of variance account for about 50% of total variance in seed mass in wild populations. In addition, this manuscript provides the first quantitative test of the prediction that, among genotypes sampled from field populations of wild plant species, narrow-sense heritability estimates for seed size derived from diallel breeding designs are, on average, significantly lower than those derived from nested designs. These results are based on published values of the variance components of seed size in wild populations, spanning 41 species and 17 families in 39 studies, and tested for the effects of several factors (breeding design, publication, and number of genotypes sampled) on estimates of the genetic and environmental determinants of seed size and on its narrow- and broad-sense heritabilities.
Heritability estimates of wild species that are neither economically important nor of conservation concern are often based on data sets that are compromised by relatively low sample sizes or by a limited number of environmental treatments. These limitations may be common primarily because such studies are labor- and time-intensive to perform, and funding to support studies of wild species is limited. Given the potential sensitivity of heritability estimates to sample size, the number of genotypes sampled, and environmental conditions (as evidenced by the genotype x environment interactions affecting seed size detected by Zas and Sampedro (2015), our ability to test hypotheses of the types addressed in this paper will similarly be limited in the absence of greater data availability.
Heritability of seed size
Consistent with evolutionary theory, h2 in seed size tended to be low (μ = 0.13, 95% CI = 0.08–0.20), Table 2). This estimate of h2, however, does not on its own shed light on the underlying causes of such low apparent evolutionary potential of seed size because several variance components other than additive genetic variance—including variance due to maternal genetic and environmental effects, dominance, and epistasis—can inflate total phenotypic variance, lowering the estimate of h2. In short, either low additive genetic variance (which comprises the numerator of the ratio that defines h2) or high total phenotypic variance (the denominator) will diminish (narrow-sense) heritable variation in a focal trait. The current study, however, reveals that, in wild plant species, maternal and environmental sources of variance in seed size are on average much larger than additive genetic variance (Fig. 1), indicating that the low narrow-sense heritability of seed size reported here can be attributed to both low additive genetic variance and high maternal and environmental variance.
Parental and environmental sources of variance in seed size
Maternal and environmental variances were the highest among all of the alternative sources that comprised phenotypic variance in seed size (Fig. 1, Table 2). Maternal genetic effects are widely recognized as a pervasive source of seed size variation in many plant species (Antonovics and Schmitt 1986; Biere 1991; Byers et al. 1997). Three independent sources of maternal genetic effects can influence seed size: the maternal haploid nuclear contribution to female gametes and the endosperm, the maternal sporophyte’s (diploid) genotypic effect on resource allocation per seed, and cytoplasmic effects expressed by the female gametophyte, i.e., the extranuclear maternal contribution to the endosperm and the egg (Roach and Wulff 1987).
Nuclear maternal effects transmitted through the eggs can be effectively separated from the maternal sporophyte’s genotypic and cytoplasmic effects on seed size only by performing controlled crosses that estimate the maternal contribution to additive genetic variance. For example, the maternal sporophyte’s genotypic effect on seed size (i.e., the maternal genes that determine provisioning) can be isolated using a breeding design that measures seed size in both F1 and F2 sibships, requiring a three-generation breeding design (Dickerson 1947; Lynch and Walsh 1998; Shaw and Waser 1994). With such a breeding design, additive genetic variance among maternal sporophytes (independent of the effects of their gametophytes) that contributes to total phenotypic variance in seed size of the offspring can be estimated.
To our knowledge, only three studies have attempted to estimate maternal additive genetic variance in seed size in wild plant populations in this way (Byers et al. 1997; Galloway et al. 2009; Thiede 1998). In Collinsia verna, seed size was found to be influenced significantly by maternal additive genetic variance. However, this source of maternal additive genetic variance was negatively correlated with direct-maternal additive genetic variance (the contribution of nuclear genes transmitted through the gametophytes), and this correlation affected the response to selection (Thiede 1998). In Nemophila menziesii, maternal additive genetic variance exceeded direct additive genetic variance, suggesting major control over seed size by the maternal sporophyte’s genotype (Byers et al. 1997). Finally, in Campanulastrum americanum, seed size was controlled exclusively by maternal environmental effects rather than by maternal genetic effects (Galloway et al. 2009). Maternal influences on seed size are typically large because both the maternal genotype and growing conditions (the source of maternal environmental variance) determine resource allocation per offspring. The observation that, in contemporary populations, maternal genetic variance (from all sources) in seed size is higher than direct additive genetic variance in seed size suggests that natural selection on seed size should result in greater short-term evolutionary change in maternally inherited genetic sources of variation than in paternally inherited genes expressed in the offspring.
It is not possible to separate maternal genetic from maternal environmental effects on seed size unless the experimental breeding design includes replicates of maternal lines provisioning their seeds in alternative environments (Tucić and Avramov 1996; Zas and Sampedro 2015), including distinct competitive conditions (Mazer and Wolfe 1992; Platenkamp and Shaw 1993); or where the design includes seeds sourced from alternative populations grown in a common garden (Lipow and Wyatt 1999; Mitchell-Olds 1986; Rice and Mack 1991; Volis 2007; Wolff 1988). Having unambiguous and independent estimates of maternal genetic and maternal environmental effects on seed size allows for the estimation of G × E interactions, which can be used to estimate phenotypic plasticity due to maternal and zygotic effects on seed size (Lynch and Walsh 1998). Environmental variance in seed size that is not due to maternal environmental effects refers to unexplained sources of variance (residual variance) in statistical models. Such unexplained variance may originate from different elements of the physical environment during seed provisioning, including position effects on seed size within or among fruits or stems.
Non-additive sources of variance such as dominance and epistasis originate from interactions between alleles at the same locus or at different loci, respectively. Theoretical predictions consistently agree that populations should retain high levels of non-additive genetic variance because natural selection is less effective in purging populations of non-additive relative to additive genetic variance. For example, high dominance variance (particularly overdominance) has been hypothesized to characterize fitness-related traits in wild species based on the argument that, because such dominance is not eroded by directional selection, it should predominate in the genetic architecture (Crnokrak and Roff 1995). Epistatic variance is difficult to estimate from experimental breeding designs, but is subject to the same considerations as dominance. As the number of interacting loci increases, the number of possible interactions increases exponentially (Lynch and Walsh 1998). Merilä and Sheldon (1999) propose that epistatic variance should remain high in quantitative traits because multiple loci contribute to variance in such traits, generating many possible interactions that influence phenotype. In addition, it is difficult for natural selection to purge populations of epistatic variance because, due to recombination, the interactions are not stably transmitted between generations and can continue to appear following selective events. In certain circumstances, such as in population bottlenecks, epistatic variance has been demonstrated to have an important role in plant adaptation due to the ability of epistatic variance to become a source of additive genetic variance (Goodnight 1988), facilitating the formation of ‘adaptive gene complexes’ (Goodnight 1995). Despite these theoretical predictions, non-additive sources of genetic variance in seed size in our study did not differ significantly from zero (Table 2).
Effects of breeding design and the number of genotypes on heritability estimates
Estimates of h2 and m2 are more sensitive to the type of breeding design used than are environmental variance ratios in seed size. Both estimates of additive genetic variance and extranuclear genetic maternal effects derived from diallel designs were lower than estimates derived from other designs (Fig. 2A), likely because the analysis of diallels provides the greatest potential for the effective partitioning of additive from non-additive sources of genetic variance in seed size. This feature is due to the inclusion of reciprocal crosses in the diallel design, which allows not only the detection of interactions between the nuclear contributions of sires and dams that affect seed size (e.g., due to dominance), but also the detection of the effects of the direction of the cross on seed mass (due to cytoplasmic or to maternal sporophytic effects). While pairwise comparisons of estimates of h2 and m2 across breeding designs only approached significance (Fig. 2), we note that the effect sizes of diallel crosses were substantial, and that their lack of significance might be due to the relatively low statistical power afforded by the small number of estimates available for each breeding design.
If the diallel design could be superior for the effective estimation of narrow-sense heritability and genetic maternal effects, why is it not more widely used in quantitative genetics? The challenge in using a diallel design derives from the fact that, to sample and to test the genetic contributions to offspring phenotype of a given number of genotypes (n), one must conduct n2 unique crosses (if one includes self-pollinations). By contrast, a nested design in which maternal plants are nested within pollen donors and in which n paternal genotypes are tested requires only n × x unique crosses, where x equals the number of maternal plants pollinated by each pollen donor. So, the feasibility of employing a diallel breeding design for a particular species depends on the ease with which individual pollinations may be conducted and the number of seeds created per pollination, as well as by the ability to keep track of the paternal genotype that was used to pollinate each flower or inflorescence.
But every breeding design has its disadvantages. As noted by Hill et al. (2012), diallel designs are not ideal for outbreeding species that produce very few seeds per flower because they require a large number of replicate pollinations to achieve sufficient seed production for phenotyping the offspring generation. Moreover, if the goal is to sample a large number of maternal (and paternal) genotypes when using a diallel design to estimate components of variance, it is necessary to create and to pollinate multiple diallels. Replication of both pollinations and diallels can be prohibitively labor intensive. By contrast, when testing a given number of paternal genotypes, the number of pollinations needed to estimate additive genetic variance using a nested design is much lower than for a diallel design, but precludes the estimation of interactions that can be detected by comparing the outcome of reciprocal crosses. The analysis of clonally propagated genotypes is particularly problematic as it does not allow for the isolation and estimation of paternal vs. maternal effects (or their interaction) on offspring phenotype. The use of clones for the estimation of genetic and environmental sources of variation in seed size has been applied in only a few cases (Zas and Sampedro 2015; Table 1S). For example, Zas and Sampedro (2015) used clonal lines to estimate the narrow-sense heritability of seed provisioning among maternal (sporophytic) genotypes (but not the additive genetic effects of the paternal genotype on seed size transmitted by its pollen). They replicated maternal genotypes within and among orchards cultivated in contrasting environments, which enabled them to test for G × E interactions affecting seed size.
In conclusion, h2 for seed size is generally low in wild plant species, but non-additive genetic sources (dominance and epistasis) tend to be even lower than additive genetic variance in seed size (Table 2). Genetic and environmental maternal effects on seed size contribute to low values of heritable variation, primarily by inflating total phenotypic variance. The importance of such genetic maternal effects suggests that the opportunity for natural selection to drive intergenerational change in mean seed size is higher among maternal sporophytes than among the nuclear genotypes of seeds. Our findings indicate that, when aiming to detect and to measure the potential for seed size to evolve through nuclear, extra-nuclear, or sporophytic genetic influences, the diallel breeding design is the most powerful option whenever time, trained labor, and plant material are sufficient to generate pedigreed seeds; otherwise, the nested breeding design is recommended to maximize the number of maternal and paternal genotypes to be tested when these resources are more limited.
Data availability
Database and R code are available at the Dryad data repository https://doi.org/10.25349/D9660G.
References
Andersson S (1992) Quantitative genetic variation in a population of Crepis tectorum subsp. pumila (Asteraceae). Biol J Linn Soc 44(4):381–393
Antonovics J, Schmitt J (1986) Paternal and maternal effects on propagule size in Anthoxanthum odoratum. Oecologia 69(2):277–282
Biere A (1991) Parental effects in Lychnis flos-cuculi. I: seed size, germination and seedling performance in a controlled environment. J Evolut Biol 4(3):447–465
Bonfil C (1998) The effects of seed size, cotyledon reserves, and herbivory on seedling survival and growth in Quercus rugosa and Q. laurina (Fagaceae). Am J Bot 85(1):79–87
Byers DL, Platenkamp GAJ, Shaw RG (1997) Variation in seed characters in Nemophila menziesii: evidence of a genetic basis for maternal effect. Evolution 51(5):1445–1456
Caignard T, Delzon S, Bodénès C, Dencausse B, Kremer A (2018) Heritability and genetic architecture of reproduction-related traits in a temperate oak species. Tree Genet Genomes 15(1):1
Cain ML, Kahn B, Silander Jr JA, Reynolds HL (1995) Genetic variability and tradeoffs among reproductive traits in white clover (Trifolium repens). Can J Bot 73(3):505–511
Castellanos MC, Medrano M, Herrera CM (2008) Subindividual variation and genetic versus environmental effects on seed traits in a European Aquilegia. Botany 86(10):1125–1132
Cideciyan MA, Malloch AJC (1982) Effects of Seed Size on the Germination, Growth and Competitive Ability of Rumex Crispus and Rumex Obtusifolius. J Ecol 70(1):227–232
Cockerham CC, Weir BS (1977) Quadratic analyses of reciprocal crosses. Biometrics 33(1):187–203
Crnokrak P, Roff DA (1995) Dominance variance: associations with selection and fitness. Heredity 75(5):530–540
Dalling JW, Hubbell SP (2002) Seed size, growth rate and gap microsite conditions as determinants of recruitment success for pioneer species. J Ecol 90(3):557–568
Dickerson G (1947) Composition of hog carcasses as influenced by heritable differences in rate and economy of gain. Iowa Agric Exp Station Res Bull 354:489–524
Falconer DS, MacKay TFC (1996) Introduction to Quantitative Genetics, 4th ed. edn. Prentice Hall, Harlow, UK
Fox J, Weisberg S (2019) Vol. 3rd. Sage, Thousand Oaks, California
Galloway LF, Etterson JR, McGlothlin JW (2009) Contribution of direct and maternal genetic effects to life-history evolution. N. Phytologist 183(3):826–838
Goodnight CJ (1988) Epistasis and the effect of founder events on the additive genetic variance. Evolution 42(3):441–454
Goodnight CJ (1995) Epistasis and the increase in additive genetic variance: implications for phase 1 of Wright’s shifting-balance process. Evolution 49(3):502–511
Gottlieb LD (1977) Phenotypic Variation in Stephanomeria exigua ssp. Coronaria (Compositae) and Its Recent Derivative Species “Malheurensis”. Am J Bot 64(7):873–880
Gustafsson L (1986) Lifetime reproductive success and heritability: empirical support for Fisher’s fundamental theorem. Am Naturalist 128(5):761–764
Hallauer AR, Carena MJ, Filho JBM (2010) Quantitative Genetics in Maize Breeding. Springer, New York
Halpern SL (2005) Sources and consequences of seed size variation in Lupinus perennis (Fabaceae): adaptive and non-adaptive hypotheses. Am J Bot 92(2):205–213
Hill J, Becker HC, Tigerstedt PM (2012) Quantitative and Ecological Aspects of Plant Breeding. Springer, Netherlands
Larios E, Venable DL (2015) Maternal adjustment of offspring provisioning and the consequences for dispersal. Ecology 96(10):2771–2780
Larios E, Venable DL (2018) Selection for seed size: The unexpected effects of water availability and density. Funct Ecol 32(0):2216–2224
Lipow SR, Wyatt R (1999) Diallel crosses reveal patterns of variation in fruit-set, seed mass, and seed number in Asclepias incarnata. Heredity 83(3):310–318
Lynch M, Walsh B (1998) Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland, MA, USA
Madden LV, Piepho H-P, Paul PA (2016) Stat Models Methods Netw Meta-Anal Phytopathol® 106(8):792–806
Mammen E, van de Geer S (1997) Penalized quasi-likelihood estimation in partial linear models. Ann Stat 25(3):1014–1035. 1022
Mazer SJ (1987) Parental effects on seed development and seed yield in Raphanus raphanistrum: implications for natural and sexual selection. Evolution 41(2):355–371
Mazer SJ, Wolfe LM (1992) Planting Density Influences the Expression of Genetic Variation in Seed Mass in Wild Radish (Raphanus sativus L.: Brassicaceae). Am J Bot 79(10):1185–1193
Merilä J, Sheldon BC (1999) Genetic architecture of fitness and nonfitness traits: empirical patterns and development of ideas. Heredity 83(2):103–109
Merilä J, Sheldon BC (2000) Lifetime Reproductive Success and Heritability in Nature. Am Naturalist 155(3):301–310
Mitchell-Olds T (1986) Quantitative genetics of survival and growth in Impatiens capensis. Evolution 40(1):107–116
Mojonnier L (1998) Natural Selection on Two Seed Size Traits in the Common Morning Glory Ipomoea purpurea (Convolvulaceae): Patterns and Evolutionary Consequences. Am Naturalist 152(2):188–203
Moles AT, Westoby M (2004) Seedling survival and seed size: a synthesis of the literature. J Ecol 92(3):372–383
Mousseau TA, Roff DA (1987) Natural selection and the heritability of fitness components. Heredity 59(2):181–197
Platenkamp GAJ, Shaw RG (1993) Environmental and Genetic Maternal Effects on Seed Characters in Nemophila menziesii. Evolution 47(2):540–555
Rice KJ, Mack RN (1991) Ecological genetics of Bromus tectorum. Oecologia 88(1):77–83
Ripley B, Venables B, Hornik K, Gebhardt A, Firth D (2013). MASS: Support Functions and Datasets for Venables and Ripley’s MASS. Modern Appl Stat S.
Roach DA, Wulff RD (1987) Maternal efects in plants. Annu Rev Ecol Syst 18(1):209–235
Schielzeth H, Dingemanse NJ, Nakagawa S, Westneat DF, Allegue H, Teplitsky C et al. (2020) Robustness of linear mixed-effects models to violations of distributional assumptions. Methods Ecol Evol 11(9):1141–1152
Searle SR, Speed FM, Milliken GA (1980) Population Marginal Means in the Linear Model: An Alternative to Least Squares Means. Am Stat 34(4):216–221
Seiwa K, Kikuzawa K (1996) Importance of seed size for the establishment of seedlings of five deciduous broad-leaved tree species. Vegetatio 123(1):51–64
Shaw RG, Waser NM (1994) Quantitative Genetic Interpretations of Postpollination Reproductive Traits in Plants. Am Naturalist 143(4):617–635
Stanton ML (1984) Seed Variation in Wild Radish: Effect of Seed Size on Components of Seedling and Adult Fitness. Ecology 65(4):1105–1112
Thiede DA (1998) Maternal inheritance and its effect on adaptive evolution: a quantitative genetic analysis of maternal effects in a natural plant population. Evolution 52(4):998–1015
Tucić B, Avramov S (1996) Maternal effects on early juvenile traits in Iris pumila (Iridaceae). Plant Syst Evol 201(1):179–197
Volis S (2007) Correlated patterns of variation in phenology and seed production in populations of two annual grasses along an aridity gradient. Evolut Ecol 21(3):381–393
Weber A, Kolb A (2014) Differences in heritable trait variation among populations of varying size in the perennial herb Phyteuma spicatum. Conserv Genet 15(6):1329–1337
Winn AA (1988) Ecological and evolutionary consequences of seed size in Prunella vulgaris. Ecology 69(5):1537–1544
Wolff K (1988) Genetic analysis of ecological relevant morphological variability in Plantago lanceolata L. Theor Appl Genet 75(5):772–778
Wulff RD (1986) Seed Size Variation in Desmodium Paniculatum: II. Effects on Seedling Growth and Physiological Performance. J Ecol 74(1):99–114
Zas R, Sampedro L (2015) Heritability of seed weight in Maritime pine, a relevant trait in the transmission of environmental maternal effects. Heredity 114(1):116–124
Acknowledgements
The authors would like to thank two anonymous reviewers who made insightful comments on the manuscript that helped improve the analysis and the results. Authors would also like to thank the UCMexus grant program for financial support for EL while he was in residence as a postdoctoral associate in the Department of Ecology, Evolution, and Marine Biology at UCSB. SJM is very grateful to the Yale Institute of Biospheric Studies, which provided sabbatical support during 2019-2020, when much of this work was conducted.
Author information
Authors and Affiliations
Contributions
EL and SJM designed the study and proposed hypotheses. EL extracted all values from the published literature and drafted the first version of the manuscript. EL and TRP performed statistical analyses. All authors contributed with the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Lindsey Compton.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Larios, E., Ramirez-Parada, T.H. & Mazer, S.J. Heritability and variance components of seed size in wild species: influences of breeding design and the number of genotypes tested. Heredity 130, 251–258 (2023). https://doi.org/10.1038/s41437-023-00597-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-023-00597-7