Abstract
Glacial cycles play important roles in determining the phylogeographic structure of terrestrial species, however, relatively little is known about their impacts on the distribution of marine biota. This study utilised modern (n = 350) and ancient (n = 26) mitochondrial genomes from Australasian snapper (Chrysophrys auratus) sampled in New Zealand to assess their demographic and phylogeographic history. We also tested for changes in genetic diversity using the up to 750-year-old mitochondrial genomes from pre-European archaeological sites to assess the potential impacts of human exploitation. Nucleotide diversity and haplotype diversity was high (π = 0.005, h = 0.972). There was no significant change in nucleotide diversity over the last 750 years (p = 0.343), with no detectable loss of diversity as a result of indigenous and industrial-scale fishing activity. While there was no evidence for contemporary population structure (AMOVA, p = 0.764), phylogeographic analyses identified two distinct mitochondrial clades that diverged approximately 650,000 years ago during the mid-Pleistocene, suggesting the species experienced barriers to gene flow when sea levels dropped over 120 m during previous glacial maxima. An exponential population increase was also observed around 8000 years ago consistent with a post-glacial expansion, which was likely facilitated by increased ocean temperatures and rising sea levels. This study demonstrates that glacial cycles likely played an important role in the demographic history of C. auratus and adds to our growing understanding of how dynamic climatic changes have influenced the evolution of coastal marine species.
Similar content being viewed by others
Introduction
Phylogeographic studies have provided key insights into past demographic changes such as population expansion, contraction and fragmentation. Geological processes such as continental drift, mountain formation, climatic changes, shifting sea currents, and Pleistocene glacial-interglacial cycles have been shown to play important roles in the evolution and phylogeographic structuring of biodiversity (Teske et al. 2011; Wallis and Trewick 2009). Glacial cycles are often considered to have had regular and significant impacts on the phylogeographic structure and population demographics of many species (Hewitt 2000). Long-term extrinsic barriers to gene flow typically produce deep divergences in gene trees, which persist for long periods of time in non-recombining genetic markers, such as the mitochondrial genome (Harpending et al. 1998). In terrestrial ecosystems, the formation of glaciers and ice sheets has led to widespread habitat loss and disruption to gene flow among populations, which resulted in phylogeographic structure that is often similar among a range of taxa (Wallis et al. 2016). For example, common phylogeographic patterns observed between terrestrial species in North America were attributed to isolation in refugia during the Last Glacial Maximum (LGM) (Shafer et al. 2010). Similarly, red deer in Europe were observed to have three divergent mitochondrial lineages, suggestive of at least three glacial refugia (Skog et al. 2009).
Glacial cycles also have strong impacts on marine ecosystems, and the phylogeography of species within them (Hewitt 2000). Pleistocene glaciations resulted in the lowering of global sea-level by approximately 120 m (Dlabola et al. 2015; Peltier and Fairbanks 2006), which altered coastal marine habitats, disrupted oceanographic currents, and closed off seaways and straits (Bowen et al. 2016). These processes have had pronounced impacts on the connectivity of marine faunal populations, with distinct mitochondrial lineages in the four-eyed sleeper fish Bostrychus sinensis (Ding et al. 2018; Qiu et al. 2016), Kentish plover Charadrius alexandrines and white-faced plover C. dealbatus (Wang et al. 2019), directly attributable to reductions in sea levels and the formation of land bridges between Taiwan and China. Moreover, sea-level change in the Qiongzhou Strait during glacial periods has been linked to population divergence in small-head hairtail Lepturacanthus savala (Gu et al. 2021), lipped periwinkle Monodonta labio (Zhao et al. 2017), lacustrine goby Gobiopterus lacustris (Wang et al. 2018), and silver pomfret Pampus argenteus (Sun et al. 2019).
Australasian snapper (Chrysophrys auratus) is a common coastal fish species in the waters around the North Island, and the northern South Island of New Zealand (Fig. 1). The species also occurs in Australian waters, but shows clear phylogenetic separation suggesting absence of gene flow between the two regions (Tabata and Taniguchi 2000). Chrysophrys auratus inhabits waters down to depths of 50 m (max 200 m), and its distribution is primarily determined by water temperature (Parsons et al. 2014), which has a large impact on their growth and survival rate (Wellenreuther et al. 2019). It is predicted that climate change will lead to range expansions as water temperature increase (Brooks 2019). However, little is known regarding their response to historic changes in global climate. Glacial cycles likely had a significant impact on C. auratus as reductions in sea levels and ocean temperatures during glacial periods would have affected their suitable habitat (Dlabola et al. 2015; Peltier and Fairbanks 2006). Such environmental changes combined with New Zealand’s isolated geographic location, and its shores running close the edge of the continental shelf, would have led to range contractions and reductions in population size. Phylogeographic signatures of such demographic events will likely be detectable in the contemporary populations.
Red diamonds indicate sites where ancient mitochondrial genomes were obtained, numbers behind site names indicate number of successfully sequenced mitochondrial genomes. See Fig. S1 for a map showing all ancient sampling sites and Table S1 for location information. The table on the top right shows the estimated age of the ancient sampling sites, and samples regions to which they are allocated.
In recent history, extractive fisheries have had a significant impact on fish populations, particularly in instances of prolonged and intense size-selective fishing (Heino et al. 2015). Artisanal fishing for C. auratus began approximately 750 years ago when the East Polynesian ancestors of Māori arrived in New Zealand (Leach 2006; Leach and Davidson 2000). Based on the presence of C. auratus bones in archaeological midden (i.e. prehistoric rubbish dumps) deposits (Leach 2006; Seersholm et al. 2018; Smith 2013), C. auratus numbers appeared to decline over the ~500 year period of Māori occupation of New Zealand prior to European arrival (Leach 2006). This decline may be related to an increase in catch per unit of effort. There was also a period of climate cooling and environmental unpredictability between 1500–1850 AD (Little Ice Age) (Lorrey et al. 2014; Waters et al. 2017), which may have also influenced fish distributions or recruitment patterns, and therefore fishing success. Morphometric data showed that the average size of C. auratus dentaries significantly decreased over this time period (Leach and Davidson 2000). These findings suggest that C. auratus population sizes fluctuated over the last 500 years, however, it is unknown to what extent environmental changes and the impacts of artisanal fishing drove these changes (Waters et al. 2017).
Since European arrival, it is well-documented that industrial scale harvesting (beginning in the 1920’s) has contributed to large reductions in population size of New Zealand C. auratus, with wild fishery biomass dropping below the soft limit (i.e. 20% of the original biomass) (Fisheries New Zealand 2018). Stocks currently appear to be recovering but it is unclear whether large reductions in abundance caused by intense fishing effort also led to a loss of genetic variation. Previous genetic studies on C. auratus in New Zealand utilising nuclear microsatellite loci, reported local losses of genetic variation in a single population with a small effective population size (Hauser et al. 2002).
Recent studies have shown great potential for the retrieval of ancient DNA from fish bones (Ferrari et al. 2021; Oosting et al. 2019), allowing for direct inference of changes in genetic diversity and phylogeographic structure through time. Here we analyse mitochondrial genomes obtained from modern and ancient individuals to assess the phylogeographic structure and demographic history of C. auratus throughout New Zealand. Specifically, the following questions are addressed: (1) is there evidence of phylogeographic structure and demographic change associated with Pleistocene glacial cycles; and (2) can past changes in abundance be detected in patterns of genetic diversity, including a potential loss of genetic diversity caused by high fishing pressure since humans colonized New Zealand?
Methods
Collection modern samples, extraction and sequencing
Fin clips were collected from 350 modern specimens, obtained from commercial and recreational fishing sources (Fig. 1, Table S1). DNA was extracted using a high-salt extraction protocol adapted from Aljanabi and Martinez (1997), as described in Oosting et al. (2020). Library preparation and high-throughput sequencing were performed by Australian Genome Research Facility (Melbourne, Australia). Double-stranded library preparation was conducted using the Nextera flex protocol with unique dual indexing. High-throughput sequencing was performed on an Illumina NovaSeq 6000 S4.
Collection ancient samples, extraction and sequencing
Dentaries from ancient C. auratus were loaned from the Museum of New Zealand Te Papa Tongarewa, Wellington (Fig. S1). We sampled 124 dentaries from 18 pre-European Māori archaeological middens (Fig. S2 and Table S1). To avoid sampling the same individual multiple times, only left or right dentaries were sampled from each site. Middens were classified as either early (1280–1450 CE) or late (1650–1800 CE) based on what is known regarding Māori subsistence practices during those time periods (Fig. S2) (Smith and James-Lee 2010).
Pre-processing and DNA extraction of ancient dentaries were performed in a dedicated ancient DNA facility (Otago Palaeogenetics Laboratory, Department of Zoology, University of Otago, New Zealand). Portions of the tooth-row were removed in a sterile extraction hood using a Dremel cutting disk (whilst preserving morphometric landmarks) and powdered using a sterile mortar and pestle. Ancient DNA was extracted following Boessenkool et al. (2017), in accordance with standard aDNA protocols (Cooper and Poinar 2000; Knapp et al. 2012), including the use of negative extraction controls.
Library preparation and high-throughput sequencing were performed in a dedicated ancient DNA facility at the University of Oslo (Norway). Double-stranded, double-indexed sequencing libraries were constructed from USER-treated DNA template (Kircher 2012; Meyer and Kircher 2010). Quality of the individual libraries was assessed using an Agilent 5200 fragment analyser using the high sensitivity (HS) assay (DNF-473), following manufacturer’s instructions. Libraries were pooled with the final concentration above 2.0 ng/µl, and sequenced on an Illumina Hi-Seq4000. Sequences were demultiplexed allowing for zero mismatches in the index, yielding approximately 10 million reads per sample (skim sequencing). Endogenous DNA content was determined by mapping sequenced reads to the nuclear genome of C. auratus (Catanach et al. 2019). This information was used to identify suitable samples for additional sequencing. Library concentrations for selected samples were adjusted to generate the desired number of reads that allowed the retrieval of mitochondrial genomes with an average coverage of five.
Read alignment
Sequenced reads from modern and ancient samples were processed using the bam_pipeline in PALEOMIX v1.2.13.3 (Schubert et al. 2014). First, adapter removal v2.2.3 was used to remove the Illumina adapters; using a mismatch rate of 33% (--mm 3) and a minimum read length of 25 bp (--minlength 25). Ambiguous nucleotides (N’s) and low-quality bases were trimmed from 5’ and 3’ read ends. Overlapping paired-end reads from ancient libraries were collapsed into a single read. Burrows-Wheeler Aligner (BWA) v0.7.15 (bwa-mem algorithm) was used to align reads. Reads were simultaneously aligned to the nuclear and mitochondrial genome to minimize false alignment (Ashton 2013; Catanach et al. 2019), creating separate bam files. Unmapped reads and reads with a mapping quality of <25 were removed. PCR duplicates generated in amplification steps during library construction were removed using MarkDuplicates, Picard tools v2.18.20 (Broad Institute 2020). Finally, local realignment was performed using the indel realigner in GATK v4.0.8.1 (McKenna et al. 2010). MapDamage v2.0.9 was used to assess post-mortem DNA damage patterns (elevated number of C-T and A-G misincorporations) to authenticate ancient DNA. (Jonsson et al. 2013). Finally, all hard and soft-clipped reads were removed from BAM files, and indexed using Samtools v1.9.
SNP calling
Variant calling was performed using GATK v4.1.4.1. First, genomic Variant Call Format (g.vcf) files were generated from the BAM files. This was done using GATK’s HaplotypeCaller, using the mitochondrial genome of C. auratus as a reference (Ashton 2013), setting ploidy to 1 (-ploidy 1) and the reference confidence mode set the gvcf (-ERC GVCF). A g.vcf file was generated for each individual and used for joined genotyping. G.vcf files were subsequently merged using GATK’s CombineGVCFs function (default settings). This deviated from GATK’s best practices and was chosen because that function allows the user to specify the ploidy of the sample. Genotypes from both modern and ancient samples were jointly called using GenotypeGVCFs using the following settings: -G StandardAnnotation; -ploidy 1; -new-qual. The vcf file was filtered using bcftools v1.9, and vcftools v0.1.16. In bcftools sites we filtered on possible strand bias (FS < 60.0, SOR < 4), mapping quality (MQ > 30), and quality by depth (QD > 2.0). In vcftools, sites were filtered on minimum genotype quality (--minGQ 15) and minimum sequencing depth (--minDP 3). FASTA sequences were generated for each individual using GATK’s FastaAlternateReferenceMaker and merged into a single alignment.
Generating mitochondrial sequences of the Pagrus major (outgroup)
Ten whole-genome-sequences from wild-caught red seabream (Pagrus major) from Japan were used as the outgroup sequences (Nam et al. 2019). Similar to the alignment C. auratus reads, high-throughput sequencing data was aligned to both the nuclear (Shin et al. 2018), and mitochondrial genomes from the same species (Miya et al. 2001). FASTA sequences were visually aligned to those of C. auratus. One indel and three invariant sites on the end of the sequences were removed so all sequences were the same length (16,724 base pairs).
Partitioning and substitution model selection
The mitochondrial genome was annotated in Geneious v11.0.3 by searching for homologous sequences (https://www.geneious.com). The annotated mitochondrial genome of C. auratus was compared for consistency to the annotation of the red seabream, obtained from the Mitochondrial Genome Database of Fish, MitoFish (Sato et al. 2018). The mitochondrial genome was split into the individual protein-coding genes, 12 S rRNA, 16 S rRNA, control region (or D-loop), and the 22 tRNA sequences. Intergenic regions were added to the tRNA sequences. Protein coding genes were subsequently partitioned into the 1st, 2nd, and 3rd codon positions, and concatenated to create 1st, 2nd, and 3rd codon alignments. Genes with reversed reading frames (from ‘3 to ‘5, e.g. ND6) were grouped as if the 3rd codon was the first. PartitionFinder2 identified four distinct partitions that followed different substitution models (Table S2) (Lanfear et al. 2016), based on the mitochondrial annotation (Fig. S3). First codon positions were grouped with ribosomal and transfer-RNA sequences, while 2nd codon, 3rd codon and D-loop had their own partition model. Substitution models for each partition were estimated using Jmodeltest2 (Darriba et al. 2012). Model selection was evaluated using information criteria (i.e. AIC, aAIC, and BIC), giving the most weight to BIC (Table S3).
Mitochondrial genome variation
The number of unique haplotypes, number of polymorphic sites, number of sites with missing nucleotides, nucleotide diversity (π), haplotype diversity (h), genetic diversity (θ), were estimated using the R-package pegas (Paradis 2010). Tajima’s D was estimated using DNAsp (Rozas et al. 2017). Tests for significant differences in nucleotide diversity between sampling locations and sample types (modern vs ancient) were performed using a custom script published by Alexander et al. (2016) using 10,000 permutations. An hierarchical AMOVA (1000 permutations) was performed using the R-package apex (Jombart et al. 2017), to test for genetic structure.
Phylogenetic and demographic history
A maximum-likelihood tree was estimated in IQTREE (Nguyen et al. 2015), using default parameters and 1,000 bootstrap replicates. The consensus tree was used to produce a haplotype genealogy in Fitchi (Matschiner 2016). Fitchi was run using default parameters, except the aesthetics value for node size (-m 0.3) and specifying the ploidy (--haploid). The output of Fitchi was used to group samples into clades. We tested for changes in the relative haplotype abundance between different clades to determine whether certain clades had a potential selective advantage over the past 750 years. Fisher’s exact test was performed to test for significant changes in haplotype distribution over time. To ensure unbiased comparison, the modern data set was randomly subsampled down to the same number of observations as the ancient DNA data set (i.e. 26). Subsampling was bootstrapped 1,000 times to obtain confidence intervals. Calculations were performed using a custom Rscript (R Core Team 2013).
Bayesian analyses were performed using BEAST2 v2.6.2 (Bouckaert et al. 2014). Data partitioning and nucleotide substitution models were based on the results obtained from PartitionFinder2 and Jmodeltest2 (Table S3). A strict clock rate was used for all partitions. Different clock rates were used for the control region and the remainder of the mitochondrial genome, 5.0e−8 per site per year (site−1/year−1), and 3.28e−9 site−1/year−1, respectively (Bowen et al. 2006; Gillooly et al. 2005). The latter estimate was based on the average clock rate of nine fish species presented in Gillooly et al. (2005). The substitution rate was allowed to vary between Codon1+RNA, Codon2, and Codon3 partitions (Fig. S4). Base frequencies were estimated empirically from the data. The phylogeny of C. auratus was estimated by applying the Coalescent Exponential Population prior. A second phylogeny was estimated using the Yule prior to assess the divergence time between C. auratus and P. major. Finally, changes in population size of C. auratus through time were assessed using Extended Bayesian Skyline Plot prior. Chain lengths for each of the analyses varied between 50 and 500 million steps, depending on how long it took for runs to converge. All runs incorporated a 10% burn-in. Trace profiles were visually checked for convergence using Tracer v1.7 (Rambaut et al. 2018), and individual parameters had an effective sample size over 200. Phylogenetic trees estimated in BEAST2 were summarized in TreeAnnotator v2.6.2 (Bouckaert et al. 2014), generating a maximum clade credibility tree. A custom script was used to extract heights (age) for each node, including 95% confidence intervals. Demographic trends based on the Extended Bayesian Skyline Plots were generated and visualised in R.
Results
Sequencing and analysis of mitogenomes
Modern mitochondrial genomes of C. auratus had an average coverage of 286× (151–1,704×), and P. major mitogenomes had an average coverage of 270× (38×–1,131−). We generated and skim-sequenced 62 libraries out of the 90 successfully extracted ancient specimens. For these libraries levels of endogenous DNA ranged between 0.003% and 58% (Fig. S5 & Table S4). Based on this initial screening, we sequenced additional reads for 26 libraries, and obtained 26 ancient mitogenomes with an average coverage of 5.02×–55.7× (Fig. S6). MapDamage profiles of sequences obtained from ancient specimens showed the usual fragmentation patterns and elevated number of C-T and A-G misincorporations at either read-end that are associated with authentic ancient DNA (Fig. S7). The majority (n = 22) of the mitochondrial genomes originated from middens associated with early Māori period (1280–1450), with four mitogenomes from the late Māori period (1450–1800) (Table S4).
Mitochondrial genome variation
In total, 472 polymorphic sites resulted in 233 unique haplotypes (Table 1). The overall haplotype diversity was high (h = 0.972) and ranged between 0.932 and 0.989 among populations. High haplotype diversity was also observed in the rarefaction curve (Fig. S8). Nucleotide diversity (π = 0.0042–0.0055) and genetic diversity (θ = 54.47–64.35) of modern sequences was consistent across populations (Table 1). We did not detect significant differences in nucleotide diversity between sampled populations (Table S5). Tajima’s D was positive for the total dataset and showed varied between sample locations (Table 1). Diversity estimates were similar compared to the total data and modern dataset (Table 1), with no significant difference in nucleotide diversity between modern and ancient sequences (p = 0.343).
Phylogeographic structure and demographic history
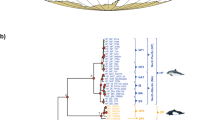
The haplotype genealogy showed that mitochondrial sequences clustered in two major mitochondrial clades (Figs. 2 and S9), with a sequence divergence (DA) of 0.84%. Eight minor clades were subsequently identified (Fig. 2). The hierarchical AMOVA showed no significant differences between sampled populations (p = 0.764). There was also no significant mitochondrial structure observed between two genetic clusters identified using whole-genome data obtained from the high-throughput sequences used for this study (p = 0.104) (Northern cluster: Northland, Hauraki, Bay of Plenty, East Cape, Gisborne, and Southern cluster: Hawke’s Bay, Tasman Bay, Karamea Bight, Kapiti Coast, and West Coast) (Fig. S9) (Oosting et al. in prep.). Less than 0.01% of the variation observed between sample locations and nuclear clusters, implying all samples could be treated as single population. There was also no change in the relative abundance of haplotypes between clades over time, p = 0.723 and p = 0.737 for the major and minor clades respectively.
Solid black circles show two major clades, and dashed circles show eight minor clades. Colours indicate sample location. Each grey circle represents a single nucleotide change. The black bars indicate a section between the major clades representing 108 nucleotide changes. See Fig. 3 for an unmodified version of the figure that shows the distance between the two major clades.
Phylogenetic trees estimated using the Yule and Coalescent Exponential Population models also showed the presence of two highly diverged clades within C. auratus (Fig. 3 and Table 2). Branches older than 40,000 years were highly resolved (posterior > 0.99), while younger branches were poorly supported (posterior < 0.5). Based on the Coalescent Exponential Population model, the two major clades separated approximately 650,000 years ago (T1) during the mid-Pleistocene (Fig. 3 and Table 2). The Yule model suggested that C. auratus and P. major (outgroup) diverged around 727,000 years ago (T0, 95%CI = 591,000–865,000), which was not significantly different from when the major clades observed in C. auratus diverged. Nodes T2 and T3 were estimated at ~150,000 years ago. A large radiation of new lineages was observed in all models between 7,000 and 35,000 years ago (T4) (Fig. 3 and Table 2), consistent with an expansion (Slatkin and Hudson 1991). A population expansion was further supported by the Coalescent Exponential Population model where the growth rate was significantly different from 0 (mean = 3.7238e−6, CI95% = 1.3998e−6–6.2791e−6). The Extended Bayesian Skyline Plot analysis also showed that C. auratus experienced an exponential population increase approximately 8,000 years ago (Fig. 4). The analyses estimated that the effective female population size increased by a factor of hundred somewhere after the LGM.
See Table 2 for time estimates for each node. This figure is produced using the Yule prior using modern, ancient, and outgroup samples. Phylogenetic trees generated using the other priors showed the same results, where the older nodes were consistently resolved, and thus shown in a single figure. T0 was not estimated for models without an outgroup. The haplotype genealogy is the unmodified version of Fig. 2.
Discussion
We used contemporary and ancient mitochondrial genomes to investigate the phylogeographic and demographic history of C. auratus. Below, we discuss how glacial cycles had a major impact on the phylogeographic structure and population abundance of C. auratus. Finally, we will discuss the effect of historic structure on the contemporary estimates of genetic diversity, and the potential effect of exploitation on genetic diversity.
Phylogeographic structure
The most prominent observation was the presence of two distinct mitochondrial clades among C. auratus individuals in New Zealand (Fig. 2). These clades date to a shared common ancestor approximately 650,000 years ago, which is during the mid-Pleistocene (Table 2). The large confidence intervals around the date of the split and uncertainty around clock rates make it difficult to accurately determine the time point of this event (Table 2). It is important to note that that time to the last known common ancestor indicates the maximum divergence time for the two clades. Barriers to gene flow can become established much later than the mutation and divergence process of two lineages, particularly if the ancestral population was large or highly structured populations (Avise 2000). While this phylogenetic split suggests a history of strong population structuring, we did not detect any significant population structure between sample locations, (AMOVA, p = 0.764). This two-clade phylogeographic structure typically arises when populations have a significant level of genetic divergence after been in isolation for long periods of time, and have subsequently mixed when barriers to gene flow disappeared (Avise et al. 1987). The subsequent mixing of individuals from both two clades is supported by nuclear data (Oosting et al. in prep.). Glacial cycles throughout the mid-Pleistocene most likely disrupted gene flow with physical barriers to migration (Elderfield et al. 2012). During glacial periods the marine environment would have experienced drastic changes as reductions in sea levels resulted in the formation of land bridges isolating once connected populations (Bowen et al. 2016). This would have had a substantial impact on C. auratus populations and gene flow, and was likely an important factor in the formation of the distinct mitochondrial clades (Fig. 2). It is less clear where the geographical separation of these two populations may have occurred. There are various demographic scenarios that could explain the observed ice-age phylogeographic pattern.
Haplotypes from both clades have been detected in Australian samples using mitochondrial control region sequences (unpublished data, Fig. S10), showing trans-Tasman dispersal has occurred. It is possible that the clades diverged on one side of the Tasman Sea and then dispersed to the other side. Both landmasses have coastal environments that have changed significantly as sea level changes formed land bridges over the last 650,000 years, providing the opportunity for population separation and independent lineage formation on either side of the Tasman Sea (Dlabola et al. 2015; Peltier and Fairbanks 2006; Williams et al. 2018). Other possible explanations for the presence of both clades in New Zealand and Australia are more complex, involving repeated bidirectional dispersal across the Tasman Sea. However, we argue that is most likely the two clades were established prior to the arrival of C. auratus to New Zealand and Australia, and diverged in the Indo-Pacific region. First, the estimated age of the split between the clades was not significantly different from the 727,000 year old divergence estimate between C. auratus and P. major (Table 2) (Supplementary information 1), a species that occurs in the South Chinese Sea (Tabata and Taniguchi 2000). The Indo-Pacific is a likely location for where the common ancestor of both species occurred. Second, similar patterns of phylogeographic structure have been observed in other species occurring in the Indo-Pacific region, where glacial cycles reduced sea levels by approximately 120 m, exposing land and thereby separating bodies of water (Bowen et al. 2016; Dlabola et al. 2015; Peltier and Fairbanks 2006). For example, populations of scalloped hammerhead Sphyrna lewini, bigscale soldierfish Myripristis berndti, and peacock hind Cephalopholis argus are thought to have been separated on either side of the Indo-Pacific barrier (what is now Indonesia), after which distinct mitochondrial lineages intermixed during secondary contact (Craig et al. 2007; Duncan et al. 2006; Gaither et al. 2011). Such population fragmentation in the Indo-Pacific ancestral species would pre-date the colonisation and speciation of C. auratus in New Zealand and Australia. Overall, this appears to be the most parsimonious explanation for the observed pattern of distinct mitochondrial haplotypes in C. auratus.
The haplotype genealogy and phylogenetic trees also showed the presence of eight smaller lineages (minor clades in Fig. 2), which likely diverged over the last 140,000–170,000 years (Fig. 3 and Table 2). Following the same logic as above, these lineages could have diverged through multiple range expansions and contractions throughout the Pleistocene glacial-interglacial cycles as sea levels and temperatures fluctuated over this period (Barrows et al. 2007). Reductions in sea levels and ocean temperatures likely formed isolated pockets of suitable habitat for inshore species such as C. auratus (Dlabola et al. 2015). It is possible that these lineages were established when C. auratus had already dispersed to New Zealand. Strong phylogeographic structuring has also been observed in New Zealand gastropods (Cominella spp), a species with limited dispersal capabilities around New Zealand (Dohner et al. 2018; Fleming et al. 2018). In contrast, species with a broader range of suitable habitats such as tarakihi (Nemadactylus macropterus) do not show evidence of phylogeographic structuring (Papa et al. 2021). Range contractions and phylogeographic structuring during the Otiran glacial cycles (74–14 Kya) have also been reported for a wide variety of terrestrial species in New Zealand (Rawlence et al. 2012; Wallis and Trewick 2009). Mitochondrial genomes from Australian samples could be used in the future to corroborate whether these minor clades were established in New Zealand waters. If the minor clades were established in New Zealand, we would expect to see a different phylogeographic pattern in the Australian samples, suggestive of separate demographic histories. It should be noted that while specific physical barriers to gene flow are often used to explain distinct phylogeographic lineages, genetic differentiation in species with large effective population sizes can also arise in populations with either a widespread distribution or limited spawning opportunities (Hogner et al. 2012; Webb et al. 2011).
Exponential population expansion after the last glacial period
Demographic trends suggest that C. auratus went through an exponential population size increase approximately 8000 years ago, which generally coincidences with the Pleistocene-Holocene boundary (Fig. 4) (Walker et al. 2009). The presence of historic structure violates a model assumption of panmixia under which the Extended Bayesian Skyline Plot analyses is performed (Villanea et al. 2020). However, the historic structure has had no significant effect on the analyses as runs performed on each clade separately showed the same demographic trend (Fig. S11). We also confirmed that excluding the ancient sequences from the analyses did not influence the observed patterns in population abundance through time (Fig. S12). Given the uncertainty that can be involved in estimating mutation rates (Ho and Phillips 2009), and the estimating the timing of demographic events (Dos Reis and Yang 2013), the maximum 8000 year-old population increase appears to coincide with a post LGM expansion (29–19 Kya). We hypothesize that the population abundance of C. auratus exponentially increased somewhere after the LGM as sea levels rose and the climate warmed (Blunier and Brook 2001; Clark et al. 2009; Newnham et al. 2013; Pahnke et al. 2003). This population expansion was likely facilitated by an increase in suitable habitat, as sea level rise would have extended the available habitat on the continental shelf. Chrysophrys auratus most commonly occupy waters up to 50 m, with 200 m considered as their maximum depth (Parsons et al. 2014). Suitable habitat would have been limited and most likely more fragmented during the LGM because the edge of the continental shelf runs relatively close to parts of the New Zealand coast. This can be observed using a rough extrapolation of the suitable habitat using the modern-day bathymetry of New Zealand and lowering the sea level by 120 m (Fig. S13). As sea levels have risen over 120 m in the last 20,000 years (Dlabola et al. 2015; Peltier and Fairbanks 2006), a large proportion of the continental shelf would have become submerged (Fig. S13). Second, the average sea temperature around New Zealand increased by approximately 6 °C (rising from ~10 °C to ~16 °C) after the LGM (Barrows et al. 2007). Increased sea temperature could have facilitated ecological changes (e.g. increased primary production) which may have had a cascading effect through the food web (Cabrera et al. 2022), contributing to the population expansion observed in C. auratus. The southern range of contemporary populations of C. auratus are primarily dictated by temperature (Parsons et al. 2014). It is logical to assume that the post-LGM increase in ocean temperature would have resulted in a southward expansion of C. auratus. With sea surface temperatures expected to rise with 0.8–2.5 degrees in New Zealand by 2100 (Law et al. 2018), C. auratus will likely continue to expand their distribution southwards. This idea is supported by ecological niche modelling the distribution of C. auratus under different climate scenarios (Brooks 2019), which predicts the species will expand southwards down the West Coast of the South Island under all future climate scenarios.
Mitogenomic variation
The observed high levels of genetic variation are likely the product of a large effective population size and historic genetic structure. Fish are known to maintain large effective population sizes, which means they can retain high levels of genetic diversity (Ellegren and Galtier 2016; Frankham 1996). This is consistent with exponential population increase C. auratus experienced (Fig. 4). High haplotype diversity (h = 0.972) and the rarefaction curve show that our sample size was unable to capture most of the mitochondrial diversity present in the population (Table 1) (Fig. S8). Nucleotide diversity (π = 0.005) was very high compared to Gadus morhua (π = 0.002) (Martínez-García et al. 2021), a species that has maintained high levels of genetic diversity despite being heavily exploited (Pinsky et al. 2021). This high nucleotide diversity in C. auratus had previously been reported based on analyses of mitochondrial DNA control region sequences (Fig. S14) (Papa et al. 2021).
The relatively high nucleotide diversity (π = 0.005) in C. auratus can in part be attributed to historic genetic structure. While the mitochondrial clades show no significant genetic structure between contemporary sample locations or clusters based on nuclear loci (AMOVA, p = 0.764 and 0.104 respectively), their 650,000-year-old lineages are influencing contemporary diversity estimates. Summary statistics such as nucleotide diversity, genetic diversity and Tajima’s D are estimated using the number of segregating sites. The number of segregating sites is relatively high in C. auratus because of the presence of the two highly diverged clades which do not necessarily reflect current day demographics. This may imply that diversity estimates do not correlate well with long-term effective population size (Ellegren and Galtier 2016; Frankham 1996), however, we acknowledge that a number of potentially interacting processes in addition to population size also influence the genetic diversity of wild populations (Bernatchez 2016). For example, the nucleotide diversity in G. morhua (π = 0.002) implies that the population is much smaller than that of C. auratus (π = 0.005) (Martínez-García et al. 2021). However, estimates of effective female population size in Atlantic cod show similar or larger estimates compared to C. auratus (Martínez-García et al. 2021) (Fig. 4). Other mechanisms such as selection or life history traits can influence the relation between diversity and population size (Ellegren and Galtier 2016), however we believe that the historic structure plays a significant role in the comparatively high diversity observed in C. auratus. The mitochondrial genome is also a single marker and only captures one gene tree which represents the evolutionary history of the species. The historic structure also effected Tajima’s D, making the interpretation of this summary statistic difficult. This can be observed when comparing the Tajima’s D estimate of the total population with those of the individual clades (Table S6). Here, significant negative Tajima’s D estimates for separate clades suggested a population expansion (TD = −2.179 & −1.66), which is consistent with the expansion observed in the Extended Bayesian Skyline Plot (Fig. 4).
The ancient mitochondrial genomes provided a direct test for changes in genetic diversity over the last 750 years. First, we did not find evidence for changes in the relative abundance of haplotypes (p = 0.723), suggesting there was no recent selective advantage for specific mitochondrial haplotypes. Second, nucleotide diversity has not significantly changed (p = 0.349), suggesting no detectable loss of genetic variation since the onset of fishing in New Zealand. This is despite lower temperatures during the Little Ice Age (which could have influenced population abundance) (Lorrey et al. 2014; Waters et al. 2017), the potential impacts of pre-European fishing practices (Leach 2006; Leach and Davidson 2000), and large reductions in population sizes due to industrial-scale harvesting over the last hundred years (Fisheries New Zealand 2018). The ability to detect a genetic signature from a bottleneck depends on the severity, duration, and the amount time since the event (Peery et al. 2012). This would imply that the bottleneck was not severe or long enough for it to effect genetic diversity, or that more time must pass before its effect can be observed in the population. It is also possible that the effects of commercial fishing are not yet detectable in the mitochondrial genome due to low statistical power. Loss of genetic variation was observed in the Tasman Bay using microsatellites (Hauser et al. 2002), yet microsatellites are often selected based on variability (i.e. number of alleles), which implies that heterozygosity estimates can be inflated (Brandström and Ellegren 2008; Väli et al. 2008). Their high variability could make microsatellites more sensitive for detecting reductions in genetic variation. The use of a single marker for this study has limited the power to detect the recent changes in genetic diversity. The addition of nuclear markers will be beneficial in addressing the potential effects of exploitation over the last 750 years. Unfortunately, our approach of shotgun sequencing ancient DNA libraries and low levels of endogenous DNA meant that it was difficult to obtain enough nuclear reads for re-sequencing genomes from past populations. Future efforts may improve this by using capture-based enrichment to extract and genotype nuclear markers to test for changes in genetic variation (Carpenter et al. 2013). Our findings do correspond with similar observations based on nuclear markers in Atlantic cod (G. morhua), Atlantic bluefin tuna (Thunnus thynnus), and Pacific herring (Clupea pallasi), which showed that exploitation has not yet led to detectable losses of genetic diversity (Andrews et al. 2021; Martínez-García et al. 2021; Moss et al. 2016; Pinsky et al. 2021; Speller et al. 2012). Together these studies suggest that heavily exploited species do not appear to have lost any appreciable amounts of genetic diversity. These finding suggest that as stocks are allowed to rebuild, they might have retained much of their pre-exploitation levels of diversity. If correct, this is an encouraging finding for the future of fisheries management. This is also particularly important for resilience as fisheries population adapt to climate change.
Conclusion
Our findings suggest that Pleistocene glacial cycles have played an important role in the phylogeographic history of C. auratus. The end of the last glacial cycle resulted in an exponential population expansion, likely facilitated by increased ocean temperatures and areas of suitable habitat. Current climate change is likely to continue to challenge fisheries species. In some cases, this could result in the expansion of suitable habitat, as is supported by ecological niche modelling of C. auratus (Brooks 2019). Finally, we did not find any evidence of a loss of genetic diversity based on data from the mitochondrial genome since the onset of high fishing pressure. Our study has provided new insights into our understanding the phylogeography of a fisheries species and the retention of genetic diversity despite many years of exploitation.
Data archiving
Mitochondrial genome BAM files have been deposited in the European Nucleotide Archive (PRJEB49332).
References
Alexander A, Steel D, Hoekzema K, Mesnick SL, Engelhaupt D, Kerr I et al. (2016) What influences the worldwide genetic structure of sperm whales (Physeter macrocephalus)? Mol Ecol 25(12):2754–2772
Aljanabi SM, Martinez I (1997) Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res 25(22):4692–4693
Andrews AJ, Puncher GN, Bernal-Casasola D, Di Natale A, Massari F, Onar V et al. (2021) Ancient DNA SNP-panel data suggests stability in bluefin tuna genetic diversity despite centuries of fluctuating catches in the eastern Atlantic and Mediterranean. Sci Rep. 11(1):20744
Ashton D (2013). Population Genetics of New Zealand Pagrus auratus and Genetic Variation of an Aquaculture Broodstock. Master thesis, Victoria University of Wellington, Wellington.
Avise JC (2000). Phylogeography: the history and formation of species. Harvard university press.
Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE et al. (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst 18(1):489–522
Barrows TT, Juggins S, De Deckker P, Calvo E, Pelejero C (2007) Long‐term sea surface temperature and climate change in the Australian–New Zealand region. Paleoceanography 22(2):1:17
Bernatchez L (2016) On the maintenance of genetic variation and adaptation to environmental change: considerations from population genomics in fishes. J Fish Biol 89(6):2519–2556
Blunier T, Brook EJ (2001) Timing of millennial-scale climate change in antarctica and greenland during the last glacial period. Science 291(5501):109–112
Boessenkool S, Hanghøj K, Nistelberger HM, Der Sarkissian C, Gondek AT, Orlando L et al. (2017) Combining bleach and mild predigestion improves ancient DNA recovery from bones. Mol Ecol Resour 17(4):742–751
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D et al. (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10(4):e1003537
Bowen BW, Gaither MR, DiBattista JD, Iacchei M, Andrews KR, Grant WS et al. (2016) Comparative phylogeography of the ocean planet. Proc Natl Acad Sci USA 113(29):7962–7969
Bowen BW, Muss A, Rocha LA, Grant WS (2006) Shallow mtDNA Coalescence in Atlantic Pygmy Angelfishes (Genus Centropyge) Indicates a Recent Invasion from the Indian Ocean. J Heredity 97(1):1–12
Brandström M, Ellegren H (2008) Genome-wide analysis of microsatellite polymorphism in chicken circumventing the ascertainment bias. Genome Res 18(6):881–887
Broad Institute (2020). Picard toolkit. GitHub Repository. http://broadinstitute.github.io/picard/.
Brooks A (2019). Using Ecological Niche Modelling to Predict Climate Change Responses of Ten Key Fishery Species in Aotearoa New Zealand. Master thesis, Victoria University of Wellington.
Cabrera AA, Schall E, Bérubé M, Anderwald P, Bachmann L, Berrow S et al. (2022) Strong and lasting impacts of past global warming on baleen whales and their prey. Glob Change Biol 28(8):2657–2677
Carpenter ML, Buenrostro JD, Valdiosera C, Schroeder H, Allentoft ME, Sikora M et al. (2013) Pulling out the 1%: whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am J Hum Genet 93(5):852–864
Catanach A, Crowhurst R, Deng C, David C, Bernatchez L, Wellenreuther M (2019) The genomic pool of standing structural variation outnumbers single nucleotide polymorphism by threefold in the marine teleost Chrysophrys auratus. Mol Ecol 28(6):1210–1223
Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B et al. (2009) The last glacial maximum. science 325(5941):710–714
Cooper A, Poinar HN (2000) Ancient DNA: do it right or not at all. Science 289(5482):1139–1139
Craig MT, Eble JA, Bowen BW, Robertson DR (2007) High genetic connectivity across the Indian and Pacific Oceans in the reef fish Myripristis berndti (Holocentridae). Mar Ecol Prog Ser 334:245–254
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat methods 9(8):772–772
Ding S, Mishra M, Wu H, Liang S, Miyamoto MM (2018) Characterization of hybridization within a secondary contact region of the inshore fish, Bostrychus sinensis, in the East China Sea. Heredity 120(1):51–62
Dlabola E, Wilson G, Gorman A, Riesselman C, Moy C (2015) A post-glacial relative sea-level curve from Fiordland, New Zealand. Glob Planet Change 131:104–114
Dohner M, Phillips NE, Ritchie PA (2018) Fine-scale genetic structure across a New Zealand disjunction for the direct-developing intertidal whelk Cominella maculosa (Gastropoda: Buccinidae). Biol J Linn Soc 123(3):593–602
Dos Reis M, Yang Z (2013) The unbearable uncertainty of Bayesian divergence time estimation. J Syst Evolution 51(1):30–43
Duncan K, Martin A, Bowen B, De Couet H (2006) Global phylogeography of the scalloped hammerhead shark (Sphyrna lewini). Mol Ecol 15(8):2239–2251
Elderfield H, Ferretti P, Greaves M, Crowhurst S, McCave IN, Hodell D et al. (2012) Evolution of ocean temperature and ice volume through the mid-Pleistocene climate transition. science 337(6095):704–709
Ellegren H, Galtier N (2016) Determinants of genetic diversity. Nat Rev Genet 17(7):422–433
Ferrari G, Cuevas A, Gondek-Wyrozemska AT, Ballantyne R, Kersten O, Pálsdóttir AH et al. (2021) The preservation of ancient DNA in archaeological fish bone. J Archaeological Sci 126:105317
Fisheries New Zealand (2018). Fisheries Assessment Plenary, May 2018: stock assessments and stock status. Compiled by the Fisheries Science Group, Fisheries New Zealand, Wellington, New Zealand. 1660p.
Fleming AM, Dohner MM, Phillips NE, Ritchie PA (2018) Genetic connectivity among populations of two congeneric direct-developing whelks varies across spatial scales. NZ J Mar Freshw Res 52(1):100–117
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10(6):1500–1508
Gaither MR, Bowen BW, Bordenave T-R, Rocha LA, Newman SJ, Gomez JA et al. (2011) Phylogeography of the reef fish Cephalopholis argus (Epinephelidae) indicates Pleistocene isolation across the Indo-Pacific Barrier with contemporary overlap in the Coral Triangle. BMC Evolut Biol 11(1):1–16.
Gillooly JF, Allen AP, West GB, Brown JH (2005) The rate of DNA evolution: Effects of body size and temperature on the molecular clock. P Natl Acad Sci USA 102(1):140–145
Gu S, Yi M-R, He X-B, Lin P-S, Liu W-H, Luo Z-S et al. (2021) Genetic diversity and population structure of cutlassfish (Lepturacanthus savala) along the coast of mainland China, as inferred by mitochondrial and microsatellite DNA markers. Regional Stud Mar Sci 43:101702
Harpending HC, Batzer MA, Gurven M, Jorde LB, Rogers AR, Sherry ST (1998) Genetic traces of ancient demography. Proc Natl Acad Sci 95(4):1961–1967
Hauser L, Adcock GJ, Smith PJ, Ramirez JH, Carvalho GR (2002) Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc Natl Acad Sci USA 99(18):11742–11747
Heino M, Diaz Pauli B, Dieckmann U (2015) Fisheries-Induced Evolution. Annu Rev Ecol Evol S 46:461–480
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405(6789):907–913
Ho SYW, Phillips MJ (2009) Accounting for Calibration Uncertainty in Phylogenetic Estimation of Evolutionary Divergence Times. Syst Biol 58(3):367–380
Hogner S, Laskemoen T, Lifjeld JT, Porkert J, Kleven O, Albayrak T et al. (2012) Deep sympatric mitochondrial divergence without reproductive isolation in the common redstart P hoenicurus phoenicurus. Ecol evolution 2(12):2974–2988
Jombart T, Archer F, Schliep K, Kamvar Z, Harris R, Paradis E et al. (2017) apex: phylogenetics with multiple genes. Mol Ecol Resour 17(1):19–26
Jonsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L (2013) mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29(13):1682–1684
Kircher M (2012) Analysis of high-throughput ancient DNA sequencing data. Ancient DNA. Springer, 197–228
Knapp M, Clarke AC, Horsburgh KA, Matisoo-Smith EA (2012) Setting the stage–building and working in an ancient DNA laboratory. Ann Anat-Anatomischer Anz 194(1):3–6
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2016) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evolution 34(3):772–773
Law CS, Rickard GJ, Mikaloff-Fletcher SE, Pinkerton MH, Behrens E, Chiswell SM et al. (2018) Climate change projections for the surface ocean around New Zealand. NZ J Mar Freshw Res 52(3):309–335
Leach F (2006) Fishing in pre-European New Zealand. Archaeofauna 15(15):15-+
Leach F, Davidson J (2000) Pre-European catches of snapper (Pagrus auratus) in northern New Zealand. J Archaeological Sci 27(6):509–522
Lorrey A, Fauchereau N, Stanton C, Chappell P, Phipps S, Mackintosh A et al. (2014) The Little Ice Age climate of New Zealand reconstructed from Southern Alps cirque glaciers: a synoptic type approach. Clim Dyn 42(11):3039–3060
Martínez-García L, Ferrari G, Oosting T, Ballantyne R, van der Jagt I, Ystgaard I et al (2021) Historical demographic processes dominate genetic variation in Ancient Atlantic Cod Mitogenomes. Front Ecol Evol 9(342):1:14
Matschiner M (2016) Fitchi: haplotype genealogy graphs based on the Fitch algorithm. Bioinformatics 32(8):1250–1252
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303
Meyer M, Kircher M (2010) Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc 2010(6):prot5448. pdb
Miya M, Kawaguchi A, Nishida M (2001) Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evolution 18(11):1993–2009
Moss ML, Rodrigues AT, Speller CF, Yang DYY (2016) The historical ecology of Pacific herring: Tracing Alaska Native use of a forage fish. J Archaeol Sci-Rep. 8:504–512
Nam B-H, Yoo D, Kim Y-O, Park JY, Shin Y, Shin G-H et al. (2019) Whole genome sequencing reveals the impact of recent artificial selection on red sea bream reared in fish farms. Sci Rep. 9(1):6487
Newnham R, McGlone M, Moar N, Wilmshurst J, Vandergoes M (2013) The vegetation cover of New Zealand at the Last Glacial Maximum. Quat Sci Rev 74:202–214
Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol evolution 32(1):268–274
Oosting T, Hilario E, Wellenreuther M, Ritchie PA (2020) DNA degradation in fish: Practical solutions and guidelines to improve DNA preservation for genomic research. Ecol Evol 10(16):8643–8651
Oosting T, Star B, Barrett JH, Wellenreuther M, Ritchie PA, Rawlence NJ (2019) Unlocking the potential of ancient fish DNA in the genomic era. Evol Appl 12(8):1513–1522
Pahnke K, Zahn R, Elderfield H, Schulz M (2003) 340,000-Year Centennial-Scale Marine Record of Southern Hemisphere Climatic Oscillation. Science 301(5635):948–952
Papa Y, Halliwell AG, Morrison MA, Wellenreuther M, Ritchie PA (2021) Phylogeographic structure and historical demography of tarakihi (Nemadactylus macropterus) and king tarakihi (Nemadactylus n.sp.) in New Zealand. N Z J Marine Freshwater Res 48(1):1–25
Paradis E (2010) pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26(3):419–420
Parsons D, Sim-Smith C, Cryer M, Francis M, Hartill B, Jones E et al. (2014) Snapper (Chrysophrys auratus): a review of life history and key vulnerabilities in New Zealand. NZ J Mar Freshw Res 48(2):256–283
Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet‐Bëer E, Robinson S et al. (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol 21(14):3403–3418
Peltier W, Fairbanks RG (2006) Global glacial ice volume and Last Glacial Maximum duration from an extended Barbados sea level record. Quat Sci Rev 25(23-24):3322–3337
Pinsky ML, Eikeset AM, Helmerson C, Bradbury IR, Bentzen P, Morris C et al. (2021) Genomic stability through time despite decades of exploitation in cod on both sides of the Atlantic. Proc Natl Acad Sci 118(15):e2025453118
Qiu F, Li H, Lin H, Ding S, Miyamoto MM (2016) Phylogeography of the inshore fish, Bostrychus sinensis, along the Pacific coastline of China. Mol Phylogenet Evol 96:112–117
R Core Team (2013). R: A language and environment for statistical computing.
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67(5):901
Rawlence NJ, Metcalf JL, Wood JR, Worthy TH, Austin JJ, Cooper A (2012) The effect of climate and environmental change on the megafaunal moa of New Zealand in the absence of humans. Quat Sci Rev 50:141–153
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE et al. (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol evolution 34(12):3299–3302
Sato Y, Miya M, Fukunaga T, Sado T, Iwasaki W (2018) MitoFish and mifish pipeline: a mitochondrial genome database of fish with an analysis pipeline for environmental DNA metabarcoding. Mol Biol Evolution 35(6):1553–1555
Schubert M, Ermini L, Der Sarkissian C, Jónsson H, Ginolhac A, Schaefer R et al. (2014) Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc 9(5):1056
Seersholm FV, Cole TL, Grealy A, Rawlence NJ, Greig K, Knapp M et al. (2018) Subsistence practices, past biodiversity, and anthropogenic impacts revealed by New Zealand-wide ancient DNA survey. Proc Natl Acad Sci USA 115(30):7771–7776
Shafer AB, Cullingham CI, Cote SD, Coltman DW (2010) Of glaciers and refugia: a decade of study sheds new light on the phylogeography of northwestern North America. Mol Ecol 19(21):4589–4621
Shin GH, Shin Y, Jung M, Hong JM, Lee S, Subramaniyam S et al. (2018) First draft genome for red sea bream of family sparidae. Front Genet 9(643):643
Skog A, Zachos F, Rueness E, Feulner P, Mysterud A, Langvatn R et al. (2009) Phylogeography of red deer (Cervus elaphus) in Europe. J Biogeogr 36(1):66–77
Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129(2):555–562
Smith I (2013) Pre-European Maori exploitation of marine resources in two New Zealand case study areas: species range and temporal change. J R Soc NZ 43(1):1–37
Smith I, James-Lee T (2010). Data for an Archaeozoological Analysis of Marine Resource Use in Two New Zealand Study Areas. Otago DoAAUo. http://hdl.handle.net/10523/5945.
Speller CF, Hauser L, Lepofsky D, Moore J, Rodrigues AT, Moss ML et al. (2012) High potential for using DNA from ancient herring bones to inform modern fisheries management and conservation. PLoS One 7(11):e51122
Sun D, Ge Y, Cheng Q (2019) Genetic diversity of eight wild populations of Pampus argenteus along the coast of China inferred from fifteen polymorphic microsatellite markers. Bra J Oceanogr 67:1–11
Tabata K, Taniguchi N (2000) Differences between Pagrus major and Pagrus auratus through mainly mtDNA control region analysis. Fish Sci 66(1):9–18
Teske PR, Von der Heyden S, McQuaid CD, Barker NP (2011) A review of marine phylogeography in southern Africa. South Afr J Sci 107(5-6):43–53.
Väli Ü, Einarsson A, Waits L, Ellegren H (2008) To what extent do microsatellite markers reflect genome‐wide genetic diversity in natural populations? Mol Ecol 17(17):3808–3817
Villanea FA, Kitchen A, Kemp BM (2020) Applications of Bayesian skyline plots and approximate Bayesian computation for human demography. Hum Biol 91(4):279–296
Walker M, Johnsen S, Rasmussen SO, Popp T, Steffensen JP, Gibbard P et al. (2009) Formal definition and dating of the GSSP (Global Stratotype Section and Point) for the base of the Holocene using the Greenland NGRIP ice core, and selected auxiliary records. J Quat Sci: Published Quat Res Assoc 24(1):3–17
Wallis GP, Trewick SA (2009) New Zealand phylogeography: evolution on a small continent. Mol Ecol 18(17):3548–3580
Wallis GP, Waters JM, Upton P, Craw D (2016) Transverse Alpine Speciation Driven by Glaciation. Trends Ecol Evolution 31(12):916–926
Wang X, Que P, Heckel G, Hu J, Zhang X, Chiang CY et al. (2019) Genetic, phenotypic and ecological differentiation suggests incipient speciation in two Charadrius plovers along the Chinese coast. BMC Evol Biol 19(1):135
Wang Z-D, Liao J, Huang C-Q, Long S-S, Zhang S, Guo Y-S et al. (2018) Significant genetic differentiation of Gobiopterus lacustris, a newly recorded transparent goby in China. Mitochondrial DNA Part A 29(5):785–791
Waters JM, Fraser CI, Maxwell JJ, Rawlence NJ (2017) Did interaction between human pressure and Little Ice Age drive biological turnover in New Zealand? J Biogeogr 44(7):1481–1490
Webb WC, Marzluff JM, Omland KE (2011) Random interbreeding between cryptic lineages of the Common Raven: evidence for speciation in reverse. Mol Ecol 20(11):2390–2402
Wellenreuther M, Le Luyer J, Cook D, Ritchie PA, Bernatchez L (2019) Domestication and Temperature Modulate Gene Expression Signatures and Growth in the Australasian Snapper Chrysophrys auratus. G3 (Bethesda) 9(1):105–116
Williams AN, Ulm S, Sapienza T, Lewis S, Turney CS (2018) Sea-level change and demography during the last glacial termination and early Holocene across the Australian continent. Quat Sci Rev 182:144–154
Zhao D, Li Q, Kong L, Yu H (2017) Cryptic diversity of marine gastropod Monodonta labio (Trochidae): did the early Pleistocene glacial isolation and sea surface temperature gradient jointly drive diversification of sister species and/or subspecies in the Northwestern Pacific? Mar Ecol 38(4):e12443
Acknowledgements
We thank the Royal Society of New Zealand Marsden Fund (Grant/Award Number: 16-VUW-040), European Union’s Horizon 2020 4-OCEANS Synergy grant agreement no. 951649 and the University of Otago for funding this research. We thank Museum of New Zealand Te Papa Tongarewa (Carolyn McGill) for permission to sample archaeological C. auratus bones. Also, Ministry of Primary Industries (MPI), the National Institute of Water and Atmospheric Research (NIWA), Gisborne Tatapouri Sports Fishing Club, Hawkes Bay Sports Fishing Club, Petone Working Man’s Club, and Leah Kemp for providing samples or sampling opportunities for modern C. auratus. Thank you to Ngāti Kuia, Ngāi Tai Ki Tāmaki, Ngāti Toa, and Ngāti Koata for supporting this research. We thank the Norwegian Sequencing Centre, UiO (https://www.sequencing.uio.no) for sequencing of the ancient specimens.
Author information
Authors and Affiliations
Contributions
TO was responsible for the overall design of the study, sample collection and DNA extraction of the modern samples, performing all analyses, and the lead over the writing this manuscript. LMG contributed by helping out partitioning of the mitochondrial genome and performing analyses in BEAST2. GF supervised and performed library preparation and sequencing of the ancient samples. AJFV and LS performed DNA extractions for the ancient samples. AJFV also assisted in sampling of the ancient samples. NJR was responsible for funding acquisition, the overall collection of the ancient samples, corresponded with relevant Māori iwi regarding the use of culturally significant materials, providing feedback on analyses, and helped write the manuscript. MW was responsible for funding acquisition, supervision, and provided feedback on analyses and helped write the manuscript. BS contributed by helping with the overall design of this study, supervised library preparation and sequencing of ancient samples, and helped write the manuscript. PR was responsible for funding acquisition, supervision, project administration, contributed by helping with the overall design of this study, and helped write the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Jane Hughes.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oosting, T., Martínez-García, L., Ferrari, G. et al. Mitochondrial genomes reveal mid-Pleistocene population divergence, and post-glacial expansion, in Australasian snapper (Chrysophrys auratus). Heredity 130, 30–39 (2023). https://doi.org/10.1038/s41437-022-00579-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-022-00579-1