Abstract
Silphium integrifolium (Asteraceae) has been identified as a candidate for domestication as a perennial oilseed crop and is assumed to have sporophytic self-incompatibility system—the genetic basis of which is not well understood in the Asteraceae. To address this gap, we sought to map the genomic location of the self-recognition locus (S-locus) in this species. We used a biparental population and genotyping-by-sequencing to create the first genetic linkage map for this species, which contained 198 SNP markers and resolved into the correct number of linkage groups. Then we developed a novel crossing scheme and set of analysis methods in order to infer S-locus genotypes for a subset of these individuals, allowing us to map the trait. Finally, we evaluated potential genes of interest using synteny analysis with the annual sunflower (Helianthus annuus) and lettuce (Lactuca sativa) genomes. Our results confirm that S. integrifolium does indeed have a sporophytic self-incompatibility system. Our method is effective and efficient, allowed us to map the S. integrifolium S-locus using fewer resources than existing methods, and could be readily applied to other species.
Similar content being viewed by others
Introduction
Silphium integrifolium (Michx.) (wholeleaf rosinweed or silflower) is a member of the Asteraceae family native to prairies throughout the central United States. In the early 2000s, S. integrifolium was selected to be a candidate for domestication as a perennial oilseed crop by the Land Institute in Salina, Kansas (Van Tassel et al. 2017), attracting attention for its tolerance to drought, upright growth habit, and large seeds (DeHaan et al. 2016). Subsequently, S. integrifolium has been found to have a seed oil composition similar to landrace sunflower (Reinert et al. 2019) and good winter survival and persistence in a range of climates (J.H. Price and D.L. Van Tassel, pers. obs.). In addition, the yield potential of S. integrifolium populations that have undergone relatively little selection is approximately 60% the yield of advanced sunflower hybrids (Kandel et al. 2019; Schiffner et al. 2020), indicating that significant improvement is likely with continued breeding efforts. These characteristics further encourage the domestication of this species as a new crop. In the past, domestication occurred over long periods of time, due to the largely unintentional nature of early selection (Rindos 1984). With the advantage of contemporary knowledge of genetics, genomics, and breeding techniques, the amount of time necessary to domesticate a new crop could be drastically reduced. Therefore, the development of genomic resources is a crucial step in this process (Sedbrook et al. 2014). To this end, we have developed the first genetic linkage map for S. integrifolium. Among the traits for which better genetic knowledge will accelerate the domestication of S. integrifolium is self-incompatibility. Although occasional S. integrifolium individuals have been observed to produce at least some seed when self-pollinated (Reinert et al. 2020), S. integrifolium is self-incompatible, and as a member of the Asteraceae family is assumed to have a sporophytic self-incompatibility (SSI) system (Hiscock 2000), although prior to this study this fact had not been experimentally confirmed. In sporophytic systems, self-recognition is typically controlled by a single multi-allelic locus, known as the “S-locus”, with rejection of self-pollen caused by stigma recognition of S-locus gene products found in or on the pollen. Because these products are produced in the anther, pollen acceptance or rejection is determined by the diploid genotype of the male parent, rather than the haploid genotype of a given pollen grain (Hiscock and Tabah 2003). SSI alleles are also expected to display complex dominance patterns, and dominance relationships between alleles may differ from the anther to the stigma (Hiscock and Tabah 2003).
The molecular mechanisms that underlie SSI are best described in the Brassicaceae, where the female S-phenotype is determined by a receptor kinase complex known as SRK. When pollen of the same S-phenotype lands on the stigma, this kinase binds a cysteine-rich protein found in the pollen coat, known as SP11/SCR, initiating the pollen rejection response (Fujii and Takayama 2018). These two genes are tightly linked and rarely recombine; thus, they combine to form the S-locus, which is more properly thought of as an S-haplotype (Edh et al. 2009). SRK is unable to bind or is otherwise not activated by SP11/SCR proteins produced by different haplotypes, resulting in the acceptance of non-self pollen (Fujii and Takayama 2018). Although the identity of the receptor protein varies, secreted cysteine-rich proteins are also considered a promising candidate for the male determinant of SSI in the Convolvulaceae (morning glory) family (Rahman et al. 2007), and serve as the female determinant of gametophytic self-incompatibility (GSI) in the genus Papaver (poppy) (Marshall et al. 2011).
Asteraceae systems are less well understood. In Senecio squalidis, and subsequently in other Asteraceae, SRK-like sequences have been identified and cloned. However, results from S. squalidis and chicory (Cichorium intybus) indicate that they likely are not integral to S-genotype determination (Hiscock and Tabah 2003; Gonthier et al. 2013), and that the molecular control of Asteraceae SSI is quite different from the Brassicaceae system (Allen et al. 2011). However, the general model of SSI in the Brassicaceae; a tightly bound male and female specific kinase and ligand pair, may help to guide the search for the Asteraceae genes which control SSI. Efforts to map the S-locus in chicory provided a 1.8 cM QTL region but have not yet determined a molecular basis for self-incompatibility (Gonthier et al. 2013). Although efforts have been undertaken in other species, chicory represents perhaps the only example where a true Asteraceae S-locus has been definitively mapped, as opposed to other loci contributing to breakdowns in self-incompatibility (Gandhi et al. 2005; Koseva et al. 2017).
Mapping the S-locus is important for breeding efforts and for our understanding of the genetics and evolution of this critical locus. For example, the “collaborative nonself recognition” system identified in the Solanaceae (Kubo et al. 2010) has revealed that the mechanism underlying self-incompatibility can change how new S-alleles originate (Bod’ová et al. 2018, Harkness et al. 2021) and migrate across populations by favoring locally rare pollen alleles and disfavoring stylar alleles that can limit female reproduction if locally compatible pollen is rare (Castric et al. 2008, Harkness and Brandvain 2021). Identifying the basis (or bases) of SSI in the Asteraceae would help us better understand and predict features of its evolution, including its maintenance in populations with very few (two to six) S-alleles (Brennan et al. 2006).
To map the S-locus, researchers must determine the S-locus genotype for individuals in a population large enough to conduct linkage mapping. In other species, S-genotype determination required mating large numbers of full-sibling individuals to one or a few “tester” genotypes with a known S-genotype (Camargo et al. 1997; Tomita et al. 2004). Testers may be a parent of the population, or may be obtained by mating siblings in a diallel design, grouping individuals based on their compatibility (Hiscock 2000), and then selecting one or several of these individuals as a tester for their siblings (Gonthier et al. 2013). However, this process has several limitations. For some species, pollen availability may limit the number of testcrosses that can be made, especially if clonal propagation is not used to multiply tester individuals. Additionally, because the tester is typically used as the male parent, differentiation between alleles may be difficult for SSI as dominance relationship between alleles may differ from anthers to stigma. Finally, this process often requires a large number of crosses, especially if the parents are not available. A full diallel requires n(n-1) crosses, where n is the number of individuals to be analyzed, and multiple crosses are likely required for each individual being tested. Even selecting a tester from amongst a conservative number of siblings may require a prohibitively high number of crosses. Methods of S-allele determination that address these issues may make S-locus mapping feasible for a greater number of species.
In this paper, we describe such a method, and employ it to identify a putative location for the S. integrifolium S-locus. Specifically, we developed a novel framework for inferring the S-genotypes of individuals within a population large enough for mapping. Our method does not require tester individuals with known S-genotypes, requires only three to four crosses per individual, and identifies alleles with sex-specific dominance interactions. In addition, we have constructed the first linkage map for S. integrifolium; combining this map with the S-genotypes inferred by our methods we identified a QTL likely containing the S-locus. We then identify regions in published sunflower (Helianthus annuus, con-tribal with S. integrifolium) and lettuce (Lactuca sativa) genome assemblies potentially syntenic with our putative S-locus, and use this information to assess classes of genes which could be involved with S-allele determination.
Materials and methods
Population development
A linkage map was constructed using an F1 population of 265 Silphium integrifolium (Michx.) individuals, derived from the crossing of two genotypes known as “965” and “1767”, selected at the Land Institute. These parents were chosen because they expressed agronomically favorable morphologies and differed for several phenological traits. Seventy-four seeds from this cross were grown at the Land Institute, and 191 were grown at the University of Minnesota. All 265 individuals were used for map construction, but only a subset of the 191 Minnesota individuals were used for S-allele determination. Following approximately two months of growth in a greenhouse, the Minnesota seedlings were transplanted to a field at the Minnesota Agricultural Experiment Station at the University of Minnesota St. Paul campus with 1.2 meters between each plant.
Genotyping and variant detection
Tissue was collected from seedling leaves of the 74 Land Institute progeny, and adult leaf tissue was collected from the parents and all other progeny, with all tissue lyophilized prior to DNA extraction. Dual indexed genotyping-by-sequencing (GBS) libraries were created from genomic DNA using the restriction enzymes SbfI and TaqI and sequenced on 1.5 Illumina NovaSeq 6000 lanes (1 × 100 single-end reads) (Elshire et al. 2011). All extraction, library preparation, and sequencing occurred at the University of Minnesota Genomics Center.
Demultiplexed reads were mapped to S. integrifolium genomic contigs (N50 = 6.07 Mbp, L50 = 176, contigs = 20681391) using BWA-MEM (Y. Brandvain, unpublished; Li 2013). The “ref_map” pipeline of Version 2.5 of the “Stacks” variant calling software was then used for the detection of SNP loci (Catchen et al. 2013). Loci that were missing in at least 30% of the population were excluded, as were loci missing in either parent. This resulted in 935 SNP markers.
Linkage map construction
Linkage analysis and map construction were carried out in JoinMap5 (Stam 1993). First, 715 SNP markers showing significant (Chi-square test, P > 0.01) segregation distortion were removed. This step removed both markers showing biologically meaningful segregation distortion, as well as markers which likely show apparent distortion due to genotyping errors. Markers were then grouped by LOD independence score, using a threshold score of five. Any groups that contained fewer than four markers were discarded. Map order and distances were then estimated using the maximum likelihood mapping algorithm, with default settings. One marker was removed from the end of linkage group three because it created a gap of more than 50 cM. Linkage group names were assigned based on estimated centimorgan length, with group one being the longest.
Crossing design to determine self-incompatibility genotype
Of the 191 members of this mapping population planted in Minnesota, 84 were intermated for S-allele determination. To cross, capitula (compound flower heads) to be used as a female, or both as a male and female, were covered with a cotton bag prior to anthesis to prevent pollination. Capitula to be used exclusively as a male parent were covered with mesh bags at least one day prior to crossing to reduce contamination by insects depositing pollen from other plants. After stigma emergence, pollen from a male parent capitulum was collected into a container and dusted onto stigma (found in the ray florets) with a pipe cleaner. Capitula were only pollinated if all of the ray florets had opened. Alternatively, on occasion a capitulum was removed and used to brush pollen directly onto a female parent. Each female capitulum was mated with only one male parent, and typically each cross was performed without replication. The bag was then reclosed, and capitula harvested after senescence and dried. For each capitulum, the number of filled seeds and total number of ovules were then counted, with fertilized (and therefore filled) and unfertilized ovules differentiated by visual and manual assessment. Seed set was then calculated as the ratio of fertilized to total ovules.
Individuals were crossed in a structured design, which we have named the “connected small diallel design”, illustrated in Fig. 1. This structure is based on groupings of four individuals that are mated in a diallel. These small diallels are then linked together through reciprocally mating single individuals. The purpose of this design was to maximize the amount of information that could be derived from the mating behavior of any given individual, while minimizing the number of matings for which it would need to be used. The design ensures that all individuals may be connected to one another through pairs of matings, allowing for the entire population to be used to predict the mating behavior each individual.
Each circle represents one plant, with black circles representing plants that were used for crosses, and gray plants representing other members of the mapping population that were not selected. The dashed box represents one of the small diallels that formed the basis for the design—twenty of these were used for this experiment. Arrows represent matings, with each arrow pointing from the male parent to the female parent.
The design for this experiment included 20 small diallels, comprised of 80 individuals. The design was implemented incompletely, with many of the recommended crosses not completed due to time or pollen availability constraints. To ensure that all individuals could be connected, approximately 20 more crosses connecting diallels were conducted than indicated in the design. Individuals for these crosses were selected arbitrarily, but generally paired dialleles which otherwise would not have otherwise been directly crossed with each other. Finally, several additional individuals were included in the study with only reciprocal crosses to one other individual. In total 268 crosses were performed between the 84 individuals, covering 138 different combinations of parents. Of these crosses, 126 combinations were crossed reciprocally, with both individuals used as male and female. One hundred and eighty-six crosses were conducted in 2018, and an additional 82 were conducted in 2019, primarily to increase the number of reciprocal crosses. In seven cases, multiple replications of a cross were performed. These observations were combined by summing the total number of filled and total seeds from each replication, and then calculating a seed set from the sums.
Three distinct but complementary methods were used to translate the results of the connected small diallel experiment into an S-locus map position. These methods, along with their relative strengths and weaknesses, are described in the following sections. All methods were developed using the R language (R Core Team 2019). In all three methods, each mated pair of individuals is first determined to be compatible or incompatible, based on a threshold seed set value. A threshold value of 20% was selected for this experiment, with matings that resulted in a seed set value above 20% considered compatible. This value was selected based on a previous experiment, where manually self-pollinated S. integrifolium capitula were observed to have a maximum seed set value of 22% (Reinert et al. 2020). In an SSI system, seed set should be 0% for incompatible individuals. In practice, however, some seed set may be observed either because of pollen contamination, or due to additional factors which circumvent the SSI pollen rejection (Hiscock 2000). By selecting a value of 20%, occasional accidental cross-pollinations which resulted from the imperfect exclusion of insect pollinators or rare self-pollinations were less likely to cause genetically incompatible pairings to be classified as compatible. In another study, the average seed set for mated pairs of compatible full siblings was estimated to be around 70% (Price et al. 2021), therefore a threshold much higher than 20% would likely result in the exclusion of compatible pairs if seed set was lower than average due to imperfect pollination or environmental factors.
Direct mapping with single-marker regression
The simplest method attempted to directly associate variation for compatibility with an SNP marker, without inferring the S-genotype of any particular individual. To accomplish this, a logistic regression was conducted following the formula Si = Mij + Pij + Mij:Pij, where the success S of the ith mating was predicted by the maternal genotype M, the paternal genotype P, and the interaction of those genotypes, all for the jth marker. The calculated P-value of the maternal by paternal genotype interaction was then used to determine association with cross incompatibility, and thus the S-locus. Only biallelic markers were used in this approach, with homozygotes recoded as −1 or 1 and heterozygotes as 0. Only 184 of the available SNP markers met this criterion, so a Bonferroni corrected P-value of 2.7 × 10−4 was used as the threshold for determining significance. This approach only makes limited use of available genotype data, as each SNP marker is considered individually. Additionally, this approach could not be applied to other populations where genetic marker data is not available. Finally, because it does not make inferences about the S-genotype carried by any particular individual or take potential dominance relationships into account, it is not able to fully leverage the advantages of the connected crossing design used in this study. Thus, this method’s usefulness is likely limited to confirming the results of other methods.
Inference of self-incompatibility genotype for all individuals followed by QTL mapping of the inferred S-genotype
As an alternative approach to mapping the S-locus, we develop two methods to infer every individual’s S-locus genotype from its crossing behavior, then map this inferred phenotype with traditional QTL mapping software. Both methods assumed that the two parents of the population were each heterozygous at the S-locus and did not share any S-alleles with each other, resulting in a population with four distinct S-alleles and therefore four distinct S-genotypes. If both parents shared an allele, it would have been recessive (as the two parents were able to successfully mate with each other). In this case, both methods would simply have considered this allele to be two separate alleles which behaved identically. In other words, the assumption of four distinct alleles would not necessarily need to be met for the methods to work. Each method is described in detail below. Code to replicate all methods may be found on GitHub (see data availability statement).
Inference method 1: a hill-climbing algorithm
The first S-allele determination method used a simple hill-climbing algorithm to fit genotypes, given a user-generated set of dominance relationships among alleles. To start, the user hypothesizes a set of crossing relationships, dictating which of the sixteen possible pairings of genotypes (four possible genotypes, squared) will and will not be able to successfully mate. A random genotype is then assigned to each individual in the population. One arbitrarily selected individual is always set to a predetermined genotype to enable comparison of different runs of the algorithm. Each mated pair in the dataset is then evaluated using the user-supplied crossing relationships, with the total number of matches and mismatches counted. Next, the genotype of one individual is randomly changed, and the dataset reevaluated. If the total number of matches increases, then that set of genotypes is kept; if not, the algorithm returns to the previous position.
The cycle is repeated, stopping if 1000 changes are attempted without increasing the number of matches. For this experiment, the algorithm was run 4000 times for each set of dominance relationships, with the solution to the best run considered optimal genotype assignments for a given dominance relationship. The best genotype assignments across all dominance relationships were then used for QTL mapping. Hypothetically, up to 256 dominance relationships are possible; however, we limited ourselves to the 48 which met three criteria (1) individuals sharing both alleles must be incompatible, (2) individuals sharing no alleles must be compatible, and (3) there must be at least one combination of genotypes that was asymmetrical (compatible when one individual was used as a female and incompatible when the other was used as a female), as this was observed in the crossing data. These dominance relationships are shown in Table S1.
This method has both the advantage and disadvantage of being highly parameterizable. This makes it flexible, and gives the user a high degree of control, but may take many attempts to find the right combination of parameters to develop a solution. This method may be employed using a personal computer, however, the reduction in parallelization necessary to achieve this may make it take too long to be useful. In addition, as a hill-climbing method there is no guarantee that this approach will find a global maximum.
Inference method 2: a Markov Chain Monte Carlo algorithm
The second method employed sets of extreme gradient boosting decision trees with Markov chain Monte Carlo (MCMC) algorithms to infer S-locus genotypes. For each MCMC chain, an initial set of genotypes was created by first assigning a random heterozygous S-genotype to one random individual. The genotype of an individual that had been mated to the initial individual was then determined. If the cross was incompatible, the second individual’s genotype could share one or two alleles with the initial individual’s genotype, with equal probability. If the cross was compatible, the second individual’s genotype could share either zero or one allele with the initial plant’s genotype, with equal probability. The procedure was then repeated by choosing, at random, an individual that did not yet have a genotype assigned and that was crossed to the most recent individual assigned a genotype. If all individuals that were crossed to the most recently assigned individual had an assigned genotype, a random individual that did not yet have an assigned genotype was selected and the process started again until all individual were assigned genotypes.
Maternal and paternal S-locus genotypes were then treated as predictive variables with cross success used as a binary response variable. Constraints were placed on predictive variables so that matching maternal and paternal genotypes were not allowed to interact with one another, as crosses between individuals with matching genotypes should always be incompatible. At each step of an MCMC chain, model performance was measured as error of a logistic regression for classification using an extreme gradient boosting model with four-fold cross validation with 200 iterations and maximum tree depth of four splits, using the R package “xgboost” (Chen and Guestrin 2016). A single genotype was altered at each MCMC chain step, using the same sets of probabilities used to construct the initial set. The newly proposed set of genotypes was accepted if the ratio of errors from the proposed genotype set to the former genotype set was greater than a randomly drawn number bound by zero and one. A total of 96 MCMC chains were created for this experiment.
Each MCMC chain, starting from a unique set of genotypes, was allowed to explore parameter space for 96 h (~1 million steps). The set of genotypes that produced the smallest error from each chain was then used as the starting condition for hill-climbing algorithms. These hill-climbing algorithms were included in an attempt to reach optimal genotypes sets in a smaller amount of time than would be required for the MCMC method alone. We used the same extreme gradient boosting model conditions described above, however at each step proposed genotypes were only accepted if they produced an error that was less than the previous set of genotypes. The hill-climbing algorithms each ran for 96 h. For each chain, the set of genotypes that produced the smallest error, calculated as the percentage of crosses classified incorrectly, were recorded. A cross was predicted to be a success if the estimated logistic regression probability was greater than 0.5. The lowest error models were considered the best candidates for S-genotype determination. This method has the advantage of less dependence on user decisions, making it more repeatable and facilitating the exploration of a wider space of possible solutions. However, the resources required by this method mean that it generally cannot be employed on a personal computer.
Mapping the S-locus from inferred S-locus genotypes
Assigned alleles from the two inference methods were then used as phenotypes for QTL mapping, using the “qtl” R package (Broman et al. 2003), with the population treated as a four-way testcross. Missing marker genotypes were imputed using the “sim.geno” function, and QTL were identified using the “scanone” function to conduct interval mapping using the EM algorithm. Self-incompatibility allele was treated as a binary trait, with one allele from each parent arbitrarily coded as “1” and the other as “0”, and the two parental alleles mapped separately. In other words, if the two alleles from one parent of the population are labeled S1 and S2, and the alleles from the other parent labeled S3 and S4, then the presence or absence of the of the S1 allele (absence of the S1 allele meaning presence of the S2) was treated as one trait for QTL mapping, while the presence or absence of the S3 allele was treated as a second “trait”. For the MCMC method, all 96 of the inferred genotype sets were mapped, with the maximum LOD score produced by each set serving as a criterion to differentiate the sets. Seven genotype sets produced by the hill-climbing method were also mapped. Significance thresholds were determined independently for each set of genotypes used for mapping using a permutation test; only QTL with an error probability less than 5% were considered significant. This threshold ranged from a LOD score of 3.6 to 4.2. Finally, a 95% probability Bayesian credible interval (similar to a confidence interval) around any identified QTL was calculated, using the “bayesint” function.
Synteny with related species
To help elucidate the relationship between the S. integrifolium S-locus and other Asteraceae, the genomic contigs associated with the loci that comprise this linkage map were aligned to the annual sunflower (Helianthus annuus, variety ‘HA412’, version HOv1.1) (Badouin et al. 2017) and lettuce (Lactuca sativa, variety ‘Salinas’, version 7) (Reyes-Chin-Wo et al. 2017) genome assemblies using BLASTN (Altschul et al. 1990). Syntenic regions were then identified using the R package “syntR” (Ostevik et al. 2020). For synteny analysis, all BLASTN alignments for a given S. integrifolium query sequence that had a bitscore greater than 90% of the maximum bitscore for that query sequence were used.
Evaluation of genes of interest
For regions identified as syntenic to the S. integrifolium S-locus, gene annotations for sunflower were obtained from the genome assembly HA412v1 (Badouin et al. 2017). InterPro taxonomy was searched for each gene in the region to identify plausible candidates by shared terms with S-genes in other species. The male and female determinants from Brassica oleracea (Brassicaceae) and Petunia hybrida (Solanaceae), as well as the female determinant of Papaver rhoeas (Ranunculales) were chosen as representatives for comparison (The UniProt Consortium 2019). In addition, defensin-like proteins were considered a proxy for the Ipomoea trifida (Convolvulaceae) male determinant (Rahman et al. 2007). These species were selected because their self-recognition genes are relatively well-characterized and they represent a broad range of Eudicot diversity, including both SSI and GSI systems. Any genes homologous to known cysteine-rich proteins were also considered potential genes of interest (Marshall et al. 2011). Additionally, the coding region sequence for the B. oleracea S-receptor kinase protein (Stein et al. 1991) was aligned to the lettuce and sunflower genomes using BLAST. These alignments were then compared to the lettuce and sunflower regions syntenic to the S. integrifolium S-locus.
Results
Linkage Map
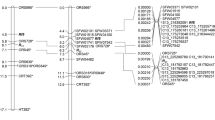
The linkage map contained 198 markers, spanning 1049 centimorgans (cM) and divided into 7 linkage groups (Fig. 2, Table S2). This was consistent with the observation that S. integrifolium has seven chromosomes (Settle 1967). The average distance between markers was 5.5 cM, with 29 gaps greater than 10 cM.
The black bar on linkage group 6 represents the putative S-locus QTL, with the circle showing the LOD peak and the extent of the bar showing a composite of the 95% Bayes credible intervals for several mapping methods. Map visualization was performed using “LinkageMapView” (Ouellette et al. 2018).
S-locus mapping
As justified in the methods, a threshold seed set value of 20% was used to classify compatible and incompatible matings, with values above 20% considered compatible. Compatibility patterns were not strongly altered by changing this threshold: Supplemental Fig. 1 and 2 illustrate compatibility patterns at several alternative thresholds. Therefore, the 20% threshold was used for all further analyses. Of the 268 crosses completed, 53% were incompatible, with a seed set value less than 20%, and 47% were compatible. These frequencies were not significantly different from an equal occurrence of compatible and incompatible crosses (df = 1, χ2 = 0.956, P = 0.328). This indicates there is a hierarchy of dominance between alleles in this population—if all alleles were codominant a ratio of 25% compatible to 75% incompatible would be expected with each genotype only able to mate with the single genotype with which it shared no alleles. This ratio was found to be extremely unlikely (df = 1, χ2 = 69.27, P < 10−16). Of the 126 reciprocal pairs of genotypes crossed, 30% were compatible in both crossing directions (both individuals could be used as male and female), 34% were incompatible, and 36% were asymmetrical, or compatible when one individual was used as a female and incompatible when the other was used as a female. The presence of asymmetrical crosses indicates that the dominance relationship between some allele pairs differs from anthers to stigma. These observations confirm that S. integrifolium has a sporophytic SI system, as dominance relationships are not observed in gametophytic systems (Breton et al. 2014). Figure 3 shows the distribution of seed set values for reciprocal crosses, numerical data for all crosses may be found in Table S3.
Each dot represents the seed set value of one plant, with lines connecting mated pairs. The individuals with the higher seed set value in the pair are placed on the left side of the chart. The horizontal line represents the threshold seed set value of 20% used to determine compatibility, see Supplemental Fig. 1 for an illustration of alternative thresholds.
Single marker regression for male/ female interaction
The interaction of male and female marker genotype was found to be a significant or near significant (P < 2.7 × 10−4) predictor of crossing success for four markers, located at two regions on the linkage map: markers “272299” and “120685” located at 18.9 and 9 cM, respectively, on linkage group six (P = 7.57 × 10−8, P = 1.46 × 10−4 df = 259 for both), and markers “185443” and “118778”, located at 126.33 cM and 120.7 cM, respectively, on linkage group two (P = 3.22 × 10−5, P = 3.31 × 10−4, df = 259 for both).
Hill-climbing algorithm method
Of the 48 dominance relationships tested using the hill-climbing algorithm, the seven with the fewest mismatches between observed and expected crossing behavior were used in QTL mapping. Of these, five produced at least one significant LOD peak, all located on linkage group 6. The highest of these (LOD = 5.3) was located at 18.9 cM, and the other four were located between 0–27.7 cM. The model which produced the highest LOD score was tied with three other models for the second-lowest mismatch score, illustrating the need to combine model and mapping metrics to identify the most accurate dominance model when using this method.
MCMC method
One of the 96 MCMC results had a model score markedly lower than the other scores (15% lower than the next best preforming model, roughly equivalent to the difference between the second-best result and the 20th result), and so was considered the best candidate for QTL mapping. However, all results were used for QTL mapping for the sake of comparison. Of these, 25 produced at least one significant LOD peak when used for QTL mapping, covering five linkage groups. Of these 25 results 20 only gave a significant QTL for one of the two sets of parental alleles, while five gave a significant QTL for both sets. The lowest-error genotype set also produced the highest LOD peak (7.56), located at 18.9 cM on linkage group 6. This genotype set produced a second significant LOD peak, at 86 cM on linkage group 6. However, a QTL model which included both of these peaks as QTL only identified the 18.9 cM QTL as significant, and so this second peak would appear either to be noise, or to be associated with the main QTL peak. Ten other of these genotype sets also produced a significant LOD peak at either 18.9 or 27.7 cM on linkage group 6, and a further five were located elsewhere between 0–31 cM on linkage group 6. Based on these observations, we conclude that the best map location from the MCMC method is 18.9 cM on linkage group six; Fig. 4 visualizes all crosses used in this study, organized by the genotype assignments from the genotype set with the lowest error and highest LOD peak. Many of the other QTL peaks are likely spurious as they are derived from genotype sets with relatively high error scores, and illustrate the necessity of using both genotype error and QTL LOD score as a criterion for selection when using this method.
Red or blue squares represent the crosses that were actually made, with red representing incompatible crosses, and blue representing compatible according to the threshold seed set value of 20%. See Supplemental Fig. 2 for an illustration of alternative thresholds. Individuals are grouped by S-locus genotype, as assigned by the best MCMC genotype inference model. “S1” and “S2” refer to the S-alleles from one parent of the population, and “S3” and “S4” to the alleles from the other. In this model, all alleles are co-dominant in the stigma, the S1 allele is recessive to S3 and S4 in the pollen, and S2 is pollen dominant over S3 and S4. These alleles which are hypothesized to be expressed are underlined in the figure.
Consensus map location
Of the three methods used to map the S-locus, the MCMC method produced the strongest association between a genomic region and S-allele. The peak of this QTL was located at 18.9 cM on linkage group six, with a 95% probability Bayes credible interval from 0 to 27.7 cM. Broadly speaking, the results of the hill-climbing method agree with this region. Additionally, the most significant single-marker regression association between the interaction of male and female genotype with cross success was found at 18.9 cM. Taken together, we conclude that the S. integrifolium S-locus is located between 0 and 27.7 cM on linkage group six, with the marker closest to the locus likely located at 18.9 cM. Supplemental Fig. 3 illustrates the QTL peak from nine of the best MCMC and hill-climbing results which led to this consensus position.
S-locus synteny with related species
Potential synteny was identified by the “syntR” package between the putative S. integrifolium S-locus region and both the sunflower and lettuce genomes. However, it is important to first note the sparseness of the S. integrifolium linkage map compared to a genome assembly, as this introduces a large amount of uncertainty into any identification of synteny. As improved genomic resources are developed for S. integrifolium, it is very likely that different syntenic regions will be identified which are more accurate. In sunflower, synteny was found with two chromosomes—the S. integrifolium 0–18.9 cM region was found to align with chromosome 17, from 230.7 megabase pairs (Mbp) to 262.67 Mbp of the HOv1.1 genome assembly, and the 18.9–31 cM region was found to align with chromosome 3, from 133.21 Mbp to 149.9 Mbp. Although the likely interval for the S-locus ends at 27.7 cM, the analysis identified this block as extending to 31 cM. In lettuce, the 0–9 cM region aligned with chromosome 4, from 18.13 Mbp to 27.29 Mbp, and the 18.9–31 cM region aligned with chromosome 9, from 144.19 Mbp to 147.34 Mbp. The syntenic region on sunflower chromosome 17 does not appear to overlap with the putative self-incompatibility breakdown QTL found on that chromosome by Gandhi et al. (2005), which we estimate was somewhere between 42 and 98 Mbp on the HOv1.1 sunflower genome, based on the alignment of SSR marker primer sequences to the sunflower genome assembly. More recent work in sunflower has found this self-incompatibility breakdown QTL to colocalize with an S-locus gene candidate identified through tissue specific expression studies, located at 176 Mb on chromosome 17 of the XRQ sunflower genome assembly (Badouin et al. 2021). More work will be required to determine conclusively if the S. integrifolium S-locus is syntenic with this sunflower region.
Evaluation of genes of interest
Of the 642 genes located within the sunflower genomic regions potentially syntenic with the S. integrifolium S-locus, 42 shared at least one InterPro term with either the male or female determinants of self-incompatibility in B. oleracea or P. hybrida, the female determinant of P. rhoeas, or the male determinant of I. trifida. Thirty of these genes were, like B. oleracea SRK, protein kinases, which is not a specific enough homology to infer a plausible gene candidate. The other 12 genes were, like the male determinant in P. hybrida, F-box genes. In addition, one gene was similar to “STIG1”, a cysteine-rich protein. The best alignment for B. oleracea SRK in sunflower was found on chromosome 11, while the best alignment in lettuce was found on chromosome 7. Neither of these alignments are near any regions likely to be syntenic with the putative S. integrifolium S-locus in either species.
Discussion
Evaluation of the putative S-locus
Combining the results of the hill-climbing and MCMC inference methods with single-marker regression, the S. integrifolium S-locus would appear to be located between 0–27.7 cM on linkage group 6, with the highest LOD peak located at 18.9 cM. Using genotype assignments produced by the lowest error MCMC genotype set, we infer that this particular S. integrifolium population contains four S-alleles, with all four alleles co-dominant in the stigma. If the two alleles from one parent of the population are labeled S1 and S2, and the alleles from the other parent labeled S3 and S4, then the S1 allele is recessive to S3 and S4 in the pollen, and S2 is pollen dominant over S3 and S4. Further work with a pseudo-F2 population derived from crosses amongst members of the F1 mapping population would be necessary to disentangle dominance relationships between the pairs of alleles which made up the original parental genotypes. This dominance model is accurate for 91% of the crosses in this experiment (Fig. 4), and it is likely that at least some of the cases in which the dominance relationship appears to inaccurately predict a crossing relationship may represent errors in the collected data. When mapping the MCMC genotype sets, often only one set of parental alleles produced a significant mapping result for a given genotype model. This may mean that crossing behavior in this population was primarily dictated by the S1 and S2 alleles, making assignments for the S3 or S4 allele less certain as they were synonymous in most potential pairings.
Our study started with the assumption that there is one S-locus in S. integrifolium which determines pollen rejection or acceptance. The development of a one-locus dominance model which explains 91% of crosses made in this experiment lends some credence to this assumption. In addition, a one QTL model was found to be the best fit for the genotype assignments which resulted from that dominance model. However, our results do leave open the possibility of more complicated models. As mentioned in the results, our single-marker regression method did identify a region on linkage group 2 which was a statistically-significant predictor of cross success, suggesting it could be associated with self-incompatibility. In addition, several hill-climbing or MCMC genotype sets which identified the linkage group 6 QTL also identified significant or near-significant QTL on a variety of other linkage groups (Supplemental Figure 3). As these other QTL were not supported by all three inference methods, they may simply be statistical noise.
Another possibility is that some of these loci represent segregating embryo lethal mutations that caused in a statistically significant reduction in seed set. A related S. integrifolium breeding population was found to carry an average of one lethal mutation per gamete (Price et al. 2021). Such an allele could measurably reduce seed set without being confused with true SSI by both the inference methods, both because the mutation would reduce but not eliminate seed set, and because it may follow a difference dominance pattern than the S-locus. It is worth noting that previous work in in the Asteraceae species Senecio squalidus suggested the possibility of a second locus which operated epistatic to the S-locus (Hiscock et al. 2003). We do not believe that our observations directly support the possibility of epistatic self-incompatibility loci in S. integrifolium; however, it is worthwhile to consider the possibility of multi-locus control of SSI as more work is done to elucidate the genetic mechanism of self-incompatibility in S. integrifolium.
The described S-locus mapping methods are effective and efficient
This study represents one of the few successful identifications of a genomic region containing the S-locus in an Asteraceae species. This result is valuable for its own sake, as the comparison of syntenic genomic regions between S. integrifolium and other species may help to answer questions about the evolution of SSI systems in other Asteraceae. In addition, this study lays the groundwork for future efforts to clone the S. integrifolium S-locus. In this experiment, we were able to predict the S-locus genotype for 84 individuals by conducting 268 crosses, with an average of 3.2 crosses per individual. This method did not require all 84 individuals to be crossed with a single tester, and so could be applied to a pre-existing full-sibling family without necessitating clonal propagation of any individuals. As a point of comparison, we estimate that the mapping of the S-locus in chicory, which combined a twenty-two plant diallel with testcrossing, required approximately 3000 crosses (Gonthier et al. 2013). That approach, which also required the clonal propagation of tester genotypes, was used to assign an S-genotype to approximately 350 individuals (multiple replications of each cross were performed). The chicory mapping effort produced a more accurate determination of S-genotype and thus a narrower S-locus QTL region than our study, but with the trade-off of requiring more crosses per individual and the clonal replication of tester genotypes. Additionally, our method requires less time to complete, as testcrossing cannot be conducted until the diallel is completed, the results interpreted, and selected testers clonally propagated. Finally, our method produced useful results using crossing data which was mostly unreplicated, suggesting that it is reasonably robust to errors in input data and future S-locus mapping studies which employed our methods would also require limited replication in the crossing portion of the experiment.
Our method should be readily applicable to any other species that meet three criteria: (1) They possess an SSI system, (2) large full sibling families can be produced, and (3) individuals can readily be used as both a male and female parent in three to five matings. Among numerous other species, self-incompatible members of the genus Helianthus could be excellent candidates for this method and could help to confirm our results. Applying this method to phylogenetically diverse Asteraceae species could evaluate the conservation of both the molecular mechanism and genomic position of SSI in the family.
Evaluation of potential S-locus genes of interest
Of the 642 annotated or predicted genes present in the sunflower genomic regions syntenic to the putative S-locus, 43 show potential similarity to known S-genes. Of these, two groups of genes were considered particularly interesting, and are discussed below. It is important to note that because virtually nothing is known about the molecular mechanisms that underlie SSI in the Asteraceae, any discussion of potential candidate genes based on annotations or similarity to S-related genes in other systems is at best informed speculation. However, we see this as an important step towards identifying the genes that control SSI in the Asteraceae, and therefore believe it is worth pursuing.
One gene of interest is the sunflower gene Ha3_00036110, which is annotated as a putative member of the “Stigma Specific Protein 1” like, or STIG1-like, group of genes. In sunflower, this gene is found on the chromosome 3 region syntenic with the 18.9–31 cM segment of the putative S. integrifolium S-locus. In lettuce, the best BLAST alignment for Ha3_00036110 is on chromosome 9, within the region syntenic to the putative S. integrifolium S-locus, implying that this region may be conserved between the three species. The best characterized member of this family, STIG1, encodes a small, cysteine-rich protein. In tomato (Solanum lycopersicum), this protein is primarily found in stigma exudate, and binds a pollen-specific kinase to promote pollen tube growth (Huang et al. 2014). More broadly speaking, because they encode cysteine-rich proteins, STIG1-like genes are members of a gene class that also includes the male determinant of SSI in the Brassicaceae and the Convolvulaceae, and the female determinant of GSI in the genus Papaver. Based on this similarity to other SI genes, the function of STIG1, and the location of this gene within the sunflower genomic region syntenic to the S. integrifolium S-locus, Ha3_00036110 may be worthy of further investigation as a potential S-gene. It is also worth noting that a serine/threonine protein kinase gene, Ha3_00036112, is located 109 Kbp downstream from Ha3_00036110. A tightly linked signaling protein and kinase is the general molecular model followed by the Brassicaceae S-locus, so investigating the genes in tandem would be worthwhile.
Another set of potential candidate genes are found on the sunflower chromosome 17 region syntenic with the 0–18.9 cM segment of the putative S. integrifolium S-locus. This region contains 12 F-box genes, resembling known gametophytic self-incompatibility (GSI) systems. For example, in Petunia, 17 tightly linked F-box genes serve as the male determinant of GSI (Williams et al. 2014). However, no genes in this sunflower region share any annotation terms with known S-RNAse genes (The UniProt Consortium 2019), which serve as the female determinant in GSI systems. However, because so little is known about the molecular basis of the Asteraceae SSI response, these genes mays still be worth of future investigation.
In addition to providing candidates for genes involved in S. integrifolium self-recognition, this study suggests that SRK-like genes are not involved in S. integrifolium SSI, as the best homologs for the B. oleracea SRK in the lettuce and sunflower genomes are not found in or near the regions of those genomes syntenic with the putative S. integrifolium S-locus. This finding adds evidence to the theory that specific genes that underlie Asteraceae SSI are different than those in the Brassicaceae (Allen and Hiscock 2008).
Implications for domestication and breeding
We expect the results of this study to facilitate the domestication of S. integrifolium. The availability of a genetic map, associated with particular restriction enzymes, supports the relatively inexpensive GBS genotyping of large numbers of progeny; both because these enzymes are now known to produce a useful number of fragments containing polymorphisms in S. integrifolium, and because it is reasonably likely that some of the same loci would be recovered if these enzymes were applied to a different population. This may support the implementation of marker-based selection, marker-based pedigree development, and genome-wide selection. Additionally, the genetic map produced in this study will assist in the anchoring and orientation of future genome assemblies.
Identifying a map location for the S-locus may also contribute to practical breeding efforts. If molecular marker data is routinely available for individuals within a breeding program, it may be possible to predict whether any two plants will be able to successfully mate, saving effort by excluding crosses that would not be successful. Perhaps more importantly, this would allow for the S-allele diversity of a given population to be monitored and maximized. It is likely that at least some S. integrifolium cultivars will take the form of synthetic populations, where a set of superior genotypes are intermated and their progeny form a distinct variety that may be reproduced for several generations. If a synthetic population is released that contains a low number of S-alleles, its long-term fecundity may be adversely affected by the limited number of individuals that are able to intermate. Alternatively, as S. integrifolium is known to express moderate to severe inbreeding depression for a number of traits (Price et al. 2021), it is possible that S-allele characterization information could be used to increase long-term productivity by limited mating among relatives for several generations within synthetic populations.
Overall, we anticipate that the availability of a genetic map and identification of the self-incompatibility locus will support efforts to domesticate S. integrifolium as a crop that will help to enable sustainable agricultural systems.
Data availability
All code and data to replicate analyses may be found on GitHub, at https://github.com/UMN-BarleyOatSilphium/SilphiumSLocus. All sequence data may be accessed under BioProject PRJNA695552.
References
Allen AM, Hiscock SJ (2008) Evolution and phylogeny of self-incompatibility systems in angiosperms. In: Franklin-Tong VE ed. Self-incompatibility in flowering plants: Evolution, Diversity and Mechanisms. Springer-Verlag, Heidelberg, DE, p 73–101
Allen AM, Thorogood CJ, Hegarty MJ, Lexer C, Hiscock SJ (2011) Pollen-pistil interactions and self-incompatibility in the Asteraceae: new insights from studies of Senecio squalidus (Oxford ragwort). Ann Bot 108(4):687–698
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Badouin, H, Boniface, MC, Pouilly, N, Fuchs, AL, Vear, F, Langlade, N et al. (2021). Pooled Single-Molecule transcriptomics identifies a giant gene under balancing selection in sunflower. bioRxiv 2021. 435796
Badouin H, Gouzy J, Grassa CJ, Murat F, Staton SE, Cottret L et al. (2017) The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546(7656):148–152
Bod’ová K, Priklopil T, Field DL, Barton NH, Pickup M (2018) Evolutionary pathways for the generation of new self-incompatibility haplotypes in a nonself-recognition system. Genetics 209(3):861–883
Brennan AC, Harris SA, Hiscock SJ (2006) The population genetics of sporophytic self-incompatibility in Senecio squalidus L. (Asteraceae): the number, frequency, and dominance interactions of S alleles across its British range. Evolution 60(2):213–224
Breton CM, Farinelli D, Shafiq S, Heslop-Harrison JS, Sedgley M, Bervillé AJ (2014) The self-incompatibility mating system of the olive (Olea europaea L.) functions with dominance between S-alleles. Tree Genet Genomes 10(4):1055–1067
Broman KW, Wu H, Sen Ś, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Camargo LEA, Savides L, Jung G, Nienhuis J, Osborn TC (1997) Location of the self-incompatibility locus in an RFLP and RAPD map of Brassica oleracea. J Heredity 88(1):57–60
Castric V, Bechsgaard J, Schierup MH, Vekemans X (2008) Repeated adaptive introgression at a gene under multiallelic balancing selection. PLoS Genet 4:e1000168
Catchen J, Hohenlohe P, Bassham S, Amores A, Cresko W (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22(11):3124–40
Chen T, Guestrin C (2016) XGBoost: A Scalable Tree Boosting System. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ‘16). New York, NY, USA: Association for Computing Machinery, 785–794
DeHaan LR, Van Tassel DL, Anderson JA, Asselin SR, Barnes R, Baute GJ et al. (2016) A pipeline strategy for grain crop domestication. Crop Sci 56(3):917–930
Edh K, Widén B, Ceplitis A (2009) Molecular population genetics of the SRK and SCR self-incompatibility genes in the wild plant species Brassica cretica (Brassicaceae). Genetics 181(3):985–995
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES et al. (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS one 6(5):e19379
Fujii S, Takayama S (2018) Multilayered dominance hierarchy in plant self-incompatibility. Plant Reprod 31(1):15–19
Gonthier L, Blassiau C, Mörchen M, Cadalen T, Poiret M, Hendriks T et al. (2013) High-density genetic maps for loci involved in nuclear male sterility (NMS1) and sporophytic self-incompatibility (S-locus) in chicory (Cichorium intybus L., Asteraceae). Theor Appl Genet 126(8):2103–2121
Harkness A, Brandvain Y (2021) Nonself-recognition-based self-incompatibility can alternatively promote or prevent introgression. N. Phytologist 231(4):1630–1643
Harkness A, Goldberg EE, Brandvain Y (2021) Diversification or collapse of self-incompatibility haplotypes as a rescue process. The American Naturalist. https://doi.org/10.1086/712424
Hiscock SJ (2000) Genetic control of self-incompatibility in Senecio squalidus L.(Asteraceae): a successful colonizing species. Heredity 85(1):10–19
Hiscock SJ, McInnis SM, Tabah DA, Henderson CA, Brennan AC (2003) Sporophytic self‐incompatibility in Senecio squalidus L.(Asteraceae)—the search for S. J Exp Bot 54(380):169–174
Hiscock SJ, Tabah DA (2003) The different mechanisms of sporophytic self-incompatibility. Philosophical transactions of the Royal Society of London. Ser B Biol Sci 358(1434):1037–1045
Huang WJ, Liu HK, McCormick S, Tang WH (2014) Tomato Pistil Factor STIG1 promotes in vivo pollen tube growth by binding to Phosphatidylinositol 3-Phosphate and the extracellular domain of the pollen Receptor Kinase LePRK2. Plant Cell 26(6):2505–2523
Gandhi SD, Heesacker AF, Freeman CA, Argyris J, Bradford K, Knapp SJ (2005) The self-incompatibility locus (S) and quantitative trait loci for self-pollination and seed dormancy in sunflower. Theor Appl Genet 111(4):619–629
Marshall E, Costa LM, Gutierrez-Marcos J (2011) Cysteine-rich peptides (CRPs) mediate diverse aspects of cell–cell communication in plant reproduction and development. J Exp Bot 62(5):1677–1686
Kandel H, Hulke B, Ostlie M, Schatz B, Aberle E, Bjerke K et al. (2019) North Dakota Sunflower Variety Trial Results for 2019 and Selection Guide. North Dakota State University Extension, Fargo, ND, USA
Koseva B, Crawford DJ, Brown KE, Mort ME, Kelly JK (2017) The genetic breakdown of sporophytic self‐incompatibility in Tolpis coronopifolia (Asteraceae). N. Phytologist 216(4):1256–1267
Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z (2010) Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330(6005):796–799
Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN].
Price JH, Brandvain Y, Smith KP (2021) Measurements of lethal and non-lethal inbreeding depression inform the de novo domestication of Silphium integrifolium. Am J Bot 108(6):980–992
Ostevik KL, Samuk K, Rieseberg LH (2020) Ancestral reconstruction of karyotypes reveals an exceptional rate of nonrandom chromosomal evolution in sunflower. Genetics 214(4):1031–1045
Ouellette LA, Reid RW, Blanchard SG, Brouwer CR (2018) LinkageMapView—rendering high-resolution linkage and QTL maps. Bioinformatics 34(2):306–307
Rahman MH, Uchiyama M, Kuno M, Hirashima N, Suwabe K, Tsuchiya T et al. (2007) Expression of stigma-and anther-specific genes located in the S locus region of Ipomoea trifida. Sex Plant Reprod 20(2):73–85
R Core Team (2019) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
Reinert S, Van Tassel D, Schlautman B, Kane N, Hulke B (2019) Assessment of the biogeographical variation of seed size and seed oil traits in wild Silphium integrifolium Michx. genotypes. Plant Genet Resour: Charact Utilization 17(5):427–436
Reinert S, Price JH, Smart BC, Pogoda CS, Kane NC, Van Tassel D et al. (2020) Mating compatibility and fertility studies in an herbaceous perennial Aster undergoing de novo domestication to enhance agroecosystems. Agron Sustain Dev 40:27
Reyes-Chin-Wo S, Wang Z, Yang X, Kozik A, Arikit S, Song C et al. (2017) Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat Commun 8(1):1–11
Rindos D (1984) The origins of agriculture: an evolutionary perspective. Academic Press, Orlando, FL, USA
Schiffner S, Jungers JM, Hulke BS, Van Tassel DL, Smith KP, Sheaffer CC (2020) Silflower seed and biomass responses to plant density and nitrogen fertilization. Agrosystems. Geosci Environ 3:e20118
Sedbrook JC, Phippen WB, Marks MD (2014) New approaches to facilitate rapid domestication of a wild plant to an oilseed crop: example pennycress (Thlaspi arvense L.). Plant Sci 227:122–132
Settle WJ (1967) The chromosome morphology in the genus Silphium (Compositae). Ohio J Sci 67(1):10–19
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J 3:739–744
Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB (1991) Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc Natl Acad Sci 88(19):8816–8820
Tomita RN, Fukami K, Takayama S, Kowyama Y (2004) Genetic mapping of AFLP/AMF-derived DNA markers in the vicinity of the self-incompatibility locus in Ipomoea trifida. Sex Plant Reprod 16(6):265–272
The UniProt Consortium (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47(D1):D506–D515
Van Tassel DL, Albrecht KA, Bever JD, Boe AA, Brandvain Y, Crews TE et al. (2017) Accelerating silphium domestication: an opportunity to develop new crop ideotypes and breeding strategies informed by multiple disciplines. Crop Sci 57(3):1274–1284
Williams JS, Der JP, dePamphilis CW, Kao TH (2014) Transcriptome analysis reveals the same 17 S-locus F-box genes in two haplotypes of the self-incompatibility locus of Petunia inflata. Plant Cell 26(7):2873–2888
Acknowledgements
Funding for this work was provided by The Perennial Agriculture project in conjunction with The Land Institute and the Malone Family Land Preservation Fund, the United States Department of Agriculture’s National Institute of Food and Agriculture Grant no. 2019-67011-29607 to JHP, the Minnesota Department of Agriculture - Forever Green Agricultural Initiative, and NSF grant #1737827 Dimensions US-China to YB. The authors thank Shannon Lee Anderson, Karen Beaubein, and Jill Ekar for their help in completing the controlled crosses for this experiment. In addition, the authors thank Dr. Kevin Dorn for assistance in developing a GBS protocol, Dr. Adam Herman for bioinformatics advice, and Dr. Owen Beisel for inspiring the hill-climbing algorithm method. Finally, the authors acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota for providing computing resources that contributed to the analysis of this study.
Author information
Authors and Affiliations
Contributions
The effort to develop a linkage map was initiated by KPS, DLVT, and YB, who also secured initial funding. DLVT created the mapping population, contributed text, and visualization, and provided feedback in writing. JHP constructed the linkage map, initiated and designed the S-locus mapping work, and developed the HC method. ARR developed the MCMC method. JHP and ARR conducted data analysis and wrote the manuscript. KPS and YB provided supervision, and feedback on analysis, writing, and visualization.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor Marc Stift.
Supplementary information
Rights and permissions
About this article
Cite this article
Price, J.H., Raduski, A.R., Brandvain, Y. et al. Development of first linkage map for Silphium integrifolium (Asteraceae) enables identification of sporophytic self-incompatibility locus. Heredity 128, 304–312 (2022). https://doi.org/10.1038/s41437-022-00530-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-022-00530-4