Abstract
The occasional westward transport of warm water of the Agulhas Current, “Agulhas leakage”, around southern Africa has been suggested to facilitate tropical marine connectivity between the Atlantic and Indian oceans, but the “Agulhas leakage” hypothesis does not explain the signatures of eastward gene flow observed in many tropical marine fauna. We investigated an alternative hypothesis: the establishment of a warm-water corridor during comparatively warm interglacial periods. The “warm-water corridor” hypothesis was investigated by studying the population genomic structure of Atlantic and Southwest Indian Ocean green turtles (N = 27) using 12,035 genome-wide single nucleotide polymorphisms (SNPs) obtained via ddRAD sequencing. Model-based and multivariate clustering suggested a hierarchical population structure with two main Atlantic and Southwest Indian Ocean clusters, and a Caribbean and East Atlantic sub-cluster nested within the Atlantic cluster. Coalescent-based model selection supported a model where Southwest Indian Ocean and Caribbean populations diverged from the East Atlantic population during the transition from the last interglacial period (130–115 thousand years ago; kya) to the last glacial period (115–90 kya). The onset of the last glaciation appeared to isolate Atlantic and Southwest Indian Ocean green turtles into three refugia, which subsequently came into secondary contact in the Caribbean and Southwest Indian Ocean when global temperatures increased after the Last Glacial Maximum. Our findings support the establishment of a warm-water corridor facilitating tropical marine connectivity between the Atlantic and Southwest Indian Ocean during warm interglacials.
Similar content being viewed by others
Introduction
The genetic structure of populations is shaped by the combined action of intrinsic (e.g., behaviour and ecology), and extrinsic processes (e.g., geographic isolation and environmental change) across space and time (Hewitt 2000). The spatio-temporal changes of the underlying processes affect the overall distribution of genetic diversity within and among individuals in different ways, from which the population history may be inferred and key processes identified (Hewitt 2000). A classic example of the latter is the isolation of populations in glacial refugia, subsequent post-glacial expansion and admixture observed in many species on the European continent (Taberlet et al. 1998). Insights into the past biological and environmental processes that shaped the contemporary genetic structure of populations are of key importance to our understanding of the evolution of biodiversity.
A few key biogeographic processes have been associated with large-scale population genetic structure and phylogeographic patterns in marine species. Many marine species show a degree of genetic divergence between Atlantic and Pacific lineages consistent with the formation of the Panama Isthmus ~2.8 million years ago (Coates et al. 1992; Lessios 2008). More recently, sea level fluctuations during the Pleistocene (Pillans et al. 1998) altered the spatial configuration and areas of different marine habitats (Ludt and Rocha 2015), in turn affecting population connectivity (Rocha 2003). For example, the lowered sea levels during glacial periods resulted in the exposure of the Sunda and Sahel shelves in the Indo-Malay Archipelago (Woodruff 2010) leading to the concurrent isolation of conspecific marine populations in the West Pacific Ocean and the Indian Ocean. Subsequent sea level rise during interglacial periods re-established connectivity and subsequent secondary contact among previously isolated populations (Gaither et al. 2011).
After the formation of the Isthmus of Panama, the waters off the southern tip of Africa became the primary marine migratory corridor between the Indian Ocean, and Atlantic Ocean (Teske et al. 2011). The cold-water Benguela Current is believed to restrict dispersal in tropical marine fauna between the two ocean basins (Bowen et al. 2016). However, the incidental leakage of warm and saline water of the Agulhas Current, or “Agulhas leakage”, from the Indian Ocean into the Atlantic Ocean (Penven et al. 2001) has been proposed to facilitate tropical marine connectivity (Bowen et al. 2016). The population genetic structure of many tropical marine fauna indicates westward gene flow from the Indian Ocean around the southern tip of Africa into the Atlantic Ocean consistent with the Agulhas leakage hypothesis. For example, analysis of mitochondrial DNA sequences suggested an Atlantic colonization by Indo-Pacific olive ridley turtles, Lepidochelys olivacea, somewhere during the last 300 thousand years (Bowen et al. 1997). Similar observations were made in reef-associated gobies of the genus Gnatholepis, for which an Atlantic colonization by Indo-Pacific lineages was inferred to have occurred 155–130 kya (Rocha et al. 2005). However, a few marine species show signatures of eastward gene flow of the Indian Ocean by Atlantic mitochondrial lineages, such as the scalloped hammerhead shark, Sphyrna lewini (Duncan et al. 2006), the silky shark, Carcharhinus falciformis (Domingues et al. 2018), the glasseye, Heteropriancanthus spp. (Gaither et al. 2015) and the green turtle, Chelonia mydas (Bourjea et al. 2007). The eastward gene flow observed in these species is inconsistent with the Agulhas leakage hypothesis, which merits exploring alternative hypotheses.
An alternative hypothesis for eastward gene flow from the Atlantic Ocean to the Indian Ocean by tropical marine fauna can be derived by extending the Agulhas leakage hypothesis with the global climate oscillations of the Pleistocene glacial cycles. The intensity of Agulhas leakage fluctuated during the last glacial cycles (Peeters et al. 2004). The transport of warm water from the Indian Ocean to the Atlantic Ocean was reduced during past glacial periods, while westward warm water transport increased during interglacial periods (Peeters et al. 2004). These observations imply that the Pleistocene glacial cycles played an important role in mediating past tropical marine connectivity between the Atlantic Ocean and Indian Ocean by influencing the intensity of Agulhas leakage. However, Pleistocene glacial cycles also caused major global shifts in the distribution of climate zones (Hewitt 2000). Tropical climate zones possibly showed greater poleward shifts during comparatively warm interglacial periods, such as the last interglacial period. Global temperatures were higher during the last interglacial period (130–115 kya) compared to present-day temperatures (Kukla et al. 2002; Bintanja et al. 2005). Comparatively large poleward expansions of tropical climate zones are supported by extensive poleward shifts in the distribution of reef corals during the last interglacial period (Kiessling et al. 2012). A combination of intensified Agulhas leakage and pronounced poleward shifts in tropical climate zones could potentially lead to the establishment of a persistent warm-water corridor around the southern tip of Africa.
Under the “warm-water corridor” hypothesis, eastward gene flow is expected to occur during interglacial periods characterized by warmer-than-present climates. Reliable dating of past colonization events requires obtaining robust estimates of demographic parameters, such as divergence times, which may be improved by basing an assessment upon a large number of unlinked, genome-wide markers (Edwards and Beerli 2000). However, most previous studies that investigated Atlantic-Indian Ocean gene flow in tropical marine fauna (Bowen et al. 1997; Rocha et al. 2005; Duncan et al. 2006; Domingues et al. 2018) were based exclusively upon analyses of mitochondrial DNA sequences, which is subject to two key limitations. First, single-locus inferences do not necessarily reflect the population history (Ball et al. 1990). Second, in most species the inferences drawn from mitochondrial DNA sequences reflect the maternal history, which in turn is affected by sex-specific migratory and dispersal characteristics (Wilson et al. 1985). However, genotyping large numbers of single nucleotide polymorphism (SNP) markers at relatively low costs is now feasible for non-model organisms (Baird et al. 2008; Peterson et al. 2012), providing an avenue for reliably dating past colonization events and testing the warm-water corridor hypothesis.
The present study assessed the warm-water corridor hypothesis by investigating the population genomic structure of Atlantic and Southwest Indian Ocean green turtles using genome-wide SNPs. The green turtle represents an excellent subject for studying the effect of past glaciations on Atlantic-Indian Ocean tropical marine connectivity using genome-wide SNP markers. The discovery of southern Atlantic mitochondrial lineages in the Southwest Indian Ocean suggested a recent eastward colonization into the Southwest Indian Ocean from the Atlantic Ocean (Bourjea et al. 2007). However, the timing of this colonization remained largely uncertain, which can be attributed to a lack of resolution due to the analysis being based on a single molecular marker (i.e., the mitochondrial control region; Bourjea et al. 2007). In addition, the green turtle is a good example of a species with highly sex-specific migratory behaviour. Adult female green turtles exhibit natal homing behaviour, returning to their natal region for mating and nesting during the breeding season (Carr et al. 1978). Accordingly, the sequence variation in the mitochondrial genome is highly structured across space in green turtles (Meylan et al. 1990). By contrast, spatial genetic structure is less apparent in nuclear diversity, suggesting male-mediated gene flow is prevalent among rookeries in green turtles (Karl et al. 1992; Roberts et al. 2004; Naro-Maciel et al. 2014), despite reports of natal homing in adult males (FitzSimmons et al. 1997; Bradshaw et al. 2018). Inferences solely based upon mitochondrial sequence variation are therefore expected to reflect the maternal population history. The application of genome-wide SNPs addresses these concerns, providing an avenue for testing the warm-water corridor hypothesis and improve our understanding of the influence of glaciations on past tropical marine connectivity between the Atlantic-, and Indian oceans.
Methods
Sample collection and DNA extraction
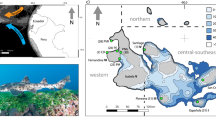
We obtained samples of foraging juvenile or sub-adult green turtles from three geographic regions (Fig. 1): the Caribbean (N = 13), East Atlantic (N = 7) and Southwest Indian Ocean (N = 8). Tissue samples comprised either a sliver of skin excised from the dorsal neck epidermal area, or skin from the front flippers. Tissue samples were excised with a sterile scalpel blade or 6 mm biopsy punch, and stored in 5 M NaCl with 25% dimethyl sulfoxide (Amos and Hoelzel 1991) or 70% ethanol. The Caribbean samples were collected from green turtles captured by hand or by netting in Lac Bay, Bonaire. A previous study showed the majority of foraging green turtles (98%) in Lac Bay originate from rookeries across the wider Caribbean (Van der Zee et al. 2019). The East Atlantic samples originated from Príncipe Island, São Tomé and Príncipe and were collected as part of an earlier study (Alfaro-Nunez et al. 2014). Most green turtles that forage in Príncipe Island originate from Guinea Bissau (77%), followed by Ascension Island (7%; Patrício et al. 2017). The Southwest Indian Ocean samples were collected off the Barren Isles, western Madagascar. Green turtles that forage off western Madagascar originate primarily from rookeries in the central (65%) and southern (34%) Southwest Indian Ocean (Jensen et al. 2020). The Caribbean, East Atlantic and Southwest Indian Ocean samples were collected in 2015–2016; 2007; and 2006–2007, respectively.
Total-cell DNA was extracted from each sample using the Gentra Puragene® Tissue Kit (QIAGEN Inc.) according to the manufacturer’s instructions and resuspended in 1XTE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). The DNA quality was assessed by agarose gel electrophoresis. DNA concentrations were estimated through fluorometric quantitation using a Qubit® 2.0 fluorometer (Life Technologies) and normalized to 20.0 ng/μL.
Library preparation
Double-digest restriction associated DNA (ddRAD) libraries were prepared following the original protocol (Peterson et al. 2012). Genomic DNA was double-digested using HindIII and MspI endo-nuclease restriction enzymes. Each DNA extraction was uniquely barcoded by ligating a unique combination of P1 and P2 adapters to the double-digested DNA fragments (Supplementary Table S1). Uniquely barcoded samples were pooled and cleaned using in-house Sera-mag SpeedBeads (1.5x ratio). Barcoded DNA fragments were size-selected (300–400 base pairs [bp] range) using a Pippin Prep™ (Sage Science Inc.). Size-selected fragments were enriched and uniquely indexed using a Phusion® High-Fidelity PCR kit (New England Biolabs). The resulting library was paired-end sequenced (100 bp) in a single lane on an Illumina HiSeq2500 in high-throughput mode at the National DNA Sequencing Centre at the University of Copenhagen (Denmark).
SNP genotyping
The full bioinformatic and analysis pipeline is described in detail in Supplementary Fig. S1. Raw sequence reads were demultiplexed and processed using process_radtags in STACKS version 1.47 (Catchen et al. 2011). Paired-end reads were aligned against a green turtle reference genome (accession number: GCF_000344595.1; Wang et al. 2013) with BOWTIE2 version 2.3.3.1 (Langmead et al. 2009) using the pre-defined settings “very-sensitive” and “end-to-end”. Discordant reads were aligned as unpaired reads. Alignments were converted to BAM format and sorted using SAMTOOLS version 1.7 (Li et al. 2009).
Genotypes were called from the aligned reads with a minimum mapping quality of 30 using the marukilow model (Maruki and Lynch 2017) implemented in gstacks (STACKS version 2.2). Loci genotyped in all three populations (p = 3) and in >80% of the samples in each population (r = 0.80) were retained using STACKS populations. We did not filter based upon Hardy-Weinberg equilibrium because our samples were from regions where genetically distinct populations overlap (Bourjea et al. 2007; Naro-Maciel et al. 2014). The maximum observed heterozygosity to process a nucleotide site at a RAD locus (hmax) was set at 0.5. All SNPs per RAD locus were retained. No minimum minor allele frequency filter was applied to avoid a potential bias in the population structure assessments (see Linck and Battey 2019). We refer to the data at this point as the “standard STACKS output” (Supplementary Fig. S1).
SNPs with a mean sequencing depth across samples below 30x or above 300x were excluded (i.e., the “depth-filtered” data; Supplementary Fig. S1) using VCFTOOLS version 0.1.15 (Danecek et al. 2011). SNPs were thinned within 100,000 bp to limit potential biases due to physical linkage (i.e., the “unlinked” SNP data; Supplementary Fig. S1) using VCFTOOLS.
Population structure assessment
Genetic differentiation among sampling locations was assessed by estimating pairwise FST from the full and downsampled unlinked data (Supplementary Fig. S1) using SCIKIT-ALLEL version 1.3.2 (Miles et al. 2017) with Python version 3.8.2. Two FST estimators were used: Weir & Cockerham’s θ based upon allele frequencies (Weir and Cockerham 1984) and Hudson’s FST estimator (Hudson et al. 1992; Bhatia et al. 2013) based upon allele counts. We chose the well-known Weir & Cockerham’s θ given it is widespread use and familiarity among researchers, and Hudson’s FST due to its relative insensitivity to sample size variance (Bhatia et al. 2013). P-values were obtained by estimating a null distribution for each test statistic (i.e., Hudson’s FST and Weir & Cockerham’s θ) using a randomization approach and calculating the proportion of values greater than or equal to the observed value. Null distributions were obtained by randomly assigning samples to a sampling location in each pairwise comparison (n = 1000 replicates) and calculating Hudson’s FST and Weir and Cockerham’s θ for each replicate.
To assess whether our sample sizes were sufficient to detect population structure at the estimated level of genetic differentiation, simulations were performed using MSPRIME version 0.7.4 (Kelleher et al. 2016) with Python version 3.8.2. For analytical convenience, we assumed a two-population model where each population was comprised of 10,000 diploid individuals. Migration among the two populations was symmetrical with migration rate m = 0.00025. We generated “RAD-like” data by simulating 20,000 loci that were 200 bp in size and a per-generation mutation rate of 1.0 × 10−8 assuming an infinite-sites mutation model (i.e., each mutation introduces a SNP at a different site). If a single in silico RAD locus contained multiple SNPs, only the first SNP was retained to ensure each SNP was unlinked. We explored a range of sample sizes per population (i.e., Ni = {2, 3, 4, 5, 7, 8, 9, 10, 15 20, 25, 50, 75, 100}; Fig. 3), and simulated 1000 datasets per sample size. For each dataset, we estimated pairwise FST using Weir & Cockerham’s θ, and Hudson’s FST with SCIKIT-ALLEL version 1.3.2 (Python version 3.8.2). The expected value of FST in a symmetrical island model was calculated according to FST ≈ 1/(1 + 4Nem) (Wright 1951) given m = 0.00025 and Ne = 10,000.
We performed model-based clustering using STRUCTURE version 2.3.4 (Pritchard et al. 2000) to detect the best-supported number of clusters K and estimate admixture proportions. STRUCTURE requires unlinked markers (Pritchard et al. 2000). Hence, the STRUCTURE analysis was conducted using the unlinked SNP data (Supplementary Fig. S1). Values of K = 1–10 were evaluated from 15 replicate assessments for each value of K. Each replicate consisted of an initial burn-in of 100,000 iterations followed by 1,000,000 iterations. The ancestry prior α was estimated separately for each sampling location (Wang 2017). The starting value of α was set to 0.25. The STRUCTURE estimations assumed correlated allele frequencies (Falush et al. 2003). The most probable number of clusters (i.e., K) was inferred from the mean likelihood of K as well as ∆K (Evanno et al. 2005). The pophelper R package (Francis 2017) was employed to estimate ∆K and visualize admixture proportions as well as merge results across replicates. In addition, we performed a principal component analysis using the ade4 R package (Dray and Dufour 2007), as well as multivariate-based clustering and a discriminant analysis of principal components (DAPC) using the adegenet R package (Jombart et al. 2010). All analyses using R were performed with R version 4.0.2 (R Core Team 2021). To facilitate a comparison with the STRUCTURE results, we performed multivariate clustering on the unlinked SNP data. We retained the number of principal components that explained 80% of the cumulative variance. In addition, the optimal number of principal components to retain in the DAPC was determined by α-score optimization. Finally, we estimated a co-ancestry matrix from the depth-filtered data (Supplementary Fig. S1) with FINERADSTRUCTURE using the default settings (Malinsky et al. 2018).
Coalescent model selection
We assessed four demographic models with different migration patterns and population divergence times (Fig. 2) using MIGRATE-N version 4.2.14 (Beerli 2006; Beerli and Palczewski 2010). Model parameter priors are listed in Supplementary Table S2. In order to reduce computational load, we conducted the MIGRATE-N assessment using five random subsets of 5000 SNPs from the unlinked SNP data (Supplementary Fig. S1). A custom Python script was used to retrieve the full DNA sequence of the RAD locus associated with each SNP in the five random subsets from the standard STACKS output (Supplementary Fig. S1). STACKS output files were converted to input files for MIGRATE-N using the stacks2mig.py script provided with MIGRATE-N. For the MIGRATE-N analysis, we assumed a per-base frequency of 0.25, a transition/transversion ratio at 2.0, a per-base sequencing error rate of 0.001, a constant mutation rate and the F84 mutation model (Felsenstein and Churchill 1996) employed by MIGRATE-N for DNA sequence data. Model parameters were evaluated via slice-sampling. We ran a single long chain with a sampling increment of 100, 1000 iterations and a burn-in of 1000. Ten replicates were run per chain. MCMC convergence was assessed by evaluating whether model parameter effective sample sizes (ESSs) were >1000, following the MIGRATE-N manual. The marginal likelihood of each model was approximated via Bezier-approximated thermodynamic integration using four chains heated to different temperatures: t1 = 1, t2 = 1.5, t3 = 3.0 and t4 = 1,000,000 (Beerli and Palczewski 2010). We calculated log-Bayes factors using the following equation: ln (prob(D | X))–ln (prob(D | Y)) where X was the model compared to the model (Y) with highest support.
(A) Model 1: an island model without divergence; (B) Model 2: CA and SWO diverged from EA; (C) Model 3: CA and EA diverged from SWO; (D) Model 4: EA and SWO diverged from CA. Arrows indicate migration. Differences in population-specific divergence times (τi) are for illustrative purposes; divergence time priors were parametrized similarly among populations (Supplementary Table S2).
Given that θ = 4Neμ and T = Neτ (i.e., population divergence times measured in effective population size times the number of generations), we obtained population divergence times in years from the estimates of θ for different generation times (τ) and mutation rates (μ). We explored values of τ between 30 and 40 years, given the unresolved status of generation times in green turtles (Bell et al. 2005; Goshe et al. 2010; Seminoff et al. 2015). A substitution rate derived from the genome-wide divergence between alligators and crocodiles (7.9 × 10−9 per site per generation) represented the lower bound of mutation rates that were explored (Green et al. 2014). As an upper bound, we used a genome-wide de novo human mutation rate estimated from pedigrees (1.2 × 10−8) per site per generation (Kong et al. 2012; Scally and Durbin 2012).
Results
Sequencing
Raw Illumina sequencing reads were obtained from 28 individual green turtles. Read and alignment statistics are summarized in Supplementary Table S1. Alignment rates were high (>94%). One sample aligned poorly (PI070009) to the green turtle reference genome sequence and was excluded from subsequent analyses, which resulted in a sample size of 27 individuals. An initial number of >156,000 loci containing between 54,000 and 100,000 SNPs were genotyped (Supplementary Table S3), with <6.5% missing data (Supplementary Table S4). Excluding SNPs with a mean sequencing depth averaged across individuals below 30x and above 300x (Supplementary Fig. S2) reduced the number of variant SNPs to ~95,200 with 36,000 to 62,000 SNPs per location (Supplementary Table S3). Depth-filtering decreased the amount of missing data per sample (<2.4%; Supplementary Table S4). Thinning SNPs within 100,000 bp windows resulted in ~12,000 unlinked SNPs (4400–7700 SNPs per sampling location; Supplementary Table S3) with low amounts of missing data (<2.4%; Supplementary Table S4). The fewest SNPs and private alleles were observed in the East Atlantic. The largest number of SNPs and private alleles was observed in the Southwest Indian Ocean (Supplementary Table S3).
Population structure
Estimates of pairwise genetic differentiation were statistically significant (P < 0.001; Supplementary Table S5), and ranged between 0.11 and 0.18 (Hudson’s FST) and 0.10–0.17 (Weir & Cockerham’s θ; Supplementary Table S5). Estimates of Weir & Cockerham’s θ were slightly lower than estimates of Hudson’s FST (Supplementary Table S5). The largest degree of genetic differentiation was observed between the East Atlantic Ocean and the Southwest Indian Ocean (FST = 0.18; θ = 0.17). The smallest genetic differentiation was observed between the Caribbean and East Atlantic (FST = 0.11; θ = 0.10). The largest number of alleles were privately shared between the Caribbean and East Atlantic (1305), followed by the Caribbean and Southwest Indian Ocean (946; Supplementary Table S5).
The simulation results are depicted in Fig. 3, and Supplementary Tables S6 and S7. The expected level of genetic differentiation was ~0.091 in our simulations, which was consistent with the empirical data. Increasing the sample size narrowed the 95% confidence interval, but the decrease in uncertainty was marginal beyond six samples per population. Approximately 2000 to 4700 SNPs were observed in silico, which was lower than the number of SNPs observed in the empirical data (Supplementary Tables S6 and S7).
Genetic differentiation was estimated as pairwise FST using (A) Hudson’s FST estimator and (B) Weir & Cockerham’s θ. The dashed line denotes the expected FST (~0.091). Filled circles denote the mean FST and error bars show the 95% confidence interval derived from 1000 replicate simulations per sample size.
The mean likelihood of K suggested the most likely number of clusters was three (K = 3; Fig. 4A), which was also supported by the multivariate clustering results (Supplementary Fig. S4 and S5). By contrast, ∆K supported K = 2 (Fig. 4B). Admixture proportions estimated for K = 2 partitioned the samples into an Atlantic and Southwest Indian Ocean cluster and indicated admixture in the Southwest Indian Ocean (Fig. 5A). Consistent with ∆K and the admixture proportions under K = 2, samples clustered according to ocean basin in the principal component analysis (Supplementary Fig. S6). The majority of variation was captured by the first principal component (Supplementary Fig. S7), which seemed to reflect inter-oceanic differentiation. For K = 3, the Atlantic cluster was further subdivided in a Caribbean and East Atlantic cluster, with considerable admixture (i.e., >30% East Atlantic ancestry) observed in all Caribbean samples (N = 13; Fig. 5B). An additional model-based clustering analysis performed on solely the Atlantic samples (i.e., Caribbean and East Atlantic) suggested the most likely number of clusters was two (K = 2) according to the mean likelihood of K and ∆K (Fig. 4C, D). In contrast with the admixture proportions estimated for the full data under K = 3, only a few Caribbean samples had East Atlantic ancestry (N = 4; Fig. 5C). The DAPC performed using a number of principal components explaining ~80% of cumulative variance (Supplementary Figs. S8, S10) indicated no admixture (Supplementary Fig. S12) but assigned one Caribbean sample (BO150013) to the East Atlantic cluster. However, α-score trajectories suggested retaining only a single principal component in the DAPC (Supplementary Figs. S9, S11). Caribbean sample BO150013 was assigned ~90% East Atlantic ancestry when a single principal component was retained in the DAPC (Supplementary Fig. S13A), but 100% East Atlantic ancestry when Southwest Indian Ocean samples were excluded (Supplementary Fig. S13B).
Pairwise co-ancestries were higher within oceans than between oceans, indicating a higher degree of genetic similarity within ocean basins consistent with hierarchical population structure (Fig. 6). Two main clades corresponding to the Atlantic (i.e., Caribbean and East Atlantic) and Southwest Indian Ocean were observed in a tree describing the relative degree of co-ancestry among samples, which were further partitioned into sub-clades corresponding to sampling locations (Fig. 6). Consistent with model-based-, and multivariate clustering, pairwise co-ancestries indicated Caribbean sample BO150013 was more genetically similar to East Atlantic samples. Two additional Caribbean samples (BO150077 and BO150141) showed a higher degree of co-ancestry shared with East Atlantic samples and were placed on a separate branch within the Caribbean sub-clade. This was in agreement with the ~20–25% East Atlantic ancestry estimated for these samples via model-based clustering (Fig. 5C).
The scale values represent the total co-ancestry, i.e., the local co-ancestries summed across RAD loci (Malinsky et al. 2018). Higher co-ancestry values imply a higher degree of genetic similarity between sample pairs. A tree depicting the relative degree of co-ancestry among samples is shown above the co-ancestry matrix. Posterior support values are shown for each node.
Coalescent model selection
Effective samples sizes were high (i.e., >1 × 109) and consistent across individual estimations (Supplementary Table S8), indicating MCMC chain convergence. The marginal likelihoods were consistent among individual estimations (Supplementary Table S9). Model 2, i.e., the Caribbean and the Southwest Indian Ocean diverged from the East Atlantic, was best-supported (Table 1). Estimates of θ were similar among the sampling locations, though the estimated value of θ in the East Atlantic (mean θEA = 0.0031) was lower than the Caribbean (mean θCA = 0.0048) and the Southwest Indian Ocean (mean θSWO = 0.0041; Supplementary Table S10). In addition, we estimated asymmetric gene flow from the East Atlantic into the Caribbean (mean MEA→CA = 800.7; mean MCA→EA = 234.1) and the Southwest Indian Ocean (mean MEA→SWO = 710.0; mean MSWO→EA = 251.4). Gene flow between the Caribbean and Southwest Indian Ocean (mean MCA→SWO = 440.5; mean MSWO→CA = 504.7) exceeded gene flow into the East Atlantic (Supplementary Table S10). The divergence time between the East Atlantic and the Southwest Indian Ocean (mean τEA→SWO = 0.0345) predated the divergence time between the Caribbean and East Atlantic (mean τEA→CA = 0.0290; Supplementary Table S10). The divergence of the Southwest Indian Ocean (Table 2A) and Caribbean (Table 2B) from the East Atlantic seemed to align with the timing of the last interglacial period (130–115 kya), but divergence times (in years) varied considerably with assumed mutation rates and generation times. The Southwest Indian Ocean divergence seemed to predate the Caribbean divergence by ~2000 years (Table 2C).
Discussion
Warm water from the Agulhas Current occasionally flows from the Indian Ocean to the Atlantic Ocean around the southern point of Africa (Penven et al. 2001), providing an avenue for tropical marine connectivity between the Indo-Pacific and Atlantic after the closure of the Isthmus of Panama (Teske et al. 2011). However, the westward direction of Agulhas leakage is inconsistent with previous reports of eastward gene flow observed in many tropical marine species in the Atlantic and Southwest Indian Ocean (Duncan et al. 2006; Bourjea et al. 2007; Gaither et al. 2015; Domingues et al. 2018). In the present study, we developed and investigated an alternative hypothesis, which differs from the Agulhas leakage hypothesis by incorporating the influence of the Pleistocene glacial cycles on connectivity between the Atlantic and Indian oceans in tropical marine fauna. The alternative hypothesis was based upon two key drivers of inter-oceanic connectivity that fluctuated between interglacial and glacial periods: (1) the intensity of Agulhas warm water leakage (Peeters et al. 2004), and (2) the distribution of climate zones (Hewitt 2000). Specifically, we hypothesized the establishment of a persistent, warm-water corridor during comparatively warmer interglacial periods due to the combined effect of an increased level of Agulhas leakage and a poleward shift in tropical climate zones. Under this warm-water corridor hypothesis, the divergence between tropical marine taxa in the Atlantic and Indian oceans is expected to coincide with the cooling period at the end of interglacial periods with comparatively higher global temperatures than present-day temperatures.
We assessed the above prediction of the warm-water corridor hypothesis by investigating the population genomic structure of Atlantic and Southwest Indian Ocean green turtles from ~12,000 genome-wide SNP markers. We detected considerable genetic divergence among the Caribbean, East Atlantic and Southwest Indian Ocean sampling locations, as well as hierarchical population structure indicating greater inter-oceanic (i.e., between the Atlantic and Southwest Indian Ocean) versus intra-oceanic (i.e., within the Atlantic) genetic differentiation. Our coalescent-based model selection identified the model in which contemporary green turtle populations in the Caribbean and the Southwest Indian Ocean diverged from the East Atlantic population as the most likely. The population divergence time of Atlantic Ocean and Southwest Indian Ocean green turtles was traced back to the last interglacial period, 130–115 kya (Dahl-Jensen et al. 2013). Our findings suggested that green turtles from the Atlantic colonized the Southwest Indian Ocean during the last interglacial period, consistent with the warm-water corridor hypothesis. The timing of the colonization of the Southwest Indian Ocean by Atlantic green turtles was uncertain in previous studies based solely on mitochondrial DNA sequence variation (Bourjea et al. 2007). The population divergence time estimates suggested that these populations became isolated at the onset of the last glaciation (Kukla et al. 2002; Clark et al. 2009; Dahl-Jensen et al. 2013). These findings suggested that tropical marine connectivity between the Atlantic and Indian oceans was disrupted when the global climate cooled during the initial phase of the last glacial period, followed by further restriction of tropical marine connectivity within the Atlantic Ocean as global cooling continued (Bintanja et al. 2005).
The warm-water corridor hypothesis postulated in the present study represented an extension of the Agulhas leakage hypothesis that is able to explain the westward and eastward colonisations around the southern point of Africa observed in many tropical marine species. For example, gobies of the Gnatholepis genus have been shown to have colonized the Atlantic Ocean from the Indian Ocean during the last interglacial period (Rocha et al. 2005). Other studies identified both west-, and eastward gene flow using mitochondrial DNA, e.g., in olive ridley turtles (Bowen et al. 1997) and scalloped hammerhead sharks (Duncan et al. 2006). However, the data employed in those studies were, in general, insufficient to date the population divergence time. The results presented here suggest that next-generation sequencing approaches, such as RAD-seq (Baird et al. 2008; Peterson et al. 2012), may possibly enable dating these events. Our study also showed that comparatively few samples (e.g., 5 to 10 per location) and a few thousand unlinked SNPs are capable of resolving genetic divergence at these levels of genetic differentiation (e.g., FST ~0.10). By shifting towards smaller sample sizes, cost-effective generation of multi-species genomic datasets becomes feasible in difficult to sample, or rare non-model organisms. Multi-species analyses are a powerful approach to further assess and interrogate the the consistency of hypotheses aiming at the influence of Pleistocene glacial cycles on tropical marine connectivity between the Atlantic and Indian oceans; such as assessing species across a range of different life history traits, e.g., migratory characteristics and age structure (Duncan et al. 2006).
The presence of three genetic clusters suggested that Atlantic and Southwest Indian Ocean green turtles were isolated in three refugia during the most recent glacial period. Our results are consistent with previous studies (Encalada et al. 1996; Reece et al. 2005; Naro-Maciel et al. 2014; Reid et al. 2019; Jensen et al. 2019). Previous studies aimed at Atlantic green turtles, employing mitochondrial DNA sequences and microsatellite genotypes, suggested the presence of two glacial refugia located in the West Caribbean and South Atlantic, and subsequent post-glacial secondary contact in the East Caribbean (Encalada et al. 1996; Naro-Maciel et al. 2014). Mitochondrial DNA lineages in the Atlantic partition into a northern clade containing West Caribbean and Mediterranean Sea individuals, and a southern clade including Caribbean and South Atlantic individuals (Encalada et al. 1996). Microsatellite variation showed a similar pattern, i.e., two Atlantic genetic clusters. A recent global population genetic analysis of green turtles, based upon mitochondrial DNA sequences, suggested the presence of glacial refugia in the Northwest Caribbean, East Atlantic and Southwest Indian Ocean, inferred from the levels of mitochondrial DNA haplotype diversity and private alleles (Jensen et al. 2019).
The admixture proportions estimated in this study suggested that green turtle populations isolated in glacial refugia underwent post-glacial secondary contact in the Caribbean and Southwest Indian Ocean, which is in agreement with previous studies (Naro-Maciel et al. 2014). Admixture proportions were higher in the Caribbean compared to the Southwest Indian Ocean, where only a small proportion of East Atlantic ancestry was detected. Including samples from the Southwest Indian Ocean appeared to increase admixture in the Caribbean (i.e., a high degree of apparent East Atlantic ancestry in Caribbean green turtles). By contrast, excluding the Southwest Indian Ocean decreased apparent admixture in the Caribbean. These findings are likely explained by hierarchical population structure and uneven sample sizes, which can lead to unpredictable clustering and erroneous ancestry inferences (Kalinowski 2011) under the island model employed by STRUCTURE (Pritchard et al. 2000). Here, we suspect that East Atlantic ancestry within the Caribbean was over-estimated when the Southwest Indian Ocean was included, because alleles could be less confidently assigned to Atlantic sub-clusters due to genetic similarities within the Atlantic.
The DAPC suggested a lack of admixture in the Caribbean and Southwest Indian compared to model-based clustering. For example, one Caribbean sample (BO150013) was assigned 100% East Atlantic ancestry, while the same sample was assigned ~64% East Atlantic ancestry via model-based clustering. We hypothesize that the lack of admixture inferred by DAPC can be attributed to over-fitting of posterior group memberships because too many principal components were retained, resulting in a “perfect” assignment of individuals to clusters (Jombart et al. 2010). Concordant with over-fitting, retaining a single principal component resulted in ~90% East Atlantic ancestry for sample BO150013 when the full data (i.e., Atlantic and Southwest Indian Ocean) was considered. We suspect that a combination of a large number of SNP markers, strong genetic differentiation among sampling locations and relatively small sample sizes contributed to the over-fitting of group memberships in our study.
Range contractions, isolation in refugia and subsequent post-glacial range expansions were likely associated with concurrent fluctuations in population sizes, a parameter not assessed in our analysis. The isolation-with-migration model framework employed in this study assumes constant population sizes (Beerli 2006). Fluctuating population sizes may bias estimates of Ne in isolation-with-migration models (Strasburg and Rieseberg 2010). In this study, divergence times were estimated in units of Ne generations. Accordingly, a downward biased estimate of Ne will lead to a downward bias in population divergence times as well. By contrast, population genetic sub-structuring appear to have negligible effects on estimates of divergence times and migration rates in isolation-with-migration models (Strasburg and Rieseberg 2010).
Obtaining estimates of divergence times in years necessitated stipulation of the generation time and mutation rates. Given the inherent uncertainty in estimating mutation rates as well as the unresolved status of generation times in green turtles, we explored a range of values for these parameters to obtain estimates of divergence times in years. Specifically, we assessed a range of mutation rates between 7.9 × 10−9 (obtained from crocodilians; Green et al. 2014) and 1.2 × 10−8 per site per generation (obtained from humans; Kong et al. 2012; Scally and Durbin 2012). Rates of molecular evolution in crocodilians and green turtles appear to be largely similar and slower than mammals (Green et al. 2014), suggesting that the “true” green turtle mutation rate may be located near the lower bound of the range of mutation rates explored in this study. However, the crocodilian mutation rate represented a substitution rate derived from the genome-wide divergence between alligators and crocodiles (Green et al. 2014). Rates of molecular evolution estimated from phylogenies, such as the crocodilian estimate, possibly underestimate the actual mutation rate since some novel mutations are expected to be removed over time via selection and genetic drift (Ho et al. 2005). An underestimate of the mutation rate, which acts as a scaling factor, implies a shift towards more recent population divergence times. Consequently, we used a genome-wide de novo mutation rate estimated from humans (Kong et al. 2012) as an upper bound. If we assume that the crocodilian substitution rate underestimates the “true” green turtle mutation rate without exceeding the human de novo mutation rate, the “true” green turtle mutation rate possibly falls within the middle of the range of explored mutation rates, consistent with a population divergence associated with the timing of the last interglacial period. In regards to the generation time, we assumed values between 30 and 40 years (Seminoff et al. 2015). However, we note that the generation time in green turtles is largely unknown (Bell et al. 2005; Goshe et al. 2010) and age in sea turtles is typically inferred indirectly from mark-recapture studies and growth rates, which depend on environmental conditions (Bjorndal et al. 2000, 2017).
An important consideration in any population genetic analysis is how many samples are required to reliably detect population genetic structure and estimate demographic parameters in evolutionary models. While our sample sizes were small, the statistical power in coalescence-based analyses is primarily determined by the number of markers (Felsenstein 2006). In addition, the effect size is an important determinant of statistical power, which when testing for population genetic structure is the magnitude of genetic differentiation among populations (Waples 1998). Pairwise genetic differentiation among sampling locations was high in our study (i.e., FST > 0.10), implying a large effect size and therefore high statistical power. This suspicion was supported by our simulation results. Only a few samples were required to characterize population genetic structure given the observed level of genetic differentiation and the number of SNP markers that were used in the present study, which is consistent with previous studies (Willing et al. 2012). Furthermore, clustering approaches are known to perform well at FST > 0.02 (Latch et al. 2006), which is considerably lower than the level of genetic differentiation observed in the present study. Finally, minimizing the number of samples required to answer a research question can be warranted for practical, economical, biological and ethical reasons. In particular for endangered species, such as the green turtle (Seminoff 2004).
Our data was comprised of individuals sampled at feeding grounds, which are typically shared by different rookeries in sea turtles (Lahanas et al. 1998). Long-distance dispersal from rookeries to feeding grounds has been reported in juveniles (Bowen et al. 1995; Boyle et al. 2009; Monzón-Argüello et al. 2010). Consequently, the three Caribbean samples inferred to possess East Atlantic ancestry possibly were immigrants from distant rookeries located outside the Caribbean. However, West Caribbean rookeries seem to account for the majority of juveniles (80%) at the Lac Bay feeding ground, while fewer individuals seem to originate from East Caribbean (18%) and South Atlantic (2%) rookeries (Van der Zee et al. 2019). Here, South Atlantic represented a grouping of Southwest Atlantic (e.g., Brazil) and East Atlantic rookeries based upon similarities in mitochondrial and nuclear DNA variation (Naro-Maciel et al. 2014; Patrício et al. 2017). Given that dispersal from the South Atlantic was estimated to be relatively rare (Van der Zee et al. 2019), we suggest that our sample may have included immigrants from East Caribbean rookeries that are possibly recent descendants of migrants from South Atlantic rookeries. Unfortunately, a lack of samples from the Southwest Atlantic precluded investigating this hypothesis.
Our study represents one of the first studies applying large numbers of SNPs obtained from next-generation sequencing to assess sea turtle population genomic structure. Recently, a Rapture-based (Ali et al. 2016) framework was published employing ~2000 oligonucleotide baits (from leatherback turtle, Dermochelys coriacea, RAD loci) to genotype SNPs using DNA capture in hard-shell sea turtles, such as the green turtle (Komoroske et al. 2019). Our genomic resources are freely available to researchers and can similarly be used to design oligonucleotide probes for DNA capture-based sequencing (Gnirke et al. 2009). For researchers interested in genotyping large numbers of unlinked genome-wide SNPs in Atlantic Ocean and Southwest Indian Ocean green turtles, the genomic resources in the present study may be of interest. While ~11,000 SNPs were identified in a global sample of green turtles using the Rapture-based framework, these were genotyped using ~2000 oligonucleotide probes and suggested many of the SNPs identified were located within close proximity (Komoroske et al. 2019). It is clear, however, that the use of next-generation sequencing tools in sea turtles is emerging and can play a major role in sea turtle research in the coming decade. Next-generation sequencing technologies have the potential to greatly improve the detection of population structure, which represents one of the key open questions in sea turtle research (Hamann et al. 2010) and of critical importance for determining the spatial scales of management efforts for marine species (Wallace et al. 2010). The fine-scale population structure demonstrated in the present study, along with the decreasing costs of sequencing, highlights the potential of SNPs for in-depth studies of population structure and connectivity in sea turtles, and for marine species in general.
Data availability
Raw sequencing data has been deposited in Dryad (https://doi.org/10.5061/dryad.31zcrjdmd). Simulation scripts have been deposited in Zenodo (https://doi.org/10.5281/zenodo.5513679) and are also available at GitHub (https://github.com/jpvdz/green-turtle-sims).
References
Alfaro-Nunez A, Bertelsen MF, Bojesen AM, Rasmussen I, Zepeda-Mendoza L, Olsen MT et al. (2014) Global distribution of Chelonid fibropapilloma-associated herpesvirus among clinically healthy sea turtles. BMC Evol Biol 14:206
Ali OA, O’Rourke SM, Amish SJ, Meek MH, Luikart G, Jeffres C et al. (2016) RAD capture (Rapture): flexible and efficient sequence-based genotyping. Genetics 202:389–400
Amos B, Hoelzel AR (1991). Long-term preservation of whale skin for DNA analysis. Rep Int Whal Comm Spec Issue 13:99–103
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA et al. (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376–8
Ball RM, Neigel JE, Avise JC (1990) Gene genealogies within the organismal pedigrees of random-mating populations. Evolution 44:360–370
Beerli P (2006) Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22:341–345
Beerli P, Palczewski M (2010) Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185:313–326
Bell CDL, Parsons J, Austin TJ, Broderick AC, Ebanks-Petrie G, Godley BJ (2005) Some of them came home: the Cayman Turtle Farm headstarting project for the green turtle Chelonia mydas. Oryx 39:137–148
Bhatia G, Patterson N, Sankararaman S, Price AL (2013) Estimating and interpreting FST: The impact of rare variants. Genome Res 23:1514–1521
Bintanja R, van de Wal RSW, Oerlemans J (2005) Modelled atmospheric temperatures and global sea levels over the past million years. Nature 437:125–128
Bjorndal KA, Bolten AB, Chaloupka MY (2000) Green turtle somatic growth model: evidence for density dependence. Ecol Appl 10:269–282
Bjorndal KA, Bolten AB, Chaloupka M, Saba VS, Bellini C, Marcovaldi MAG et al. (2017) Ecological regime shift drives declining growth rates of sea turtles throughout the West Atlantic. Glob Change Biol 23:4556–4568
Bourjea J, Lapegue S, Gagnevin L, Broderick D, Mortimer JA, Ciccione S et al. (2007) Phylogeography of the green turtle, Chelonia mydas, in the Southwest Indian Ocean. Mol Ecol 16:175–186
Bowen BW, Abreu-Grobois FA, Balazs GH, Kamezaki N, Limpus CJ, Ferl RJ (1995) Trans-Pacific migrations of the loggerhead turtle (Caretta caretta) demonstrated with mitochondrial DNA markers. Proc Natl Acad Sci 92:3731–3734
Bowen BW, Clark AM, Abreu-Grobois FA, Chaves A, Reichart HA, Ferl RJ (1997) Global phylogeography of the ridley sea turtles (Lepidochelys spp.) as inferred from mitochondrial DNA sequences. Genetica 101:179–189
Bowen BW, Gaither MR, DiBattista JD, Iacchei M, Andrews KR, Grant WS et al. (2016) Comparative phylogeography of the ocean planet. Proc Natl Acad Sci 113:7962–7969
Boyle MC, FitzSimmons NN, Limpus CJ, Kelez S, Velez-Zuazo X, Waycott M (2009) Evidence for transoceanic migrations by loggerhead sea turtles in the southern Pacific Ocean. Proc R Soc B 276:1993–1999
Bradshaw PJ, Broderick AC, Carreras C, Fuller W, Snape RTE, Wright LI et al. (2018) Defining conservation units with enhanced molecular tools to reveal fine scale structuring among Mediterranean green turtle rookeries. Biol Conserv 222:253–260
Carr AF, Carr MH, Meylan AB (1978). The ecology and migrations of sea turtles, 7. The West Caribbean green turtle colony. Bull Am Mus Nat Hist 162:1–46
Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH (2011) Stacks: building and genotyping loci de novo from short-read sequences. G3 Genes Genomes Genet 1:171–182
Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B et al. (2009) The last glacial maximum. Science 325:710–714
Coates AG, Jackson JBC, Collins LS, Cronin TM, Dowsett HJ, Bybell LM et al. (1992) Closure of the Isthmus of Panama: the near-shore marine record of Costa Rica and western Panama. GSA Bull 104:814–828
Dahl-Jensen D, Albert MR, Aldahan A, Azuma N, Balslev-Clausen D, Baumgartner M et al. (2013) Eemian interglacial reconstructed from a Greenland folded ice core. Nature 493:489–494
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Domingues RR, Hilsdorf AWS, Shivji MM, Hazin FVH, Gadig OBF (2018) Effects of the Pleistocene on the mitochondrial population genetic structure and demographic history of the silky shark (Carcharhinus falciformis) in the western Atlantic Ocean. Rev Fish Biol Fish 28:213–227
Dray S, Dufour A-B (2007) The ade4 package: Implementing the duality diagram for ecologists. J Stat Softw 22:1–20.
Duncan KM, Martin AP, Bowen BW, Couet HGD (2006) Global phylogeography of the scalloped hammerhead shark (Sphyrna lewini). Mol Ecol 15:2239–2251
Edwards S, Beerli P (2000) Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution 54:1839–1854
Encalada SE, Lahanas PN, Bjorndal KA, Bolten AB, Miyamoto MM, Bowen BW (1996) Phylogeography and population structure of the Atlantic and Mediterranean green turtle Chelonia mydas: a mitochondrial DNA control region sequence assessment. Mol Ecol 5:473–483
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Felsenstein J (2006) Accuracy of coalescent likelihood estimates: do we need more sites, more sequences, or more loci? Mol Biol Evol 23:691–700
Felsenstein J, Churchill GA (1996) A Hidden Markov Model approach to variation among sites in rate of evolution. Mol Biol Evol 13:93–104
FitzSimmons NN, Limpus CJ, Norman JA, Goldizen AR, Miller JD, Moritz C (1997) Philopatry of male marine turtles inferred from mitochondrial DNA markers. Proc Natl Acad Sci 94:8912–8917
Francis RM (2017) pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour 17:27–32
Gaither MR, Bernal MA, Fernandez-Silva I, Mwale M, Jones SA, Rocha C et al. (2015) Two deep evolutionary lineages in the circumtropical glasseye Heteropriacanthus cruentatus (Teleostei, Priacanthidae) with admixture in the south-western Indian Ocean. J Fish Biol 87:715–727
Gaither MR, Bowen BW, Bordenave T-R, Rocha LA, Newman SJ, Gomez JA et al. (2011). Phylogeography of the reef fish Cephalopholis argus (Epinephelidae) indicates Pleistocene isolation across the Indo-Pacific barrier with contemporary overlap in the coral triangle. BMC Evol Biol 11:189
Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W et al. (2009) Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27:182–189
Goshe LR, Avens L, Scharf FS, Southwood AL (2010) Estimation of age at maturation and growth of Atlantic green turtles (Chelonia mydas) using skeletochronology. Mar Biol 157:1725–1740
Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G et al. (2014) Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346:1254449
Hamann M, Godfrey MH, Seminoff JA, Arthur K, Barata PCR, Bjorndal KA et al. (2010) Global research priorities for sea turtles: informing management and conservation in the 21st century. Endanger Species Res 11:245–269
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Ho SYW, Phillips MJ, Cooper A, Drummond AJ (2005) Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol 22:1561–1568
Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589
Jensen MP, Dalleau M, Gaspar P, Lalire M, Jean C, Ciccione S et al. (2020) Seascape genetics and the spatial ecology of juvenile green turtles. Genes 11:278
Jensen MP, FitzSimmons NN, Bourjea J, Hamabata T, Reece J, Dutton PH (2019) The evolutionary history and global phylogeography of the green turtle (Chelonia mydas). J Biogeogr 46:1–11.
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Kalinowski ST (2011) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106:625–632
Karl SA, Bowen BW, Avise JC (1992) Global population genetic structure and male-mediated gene flow in the green turtle (Chelonia mydas): RFLP analyses of anonymous nuclear loci. Genetics 131:163–173
Kelleher J, Etheridge AM, McVean G (2016) Efficient coalescent simulation and genealogical analysis for large sample sizes. PLOS Comput Biol 12:e1004842
Kiessling W, Simpson C, Beck B, Mewis H, Pandolfi JM (2012) Equatorial decline of reef corals during the last Pleistocene interglacial. Proc Natl Acad Sci 109:21378–21383
Komoroske LM, Miller MR, O’Rourke SM, Stewart KR, Jensen MP, Dutton PH (2019) A versatile Rapture (RAD-Capture) platform for genotyping marine turtles. Mol Ecol Resour 19:497–511
Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G et al. (2012) Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488:471–475
Kukla GJ, Bender ML, de Beaulieu J-L, Bond G, Broecker WS, Cleveringa P et al. (2002) Last interglacial climates. Quat Res 58:2–13
Lahanas PN, Bjorndal KA, Bolten AB, Encalada SE, Miyamoto MM, Valverde RA et al. (1998) Genetic composition of a green turtle (Chelonia mydas) feeding ground population: evidence for multiple origins. Mar Biol 130:345–352
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Latch EK, Dharmarajan G, Glaubitz JC, Rhodes Jr OE (2006) Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet 7:295–302
Lessios HA (2008) The great American schism: divergence of marine organisms after the rise of the Central American Isthmus. Annu Rev Ecol Evol Syst 39:63–91
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079
Linck EB, Battey CJ (2019) Minor allele frequency thresholds strongly affect population structure inference with genomic datasets. Mol Ecol Resour 19:639–647
Ludt WB, Rocha LA (2015) Shifting seas: the impacts of Pleistocene sea-level fluctuations on the evolution of tropical marine taxa. J Biogeogr 42:25–38
Malinsky M, Trucchi E, Lawson DJ, Falush D (2018) RADpainter and fineRADstructure: population inference from RADseq data. Mol Biol Evol 35:1284–1290
Maruki T, Lynch M (2017) Genotype calling from population-genomic sequencing data. G3 Genes Genomes Genet 7:1393–1404
Meylan AB, Bowen BW, Avise JC (1990) A genetic test of the natal homing versus social facilitation models for green turtle migration. Science 248:724–727
Miles A, Murillo R, Ralph P, Harding N, Pisupati R, Rae S et al. (2017). cggh/scikit-allel: v1.3.2. Zenodo
Monzón-Argüello C, López-Jurado LF, Rico C, Marco A, López P, Hays GC et al. (2010) Evidence from genetic and Lagrangian drifter data for transatlantic transport of small juvenile green turtles. J Biogeogr 37:1752–1766
Naro-Maciel E, Reid BN, Alter SE, Amato G, Bjorndal KA, Bolten AB et al. (2014) From refugia to rookeries: phylogeography of Atlantic green turtles. J Exp Mar Biol Ecol 461:306–316
Patrício AR, Formia A, Barbosa C, Broderick AC, Bruford M, Carreras C et al. (2017) Dispersal of green turtles from Africa’s largest rookery assessed through genetic markers. Mar Ecol Prog Ser 569:215–225
Peeters FJC, Acheson R, Brummer G-JA, De Ruijter WPM, Schneider RR, Ganssen GM et al. (2004) Vigorous exchange between the Indian and Atlantic oceans at the end of the past five glacial periods. Nature 430:661–665
Penven P, Lutjeharms JRE, Marchesiello P, Roy C, Weeks SJ (2001) Generation of cyclonic eddies by the Agulhas Current in the lee of the Agulhas Bank. Geophys Res Lett 28:1055–1058
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135
Pillans B, Chappell J, Naish TR (1998) A review of the Milankovitch climatic beat: template for Plio–Pleistocene sea-level changes and sequence stratigraphy. Sediment Geol 122:5–21
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Reece JS, Castoe TA, Parkinson CL (2005) Historical perspectives on population genetics and conservation of three marine turtle species. Conserv Genet 6:235–251
Reid BN, Naro‐Maciel E, Hahn AT, FitzSimmons NN, Gehara M (2019) Geography best explains global patterns of genetic diversity and post-glacial co-expansion in marine turtles. Mol Ecol 28:3358–3370
Roberts MA, Schwartz TS, Karl SA (2004) Global population genetic structure and male-mediated gene flow in the green sea turtle (Chelonia mydas): analysis of microsatellite loci. Genetics 166:1857–1870
Rocha LA (2003) Patterns of distribution and processes of speciation in Brazilian reef fishes. J Biogeogr 30:1161–1171
Rocha LA, Robertson DR, Rocha CR, Van Tassell JL, Craig MT, Bowen BW (2005) Recent invasion of the tropical Atlantic by an Indo-Pacific coral reef fish. Mol Ecol 14:3921–3928
Scally A, Durbin R (2012) Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet 13:745–753
Seminoff JA (2004). Chelonia mydas. IUCN Red List Threat Species 2004: e.T4615A11037468
Seminoff JA, Allen CD, Balazs GH, Dutton PH, Eguchi T, Haas HL et al. (2015). Status review of the green turtle (Chelonia mydas) under the U.S. Endangered Species Act. NOAA Technical Memorandum, NOAA-NMFS-SWFSC-539. 571pp
Strasburg JL, Rieseberg LH (2010) How robust are ‘isolation with migration’ analyses to violations of the im model? A simulation study. Mol Biol Evol 27:297–310
Taberlet P, Fumagalli L, Wust‐Saucy A-G, Cosson J-F (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Teske PR, Von Der Heyden S, McQuaid CD, Barker NP (2011) A review of marine phylogeography in southern. Afr South Afr J Sci 107:43–53
Van der Zee JP, Christianen MJA, Nava M, Velez-Zuazo X, Hao W, Bérubé M et al. (2019) Population recovery changes population composition at a major southern Caribbean juvenile developmental habitat for the green turtle, Chelonia mydas. Sci Rep 9:14392
Wallace BP, DiMatteo AD, Hurley BJ, Finkbeiner EM, Bolten AB, Chaloupka MY et al. (2010) Regional management units for marine turtles: a novel framework for prioritizing conservation and research across multiple scales. PLoS ONE 5:e15465
Wang J (2017) The computer program structure for assigning individuals to populations: easy to use but easier to misuse. Mol Ecol Resour 17:981–990
Wang Z, Pascual-Anaya J, Zadissa A, Li W, Niimura Y, Huang Z et al. (2013) The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet 45:701–706
Waples RS (1998) Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J Hered 89:438–450
Weir BS, Cockerham CC (1984) Estimating F-Statistics for the Analysis of Population Structure. Evolution 38:1358–1370
Willing E-M, Dreyer C, van Oosterhout C (2012) Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 7:e42649–7
Wilson AC, Cann RL, Carr SM, George M, Gyllensten UB, Helm-Bychowski KM et al. (1985) Mitochondrial DNA and two perspectives on evolutionary genetics. Biol J Linn Soc 26:375–400
Woodruff DS (2010) Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today’s patterns and the future of the remaining refugial-phase biodiversity. Biodivers Conserv 19:919–941
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Acknowledgements
We thank all volunteers of Sea Turtle Conservation Bonaire who contributed to sample collection in Bonaire, in particular Hans and Jannie Koning, and Gielmon “Funchi“ Egberts. We thank Géraud Leroux for providing samples from the Barren Isles, Madagascar, and thank Nuno dos Santos for providing samples from Príncipe Island, São Tomé and Príncipe. This study was carried out as part of the project “Ecology and conservation of green and hawksbill turtles in the Dutch Caribbean” funded by the Netherlands Organization of Scientific Research (NWO-ALW 858.14.090).
Author information
Authors and Affiliations
Contributions
JPZ designed the study with input from PJP. JPZ, MJAC and MN collected samples in Bonaire. MN and KS provided logistical-, and field support in Bonaire. FH arranged and coordinated sample collection in Madagascar. AAN arranged and coordinated sample collection in São Tomé and Príncipe. JPZ and MB performed DNA extractions. MB created ddRAD libraries. JPZ performed bioinformatic-, and data analyses. MJAC, MN, LEB and PJP obtained funding. JPZ and PJP coordinated the writing of the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Jane Hughes.
Supplementary information
Rights and permissions
About this article
Cite this article
van der Zee, J.P., Christianen, M.J.A., Bérubé, M. et al. The population genomic structure of green turtles (Chelonia mydas) suggests a warm-water corridor for tropical marine fauna between the Atlantic and Indian oceans during the last interglacial. Heredity 127, 510–521 (2021). https://doi.org/10.1038/s41437-021-00475-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-021-00475-0
This article is cited by
-
Ecological and biogeographic features shaped the complex evolutionary history of an iconic apex predator (Galeocerdo cuvier)
BMC Ecology and Evolution (2022)
-
Population genetic diversity of green turtles, Chelonia mydas, in the Mediterranean revisited
Marine Biology (2022)