Abstract
A key piece of information for ecosystem management is the relationship between the environment and population genetic structure. However, it is difficult to clearly quantify the effects of environmental factors on genetic differentiation because of spatial autocorrelation and analytical problems. In this study, we focused on stream ecosystems and the environmental heterogeneity caused by groundwater and constructed a sampling design in which geographic distance and environmental differences are not correlated. Using multiplexed ISSR genotyping by sequencing (MIG-seq) method, a fine-scale population genetics study was conducted in fluvial sculpin Cottus nozawae, for which summer water temperature is the determinant factor in distribution and survival. There was a clear genetic structure in the watershed. Although a significant isolation-by-distance pattern was detected in the watershed, there was no association between genetic differentiation and water temperature. Instead, asymmetric gene flow from relatively low-temperature streams to high-temperature streams was detected, indicating the importance of low-temperature streams and continuous habitats. The groundwater-focused sampling strategy yielded insightful results for conservation.
Similar content being viewed by others
Introduction
How genetic structure is shaped across a landscape is an essential theme in evolutionary biology, and this information provides an empirical basis for conservation biology by elucidating habitat connectivity or predicting the effects of landscape change (Hohenlohe et al. 2021). A pattern of ‘isolation by distance’ (IBD; Wright 1943), in which genetic differentiation increases with geographic distance, is a very common pattern to explain population structure (Meirmans 2012). In addition to space, another important component of a landscape is the environment (Nosil et al. 2005; Thorpe et al. 2008). In the field of landscape genetics, many studies have investigated the interactions between genetic differentiation and landscape variables, and a couple of patterns have described the relationship between genetic structure and environment. When gene flow between populations that inhabit different environments is limited due to local adaptation or other factors, genetic differentiation increases with environmental differences and a pattern of ‘isolation by environment’ (IBE; Wang and Summers 2010) is generated. Another pattern that involves genetic structure and the environment is ‘counter-gradient gene flow’ (Sexton et al. 2014). In this scenario, gene exchange between different environments is high and forms a source-sink pattern in a landscape. Because these interactions between genetic differentiation and landscape variables are critically important for addressing classical evolutionary questions related to ecological speciation or conservation, the environment is a factor that cannot be ignored when describing the observed population structure (Orsini et al. 2013; Wang and Bradburd 2014).
However, it is very difficult to disentangle the relative strength of space (distance) and environment in observed patterns of spatial genetic differentiation because environmental differences are usually highly correlated with geographic distance (Sexton et al. 2014; Wang and Bradburd 2014). To test spatial relationships, the Mantel test and partial Mantel test are commonly used. However, the Mantel test family is strongly criticized for its inflated Type I error rate, especially under conditions where the measured variables are spatially correlated (Guillot and Rousset 2013; Harmon and Glor 2010; Meirmans 2012; Raufaste and Francois 2001; Rousset 2002). Although there are several proposed alternative methods to the Mantel test such as multiple regression on distance matrices (MRM; Lichstein 2007), Procrustes analysis, or redundancy analysis (RDA), and some of them perform much better than the partial Mantel test, previous studies have suggested that all methods fail to correctly model the relative importance of space and environment under certain patterns, which cannot be known a priori (Diniz-Filho et al. 2013; Gilbert and Bennett 2010). It is now recognized that no method has the ability to address the spatial-environmental correlation and perform hypothesis testing simultaneously (Zeller et al. 2016). These studies suggest the difficulty of separating spatial and environmental effects on genetic differences. However, once a sampling design that decouples distance and environment is developed, we will be able to clearly compare the relative strength of space and environment independent of these controversies. In the field of community ecology, Gilbert and Lechowicz (2004) used a sampling design that removed spatial autocorrelation of the environment sampled, and they analyzed the species’ spatial and environmental correlations to reject the neutral theory of biodiversity. However, subsequent attempts to partition spatial and environmental components have focused exclusively on statistical and analytical methodology (Gilbert and Bennett 2010). Especially in landscape genetics, sampling design has not received much attention (Meirmans 2015).
Here, we focused on stream ecosystems, which often exhibit heterogeneous environments (Heino et al. 2013; Uno 2016). In stream ecosystems, water temperature is the strongest factor affecting river organisms as well as flow regime (Olden and Naiman 2010), and it is increasingly being an important variable under ongoing climate change. Water temperature is determined by air temperature, riparian forests, and groundwater, among others (Caissie 2006; Nakamura and Yamada 2005). In particular, the effect of groundwater discharge on water temperature has been indicated to be very strong (Arscott et al. 2001). However, the influence of groundwater discharge on the upper sections of streams has received much less attention (Brown et al. 2007), and many ecological studies have used air temperature instead of water temperature (e.g., Almodóvar et al. 2012; Middaugh et al. 2018). Groundwater shows a very stable water temperature and flow regime; hence, groundwater discharge greatly affects the stream environment (Poff et al. 2010). Given water temperature in summer that is critical for cold-water organisms, streams with high groundwater display lower values and these streams could be an important habitat for these species. Since the amount of cold groundwater discharge vary locally within a watershed, groundwater creates spatial heterogeneity of environments within the watershed. Thus, to reveal the ecological role of water temperature, knowledge of the spatial heterogeneity caused by groundwater is thought to be useful.
As a practical study, we investigated the genetic structure of the cold-water fluvial sculpin Cottus nozawae in the upstream section of the Sorachi River, Hokkaido, Japan. Cold-water fish are very vulnerable to climate change, and it is worth studying the relationships between population structure and water temperature. Together with C. nozawae, Salvelinus malma, Salvelinus leucomaenis, Barbatula oreas, and Parahucho perryi inhabit this watershed. However, studies on S. malma and S. leucomaenis did not display strong genetic structure within this watershed due to their active migration (Koizumi 2011; Nakajima et al. 2020). Due to the lower mobility of adult C. nozawae than these species (Goto 1998; Okumura and Goto 1996), we predicted that C. nozawae should display clear genetic differentiation within the watershed. In addition, the summer water temperature has been shown to be the dominant factor determining the distribution and survival of this species (Yagami and Goto 2000), and a relationship in which low-water temperature in summer increases the density of this species has also been confirmed in our study area (Suzuki et al. 2021). Therefore, summer water temperature can be explicitly used as an important variable for C. nozawae. Hypothesis-driven studies that focus on a given variable have advantages over exploratory studies in designing sampling strategies (Richardson et al. 2016). Since ecologically important variables affecting survival and population density could cause local adaptation between different environments (Kawecki and Ebert 2004), we predicted that the differences in summer water temperature cause local adaptations and lead to the IBE pattern within the watershed.
The aims of this study are (i) to minimize the correlation of geographic distance and water temperature differences via a groundwater-focused sampling strategy, (ii) to investigate the local genetic structure of C. nozawae and its determinant factors, and (iii) to discuss the relationship between water temperature and the population structure of cold-water fish. Although recent studies have frequently used adaptative genetic markers to detect local adaptation, barriers to gene flow imposed by selection and local adaptation between populations can be detected with neutral markers (Sexton et al. 2014). Given that the use of adaptive markers is still challenging, the evaluation of current genetic differentiation and connectivity based on neutral genetic markers is still informative and can assist in allocating conservation units to preserve local genetic variation (Tsuda et al. 2015). To assess the genetic structure, we used putatively neutral genome-wide SNPs on the inter simple sequence repeat (ISSR) region and analyzed genetic differentiation and gene flow patterns.
Materials and methods
Study sites and sampling
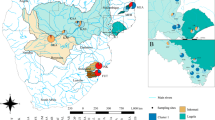
This study was conducted in the upper section of the Sorachi River, Hokkaido, Japan (Fig. 1; Table 1). To conduct sampling across a heterogeneous landscape, we used an approach based on watershed geology, which is known to cause significant variations in water temperature through groundwater discharge (Caissie 2006; Nagasaka and Sugiyama 2010; Tague et al. 2007). In the Sorachi River, many small spring-fed inputs are found in the Quaternary volcanic region (Koizumi and Maekawa 2004; Watz et al. 2019). Thus, the sampling sites were selected so that tributaries with volcanic watersheds and nonvolcanic watersheds were spatially intermingled. We also chose sites interspersed throughout the watershed such that large spatial gaps among populations were eliminated as much as possible. Watershed geology was assessed based on the Seamless Digital Geological Map of Japan V2 from the National Institute of Advanced Industrial Science and Technology. There is a large dam (Kanayama Dam) in the watershed. While the area upstream of Kanayama Dam represents one of the most continuous river habitats in Japan and there are no barriers between populations, the downstream region is slightly more influenced by anthropogenic impacts on streams. For example, one population (Pop13) is located upstream of a check dam, and another population (Pop19) is located in a tributary with a downstream portion that runs through an agricultural landscape.
A Study area. The fill colors of the sampling site depend on the maximum water temperature. Site labels correspond to population IDs in the text and Table 1. B Genetic structure inferred by STRUCTURE. The number indicates the population IDs.
From 20 sites in the Sorachi River, 531 individuals of C. nozawae were caught for genetic analysis. The fish were caught by electrofishing (model 12-B Backpack Electrofisher; Smith-Root Inc.) or with hand nets. Small pieces of fin tissue were clipped, placed in 99.5% ethanol, and stored at −20 °C in the laboratory until DNA extraction. Total DNA was extracted using the QIAGEN DNeasy Blood and Tissue Kit (QIAGEN Inc.) with a combination of Genomic DNA Extraction Column (FAVORGEN Inc.).
Water temperature
At each sampling site, the summer water temperature was recorded hourly using data loggers (Onset Computer Corp. or Gemini Data Loggers Ltd., depending on site). Water temperatures were measured during the summer of 2020, and also during the summer of 2019 at some locations. To align the data sampling periods, data from 16 July 2020 to 31 August 2020 were extracted, and the maximum water temperature in this period was identified. At one site (Pop10) where we failed to record water temperatures in some mid-summer periods, the maximum temperature in 2019 was used as an alternative. We used the maximum water temperature which significantly affects the populations of cold-water fish as a representative of summer water temperature (Yagami and Goto 2000). The correlation of water temperature differences with waterway geographic distance (hereafter referred to as geographic distance) was examined with the Mantel test (9999 permutations) using the package ECODIST 2.0.3 (Goslee and Urban 2007) in R 3.6.0 (R Core Team 2019). Then, a Mantel correlogram showing the spatial correlation of water temperature differences across multiple ranges of geographic distance was constructed. The Mantel correlogram in ten equal distance classes was assessed with 9999 permutations using the package VEGAN 2.5.6 (Oksanen et al. 2019) in R.
MIG-seq library preparation and sequencing
To obtain neutral genome-wide SNP data, we used multiplexed ISSR genotyping by sequencing (MIG-seq; Suyama and Matsuki 2015), a technique in which loci between two microsatellite regions are amplified by PCR and next-generation sequencing. MIG-seq is one of the reduced representation sequencing methods along with restriction site-associated DNA sequencing (RAD-seq), but the number of available informative loci (MIG-loci) detected by MIG-seq is less than that of RAD-seq, and most of the MIG-loci are putatively neutral. Although MIG-seq is not suitable for outlier analysis or gene identification, acquired loci are sufficient for population genetic analysis. A MIG-seq library was prepared following the protocol outlined in Suyama and Matsuki (2015), except for the minor modifications outlined below. The first PCR was conducted using eight ISSR primer sets with tail sequences at an annealing temperature of 38 °C. The second PCR was conducted using primer pairs including tail sequences, adapter sequences for Illumina sequencing, and five-base (forward) and nine-base (reverse) barcode sequences to identify each individual sample. The conditions for the second PCR were as follows: 12 cycles of denaturation at 98 °C for 10 s, annealing at 54 °C for 15 s, and extension at 68 °C for 1 min. The second PCR products of all 531 samples were mixed, and then fragments in the size range of 400–800 bp in the purified library were isolated. After library quantification, the products were sequenced on the Illumina MiSeq platform using MiSeq Reagent Kit v3 (150 cycles). Both ends of the fragments and the indexes were read by paired-end and index sequencing: 80, 80, 9, and 5 bases of sequences were determined as read 1, read 2, index-1, and index-2, respectively. The first 17 bases (SSR and anchor regions) in both reads were skipped using the DarkCycle option of the MiSeq system. Read 1 and 2 cannot overlap within each fragment, and the size range of the library was 400–800 bp. Thus, following to Suyama and Matsuki (2015), we treated reads 1 and 2 as independent reads. The final data output to the next step included sequences of 80 bases from both ends of 400–800 bp forward-reverse amplicons of various ISSR regions.
SNP detection
Read quality filtering was performed using Trimmomatic 0.39 (Bolger et al. 2014). Extremely short reads containing adapter sequence were filtered out. Then, to remove low quality reads, the reads were scanned with a four-base wide sliding window, and reads with an average quality per base below 15 were removed. After quality filtering, a total of 39,956,740 reads were obtained. SNP selection was performed using STACKS 2.41 (Catchen et al. 2013). First, reads were grouped to each locus using the ustacks, cstacks, sstacks, tsu2bam, and gstacks commands with the following parameters recommended by Paris et al. (2017): minimum depth option creating a stack (m) = 3, maximum distance between stacks (M) = 2, maximum mismatches between loci when building the catalog (n) = 2, and number of mismatches allowed to align secondary reads (N) = 4. After this process, the dataset of assembled loci (stacks dataset) was obtained. For most analyses (except for gene flow analysis), loci present at a rate of more than 80% among all samples were extracted using the populations command. Several options were included with this command: the minimum minor allele count over the entire dataset was set to three to exclude potentially artificial loci found in only one individual, sites showing excess heterozygosity (>0.6) were removed to filter potential heterozygotes resulting from artificial loci built from several paralogous genome regions, and the output was limited to one SNP per locus to exclude linkage disequilibrium among SNPs. After filtering, 489 SNPs were obtained.
Genetic diversity and differentiation
For each population, the expected heterozygosity (HE) and fixation index (FIS) were calculated using the populations command in STACKS. Significant deviations from Hardy–Weinberg equilibrium (HWE), as indicated by FIS deviating from zero, were tested by 1000 randomizations using FSTAT 2.9.4 (Goudet 1995). Genetic differentiation among populations was evaluated by calculating global/pairwise FST values (Weir and Cockerham 1984) using GenAlEx 6.5 (Peakall and Smouse 2012). To understand the spatial trend of the genetic structure, a Mantel correlogram displaying correlations between FST and geographic distance for each of the ten distance classes was constructed with 9999 permutations using the package VEGAN in R.
Population structure
The population structure was examined using STRUCTURE 2.3.4 (Pritchard et al. 2000), which implements a Bayesian clustering method using multi-locus allele frequency data. The STRUCTURE settings were the admixture and allele frequency correlated model with previous sampling location information (LOCPRIOR; Hubisz et al. 2009). The algorithm was run 20 times for each K from 1 to 14 with a burn-in of 20,000 followed by 30,000 Markov chain Monte Carlo (MCMC) replicates. The program CLUMPAK (Kopelman et al. 2015) was used to compile the results of the STRUCTURE analysis for each K. STRUCTURE HARVESTER (Earl and vonHoldt 2012) was employed to calculate the probability of the data for each K (LnP(D); Pritchard et al. 2000), the corresponding standard deviation, and Evanno’s delta K (Evanno et al. 2005).
Association of genetic variation with geographic distance and water temperature
To evaluate the effects of space and water temperature on genetic differentiation, the independent correlations of FST with water temperature differences and geographic distance were calculated by Mantel tests with 9999 permutations. To identify confounding effects between space and water temperature if present, multiple regression on distance matrices (MRM; Lichstein 2007) was used, with “water temperature differences + geographic distance” as the explanatory variable. Mantel tests and MRM were performed with the package ECODIST in R. These analyses were conducted at three scales: (a) using all 20 populations (waterway distance 0.3–70.6 km), (b) using upstream 12 populations (Pop1-12; waterway distance 0.3–24.1 km), and (c) using structured 9 populations (Pop4-12; populations assigned as one large cluster in STRUCTURE). We used scale (b) to account for the mobility of Cottus and to exclude the effects of a long spatial gap and a dam, and scale (c) was used to prevent biases when including sites displaying high genetic divergence (‘outliers’; Koizumi et al. 2006).
Some studies have considered it inappropriate to apply the Mantel test to the raw vector data and to take the difference to create a matrix (Legendre and Legendre 2012). Hence, we also used distance-based RDA, which is a constrained ordination approach that can use environmental variables as vector data and has higher power than the Mantel test (Harmon and Glor 2010; Legendre et al. 2015). We used genetic variation as the response variable and water temperature and spatial variables derived from geographic distance as explanatory variables. For genetic variation, a principal coordinate analysis of the pairwise FST matrix was conducted, and all axes were used as response variables. Spatial predictors were generated as a set of distance-based Moran’s eigenvector maps (MEMs; Griffith and Peres-Neto 2006), vectors that capture broad- to small-scale spatial structures. MEMs were generated from the geographic distance matrix using the package ADESPATIAL 0.3.8 (Dray et al. 2020) in R. To identify meaningful MEM predictors, forward selection (999 permutations) was used for generated MEM dataset. In each RDA with the explanatory variables of water temperature and geographic MEM variables, adjusted R-square values (Radj2), which penalize the increase in explanatory power due to an increase in the number of explanatory variables, were calculated. RDAs were performed on three scales, as in the Mantel tests and MRM analyses. RDAs including forward selections were performed with the package VEGAN in R.
Contemporary gene flow
To infer gene flow patterns, BA3SNP (Mussmann et al. 2019), a reconstructed version of BayesAss (Wilson and Rannala 2003) that estimates dual-direction pairwise migration rates (here migration is successful dispersal that contributes to gene flow) over a few generations, was used for the 20 populations. To improve the convergence of BayesAss, another SNP filtering criterion that increases the variance among populations was used. From the stacks dataset, loci present at a rate of more than 40% among at least two populations were extracted with a minimum minor allele count of three. As in the other analyses, sites showing excess heterozygosity (>0.6) were removed, and the output was limited to one SNP per locus. Then, 1291 SNPs were obtained. We set 20,000,000 MCMC iterations, including a burn-in period of 10,000,000. Following the proposal of Meirmans (2014), we performed ten independent runs with different seed values and chose the run with the lowest Bayesian deviance to overcome poor MCMC sampling. The inferred migration rates with 95% credible intervals that did not include zero were regarded as significant. We then defined the net immigration rate into a given population A, from another population B, as the [estimated value of B- >A minus that of A- >B]. Then, the mean net immigration rate for each population (the mean net immigration rate for a given population from all of its paired net immigration rate values between all other populations) was calculated to measure the degree to which a population is a donor or a recipient of migrants (Hänfling and Weetman 2006; Sexton et al. 2016). To investigate whether water temperature affects the source-sink structure independent of upstream-downstream dispersal, we constructed a linear model with a response variable of the mean net immigration rate and explanatory variables as maximum water temperature and elevation. The Pearson’s correlation between water temperature and elevation was −0.39; thus, no multicollinearity was considered. The independent effects of water temperature and elevation were assessed by the partial regression coefficient (β) and standardized partial regression coefficient (std β). Likelihood ratio tests were used to determine the significance of explanatory variables. This analysis was conducted using the package LMTEST (Zeileis and Hothorn 2002) in R.
Results
Heterogeneity of water temperature
Summer water temperatures varied within the watershed, and the maximum water temperature ranged from 12.5 to 23.5 °C. The correlation between geographic distance and water temperature difference was low (Mantel r = 0.08, p = 0.45). In the Mantel correlogram for water temperature, r values were not significant in all distance classes (Fig. 2A), indicating little spatial autocorrelation.
Genetic diversity, divergence, and population structure
FIS did not deviate significantly from zero except in one population (Pop20), suggesting that HWE could be assumed in most populations. The HE was similar across the watershed, but one population located in the upstream section of a check dam (Pop13) displayed a slightly lower value (Table 1). Across the 20 populations, the FST value was 0.038, indicating weak population differentiation, and pairwise FST values ranged from 0.001 to 0.127 (Table S1). In a Mantel correlogram, r values were significantly positive in up to the second distance classes (~15 km) and reached zero at a distance of 27 km (Fig. 2B).
In the STRUCTURE analysis, the probability of the data (LnP(D)) increased progressively with each K, and delta K was highest at K = 4 (Fig. S1). Distinct clusters corresponding to the geographic structure were detected up to K = 6 (Fig. 1). For K = 2, the genomes of the individuals in Pop13 were grouped into a single unique cluster. For K = 4, Pop1-3 and Pop16-19 were grouped as additional clusters. Pop14,15,20 was inferred to be an admixed cluster when K = 4 and a distinct unique cluster when K = 6. Pop3 also formed a unique cluster at K = 6. Pop4-12 was assigned to almost one cluster at even large K.
Effects of space and water temperature on genetic differentiation
Mantel tests captured significant correlations between pairwise FST and waterway geographic distance (Mantel r = 0.27, p < 0.05; Fig. 3), indicating a significant effect of IBD. Conversely, no correlation between FST and water temperature differences was detected (Mantel r = −0.10, p = 0.56). The MRM with these two matrices as an explanatory variable did not show a significant relationship, and the R2 value was not very different from that of geographic distance alone. The same pattern was obtained at different spatial scales (Table 2). The forward selection of the MEM variables identified six significant predictors and the RDA with selective MEMs was significant with an Radj2 value of 0.61 (Fig. S2). Even in RDA, water temperature did not explain the genetic variation at all.
Gene flow
A total of 11 significant migration rates were detected (Tables 3 and S2). All of them were migration from relatively low-temperature sites (mean 14.3 °C [SD 2.90]) to high-temperature sites (mean 20.0 °C [SD 1.84]). Most of them were upstream to downstream direction, but they also included migration from downstream to upstream direction (Pop4 to 5 and Pop12 to 11). The linear model indicated that water temperature had a significant effect on the mean net immigration rates (std β = 0.542; p < 0.05) but elevation did not (std β = −0.077; p = 0.70; Table 4). Populations with lower water temperatures displayed lower mean net immigration rates, indicating that they are the source populations (Fig. 4).
Discussion
Genetic differentiation
In riverscapes, lower genetic diversity upstream than downstream has been recognized as a typical pattern (Blanchet et al. 2020; Thomaz et al. 2016). However, in this study, genetic diversity was similar regardless of longitudinal position (Table 1). The relative suitability of the upstream environment for the sculpin and the source-sink gene flow that is not only in the upstream-downstream direction (Table 4) may have suppressed the decrease in genetic diversity of upstream populations.
Among the entire sampling sites, a low level of genetic differentiation was observed (FST = 0.038). A study of C. nozawae in Tohoku district displayed high genetic differentiation within one watershed (Ito et al. 2018), and previous studies investigating other fluvial Cottus species within 30–50 km2 scale have often reported high FST values (Hänfling and Weetman 2006; Junker et al. 2012; Ruppert et al. 2017; but see Peacock et al. 2016). In the present study, the degree of genetic differentiation was somewhat lower than those observed in these previous studies. Other than gene flow, the large effective population size and consequent low genetic drift from the common ancestor may result in low genetic differentiation. Nevertheless, high-resolution genome-wide SNP analysis allowed genetic differentiation within the watershed to be clearly detected.
Distinct genetic structure along the geographic structure was inferred within the watershed (Fig. 1). There was a significant correlation between genetic differentiation and geographic distance, but summer water temperature had no effect on the strength of genetic differentiation (Fig. 3). Thus, the possibility that differences in water temperature lead to genetic differentiation via local adaptation is low.
Only a handful of studies have investigated the associations between the environment and the genetic structure of sculpin. Ruppert et al. (2017) analyzed the relationship between genetic and elevation differences and displayed a pattern of IBE along elevation. Another study demonstrated genetic divergence between different habitat types (Dennenmoser et al. 2014). However, most studies have been unable to eliminate the interaction of environment and geographic distance. It is not clear whether populations in these different regions and species actually have IBE patterns, but the IBE of sculpin may not be very common in wild populations.
There may be some factors related to genetic differentiation that are not clear from this study (residuals). For example, the factors promoting the genetic divergence between Pop1-3 and Pop4-12 detected in STRUCTURE analysis are still unknown. Since we assume the IBE pattern as a hypothesis, we used only an environmental variable of each site, but the factor of genetic differentiation may also lie on the pathway between populations (e.g., streambed slope). To identify those factors contributing to the residuals of genetic differentiation, analyses using environmental data across the entire watershed (e.g., White et al. 2020) and denser sampling are necessary.
Gene flow
Another important insight into population structure was obtained via gene flow analysis. Almost all significant migration was detected in population pairs within ~15 km, the range where the spatial correlation of genetic variation was significant in the Mantel correlogram (Fig. 2B). Most of the 11 cases of significant migration were downstream direction (Tables 3 and S2), which is consistent with several previous studies about fluvial sculpins that have revealed the source-sink structure from upstream to downstream sites (Hänfling and Weetman 2006; Junker et al. 2012). However, the present study also detected migrations toward tributaries at higher elevations, from lower-temperature tributaries to higher-temperature tributaries. Of course, we should be cautious about the accuracy of BayesAss estimates (Meirmans 2014). However, the estimated pattern displayed a clear trend, and it could be concluded that suitable habitats with low summer water temperatures behave as “source” habitats in the watershed. The effect of summer water temperature on the mean net immigration rate was larger than that of elevation (Table 4) and populations with lower temperatures had a greater degree of individual supply into other populations (Fig. 4).
Although BayesAss estimates migration rates over a short period, the estimates should reflect the gene flow pattern in the studied ecosystem. In this study, significant migration was estimated between populations with different summer water temperatures at close locations (Fig. 1; Table 3). This finding implies that gene flow in the direction of across dissimilar environments is high. This type of gene flow is referred to as ‘counter-gradient gene flow’ (Sexton et al. 2014). The IBE pattern is detected when gene flow among similar environments is high; therefore, the counter-gradient gene flow is the opposite pattern to IBE and suppresses IBE. The ecological processes driving this gene flow pattern could not be identified in this study, but it may at least be related to differences in population density documented by Suzuki et al. (2021) in the same watershed as the present study (i.e., high density in low-temperature streams).
Goto (1998) used mark-recapture methods and reported that most mature individuals stay at the same site for more than 1 year, indicating almost no migration. While the mark-recapture method addresses only adult movement, genetic approaches also reflect movement during early life stages that is not easily captured by mark-recapture approaches (Lamphere and Blum 2012). If passive gene flow (i.e., the transportation of juveniles or eggs) is the driving force of gene flow, some high migration rates obtained in our study do not conflict with the results of mark-recapture methods. Furthermore, some previous studies using genetic methods on Cottus reported similar migration rates to those obtained in the present study and discrepancies between genetic and mark-recapture estimates (Junker et al. 2012; Lamphere and Blum 2012), suggesting that the detected degree of gene flow is reasonable. Still, it remains to be seen whether the estimated degree of gene flow is constantly observed. For instance, flooding is a factor that increases fish dispersal (Blondel et al. 2021; Natsumeda 2003), and it should be noted that we cannot rule out the possibility that recent large disturbances (e.g., relative large typhoon in 2016) may have temporarily promoted dispersal. To make the findings on gene flow more general, more research under different conditions and other evidence such as differences in turnover rates are needed.
Implications for future management
Because sculpins are not commercially or recreationally important fish, they are impacted relatively little by human translocations (Lucek et al. 2018). Thus, sculpin genes have a footprint of a long-term past history, and the diversity of each watershed should be protected. Within the watershed, while each tributary is characterized by a unique genetic composition, loose connectivity via gene flow between tributaries is maintained. Indeed, fragmented populations such as Pop13 show lower genetic diversity. In particular, populations in streams with less suitable environments may be maintained by immigration from low-temperature streams that are more suitable habitats. Therefore, under ongoing climate change, (i) the preservation of low-temperature streams and (ii) the prevention of making unpassable barriers between source and sink populations would be the key measures for conserving sculpin populations. Considering that cold-water fish other than benthic fish have been well studied and revealed to show high mobility between tributaries, these measures will be effective for overall cold-water fish conservation. Groundwater and its associated factors such as watershed geology are useful for identifying low-water-temperature streams and conserving possible “source” populations.
Local conservation measures can also be proposed. If the distance at which the spatial coefficient becomes nonsignificant is considered to represent the appropriate management unit size (Diniz-Filho and De Campos Telles 2002), stream networks should not be subdivided into stretches of <15 km to maintain genetic resources. However, even if a large continuous habitat is maintained, then sections consisting solely of high-temperature streams will lead to local population declines due to the absence of individual supply sources. Thus, care must be taken whenever rivers are fragmented. In the Sorachi River, many C. nozawae inhabit high density (Suzuki et al. 2021), and they are not considered to be in danger of immediate extinction. However, maintaining the abundance of general species is very important to avoid ecosystem breakdown (Baker et al. 2019). In some regions in Japan (Tohoku district), C. nozawae populations are in danger of local extinction (Ministry of the Environment Government of Japan 2020). Therefore, regardless of how large the population is, maintaining continuous stream networks and the connectivity to low-temperature sections in the networks is needed for future population persistence.
Implications for landscape/riverscape genetics
Decoupling distance and environment in a locality is a strong strategy for rigorously evaluating competing hypotheses (Gilbert and Lechowicz 2004). Through this approach, we revealed a pattern that has not been proposed for the examined genus. Freshwater populations have been identified as potentially fruitful targets for the application of a landscape genetic approach to delineate population structure (Kanno et al. 2011), and we focused on groundwater to reveal the association of water temperature with the genetic structure of cold-water fish populations.
In this study, the possibility of a source-sink structure caused by counter-gradient gene flow was suggested instead of IBE. Many source-sink structures from core populations to edge (peripheral) populations have been studied. However, local gene flow patterns are also important for discussing population structure in changing environments. The IBE pattern is easily detected when spatial-environmental correlation is present, but it is important to scrutinize whether it truly exists. Although the local source-sink pattern provides notable insights for conservation and evolution (Iles et al. 2018), a previous meta-analysis showed that counter-gradient gene flows are much less frequently reported than IBE (Sexton et al. 2014). Counter-gradient gene flow is inherently exclusive to IBE and may tend to be neglected. In population genetic studies, the results often differ from the hypotheses (Myers et al. 2019). As detecting the relationship between the environment and genetic differentiation is still challenging, we hope that the role of the environment in genetic divergence will be better understood in many studies with the help of sampling strategies.
Data availability
Raw MIG-seq reads were deposited in the DDBJ Sequence Read Archive under accession number DRA011249. The other data are available on Figshare (https://doi.org/10.6084/m9.figshare.13383245).
References
Almodóvar A, Nicola GG, Ayllón D, Elvira B (2012) Global warming threatens the persistence of Mediterranean brown trout. Glob Chang Biol 18:1549–1560
Arscott DB, Tockner K, Ward JV (2001) Thermal heterogeneity along a braided floodplain river (Tagliamento River, northeastern Italy). Can J Fish Aquat Sci 58:2359–2373
Baker DJ, Garnett ST, O’Connor J, Ehmke G, Clarke RH, Woinarski JCZ et al. (2019) Conserving the abundance of nonthreatened species. Conserv Biol 33:319–328
Blanchet S, Prunier JG, Paz-Vinas I, Saint-Pe K, Rey O, Raffard A et al. (2020) A river runs through it: the causes, consequences, and management of intraspecific diversity in river networks. Evol Appl 13:1195–1213
Blondel L, Paterson IG, Bentzen P, Hendry AP (2021) Resistance and resilience of genetic and phenotypic diversity to “black swan” flood events: a retrospective analysis with historical samples of guppies. Mol Ecol 30:1017–1028
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Brown LE, Milner AM, Hannah DM (2007) Groundwater influence on alpine stream ecosystems. Freshw Biol 52:878–890
Caissie D (2006) The thermal regime of rivers: a review. Freshw Biol 51:1389–1406
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140
Dennenmoser S, Rogers SM, Vamosi SM (2014) Genetic population structure in prickly sculpin (Cottus asper) reflects isolation-by-environment between two life-history ecotypes. Biol J Linn Soc 113:943–957
Diniz-Filho JAF, De Campos Telles MP (2002) Spatial autocorrelation analysis and the identification of operational units for conservation in continuous populations. Conserv Biol 16:924–935
Diniz-Filho JAF, Soares TN, Lima JS, Dobrovolski R, Landeiro VL, Telles MP, de C et al. (2013) Mantel test in population genetics. Genet Mol Biol 36:475–485
Dray S, Bauman D, Blanchet G, Borcard D, Clappe S, Guenard G et al. (2020) adespatial: Multivariate Multiscale Spatial Analysis. R package version 0.3-8. https://CRAN.R-project.org/package=adespatial
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Gilbert B, Bennett JR (2010) Partitioning variation in ecological communities: do the numbers add up? J Appl Ecol 47:1071–1082
Gilbert B, Lechowicz MJ (2004) Neutrality, niches, and dispersal in a temperate forest understory. Proc Natl Acad Sci USA 101:7651–7656
Goslee S, Urban D (2007) The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22:1–19
Goto A (1998) Life-history variations in the fluvial sculpin, Cottus nozawae (Cottidae), along the course of a small mountain stream. Environ Biol Fishes 52:203–212
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Griffith DA, Peres-Neto PR (2006) Spatial modeling in ecology: the flexibility of eigenfunction spatial analyses. Ecology 87:2603–2613
Guillot G, Rousset F (2013) Dismantling the Mantel tests. Methods Ecol Evol 4:336–344
Hänfling B, Weetman D (2006) Concordant genetic estimators of migration reveal anthropogenically enhanced source-sink population structure in the river sculpin, Cottus gobio. Genetics 173:1487–1501
Harmon LJ, Glor RE (2010) Poor statistical performance of the mantel test in phylogenetic comparative analyses. Evolution 64:2173–2178
Heino J, Grönroos M, Ilmonen J, Karhu T, Niva M, Paasivirta L (2013) Environmental heterogeneity and β diversity of stream macroinvertebrate communities at intermediate spatial scales. Freshw Sci 32:142–154
Hohenlohe PA, Funk WC, Rajora OP (2021) Population genomics for wildlife conservation and management. Mol Ecol 30:62–82
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Iles DT, Williams NM, Crone EE (2018) Source-sink dynamics of bumblebees in rapidly changing landscapes. J Appl Ecol 55:2802–2811
Ito N, Gotoh RO, Shirakuma T, Araki Y, Hanzawa N (2018) Genetic structure of glacial-relict populations of a freshwater sculpin, Cottus nozawae, in Yamagata area of the Tohoku district. Biogeography 20:96–102
Junker J, Peter A, Wagner CE, Mwaiko S, Germann B, Seehausen O et al. (2012) River fragmentation increases localized population genetic structure and enhances asymmetry of dispersal in bullhead (Cottus gobio). Conserv Genet 13:545–556
Kanno Y, Vokoun JC, Letcher BH (2011) Fine-scale population structure and riverscape genetics of brook trout (Salvelinus fontinalis) distributed continuously along headwater channel networks. Mol Ecol 20:3711–3729
Kawecki TJ, Ebert D (2004) Conceptual issues in local adaptation. Ecol Lett 7:1225–1241
Koizumi I (2011) Integration of ecology, demography and genetics to reveal population structure and persistence: a mini review and case study of stream-dwelling Dolly Varden. Ecol Freshw Fish 20:352–363
Koizumi I, Maekawa K (2004) Metapopulation structure of stream-dwelling Dolly Varden charr inferred from patterns of occurrence in the Sorachi River basin, Hokkaido, Japan. Freshw Biol 49:973–981
Koizumi I, Yamamoto S, Maekawa K (2006) Decomposed pairwise regression analysis of genetic and geographic distances reveals a metapopulation structure of stream-dwelling Dolly Varden charr. Mol Ecol 15:3175–3189
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across. K Mol Ecol Resour 15:1179–1191
Lamphere BA, Blum MJ (2012) Genetic estimates of population structure and dispersal in a benthic stream fish. Ecol Freshw Fish 21:75–86
Legendre P, Fortin MJ, Borcard D (2015) Should the Mantel test be used in spatial analysis? Methods Ecol Evol 6:1239–1247
Legendre P, Legendre L (2012) Numerical Ecology, 3rd edn. Elsevier, Amsterdam
Lichstein JW (2007) Multiple regression on distance matrices: a multivariate spatial analysis tool. Plant Ecol 188:117–131
Lucek K, Keller I, Nolte AW, Seehausen O (2018) Distinct colonization waves underlie the diversification of the freshwater sculpin (Cottus gobio) in the Central European Alpine region. J Evol Biol 31:1254–1267
Meirmans PG (2012) The trouble with isolation by distance. Mol Ecol 21:2839–2846
Meirmans PG (2014) Nonconvergence in Bayesian estimation of migration rates. Mol Ecol Resour 14:726–733
Meirmans PG (2015) Seven common mistakes in population genetics and how to avoid them. Mol Ecol 24:3223–3231
Middaugh CR, Kessinger B, Magoulick DD (2018) Climate-induced seasonal changes in smallmouth bass growth rate potential at the southern range extent. Ecol Freshw Fish 27:19–29
Ministry of the Environment Government of Japan (2020) Red List of Japan. http://www.env.go.jp/press/107905.html. (In Japanese)
Mussmann SM, Douglas MR, Chafin TK, Douglas ME (2019) BA3-SNPs: contemporary migration reconfigured in BayesAss for next-generation sequence data. Methods Ecol Evol 10:1808–1813
Myers EA, Xue AT, Gehara M, Cox CL, Davis Rabosky AR, Lemos-Espinal J et al. (2019) Environmental heterogeneity and not vicariant biogeographic barriers generate community-wide population structure in desert-adapted snakes. Mol Ecol 28:4535–4548
Nagasaka A, Sugiyama S (2010) Factors affecting the summer maximum stream temperature of small streams in northern Japan. Bull Hokkaido Res Inst 47:35–43. (In Japanese with English abstract)
Nakajima S, Hirota SK, Matsuo A, Suyama Y, Nakamura F (2020) Genetic structure and population demography of white-spotted charr in the upstream watershed of a large dam. Water 12:2406
Nakamura F, Yamada H (2005) Effects of pasture development on the ecological functions of riparian forests in Hokkaido in northern Japan. Ecol Eng 24:539–550
Natsumeda T (2003) Effects of a severe flood on the movements of Japanese fluvial sculpin. Environ Biol Fishes 68:417–424
Nosil P, Vines TH, Funk DJ (2005) Perspective: reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution 59:705–719
Oksanen JF, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D et al. (2019) vegan: Community Ecology Package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan
Okumura N, Goto A (1996) Genetic variation and differentiation of the two river sculpins, Cottus nozawae and C. amblystomopsis, deduced from allozyme and restriction enzyme-digested mtDNA fragment length polymorphism analyses. Ichthyol Res 43:399–416
Olden JD, Naiman RJ (2010) Incorporating thermal regimes into environmental flows assessments: modifying dam operations to restore freshwater ecosystem integrity. Freshw Biol 55:86–107
Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L (2013) Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol Ecol 22:5983–5999
Paris JR, Stevens JR, Catchen JM (2017) Lost in parameter space: a road map for STACKS. Methods Ecol Evol 8:1360–1373
Peacock MM, Gustin MS, Kirchoff VS, Robinson ML, Hekkala E, Pizzarro-Barraza C et al. (2016) Native fishes in the Truckee River: are in-stream structures and patterns of population genetic structure related? Sci Total Environ 563–564:221–236
Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Poff NL, Richter BD, Arthington AH, Bunn SE, Naiman RJ, Kendy E et al. (2010) The ecological limits of hydrologic alteration (ELOHA): A new framework for developing regional environmental flow standards. Freshw Biol 55:147–170
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raufaste N, Francois R (2001) Are partial Mantel tests adequate? Evolution 55:1703–1705
Richardson JL, Brady SP, Wang IJ, Spear SF (2016) Navigating the pitfalls and promise of landscape genetics. Mol Ecol 25:849–864
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rousset F (2002) Partial Mantel tests: reply to Castellano and Balletto. Evolution 56:1874–1875
Ruppert JLW, James PMA, Taylor EB, Rudolfsen T, Veillard M, Davis CS et al. (2017) Riverscape genetic structure of a threatened and dispersal limited freshwater species, the Rocky Mountain Sculpin (Cottus sp.). Conserv Genet 18:925–937
Sexton JP, Hangartner SB, Hoffmann AA (2014) Genetic isolation by environment or distance: Which pattern of gene flow is most common? Evolution 68:1–15
Sexton JP, Hufford MB, Bateman AC, Lowry DB, Meimberg H, Strauss SY et al. (2016) Climate structures genetic variation across a species’ elevation range: a test of range limits hypotheses. Mol Ecol 25:911–928
Suyama Y, Matsuki Y (2015) MIG-seq: an effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Sci Rep. 5:1–12
Suzuki K, Ishiyama N, Koizumi I, Nakamura F (2021) Combined effects of summer water temperature and current velocity on the distribution of a cold-water-adapted scupin (Cottus nozawae). Water 13:975
Tague CL, Farrell M, Grant G, Lewis S, Rey S (2007) Hydrogeologic controls on summer stream temperatures in the McKenzie River basin, Oregon. Hydrol Process 21:3288–3300
Thomaz AT, Christie MR, Knowles LL (2016) The architecture of river networks can drive the evolutionary dynamics of aquatic populations. Evolution 70:731–739
Thorpe RS, Surget-Groba Y, Johansson H (2008) The relative importance of ecology and geographic isolation for speciation in anoles. Philos Trans R Soc B Biol Sci 363:3071–3081
Tsuda Y, Nakao K, Ide Y, Tsumura Y (2015) The population demography of Betula maximowicziana, a cool-temperate tree species in Japan, in relation to the last glacial period: Its admixture-like genetic structure is the result of simple population splitting not admixing. Mol Ecol 24:1403–1418
Uno H (2016) Stream thermal heterogeneity prolongs aquatic-terrestrial subsidy and enhances riparian spider growth. Ecology 97:2547–2553
Wang IJ, Bradburd GS (2014) Isolation by environment. Mol Ecol 23:5649–5662
Wang IJ, Summers K (2010) Genetic structure is correlated with phenotypic divergence rather than geographic isolation in the highly polymorphic strawberry poison-dart frog. Mol Ecol 19:447–458
Watz J, Otsuki Y, Nagatsuka K, Hasegawa K, Koizumi I (2019) Temperature-dependent competition between juvenile salmonids in small streams. Freshw Biol 64:1534–1541
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
White SL, Hanks EM, Wagner T (2020) A novel quantitative framework for riverscape genetics. Ecol Appl 30:e02147
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wright S (1943) Isolation by Distance. Genetics 28:114–138
Yagami T, Goto A (2000) Patchy distribution of a fluvial sculpin, Cottus nozawae, in the Gakko River system at the southern margin of its native range. Ichthyol Res 47:277–286
Zeileis A, Hothorn T (2002) Diagnostic checking in regression relationships. R News 2:7–10
Zeller KA, Creech TG, Millette KL, Crowhurst RS, Long RA, Wagner HH et al. (2016) Using simulations to evaluate Mantel-based methods for assessing landscape resistance to gene flow. Ecol Evol 6:4115–4128
Acknowledgements
We are grateful to Jorge García Molinos for support with the data preparation. We thank the staff of the University of Tokyo Hokkaido Forest for their cooperation in selecting study sites. We also thank Suzuki K., Hotta W., Nishio D., Motosugi N., Kawai H., and Zakoh K. of Hokkaido University for their help in conducting the field sampling and laboratory work. We appreciate anonymous reviewers for their valuable comments to the earlier versions of the manuscript. This study is partly supported by the research fund for the Ishikari and Tokachi Rivers provided by the Ministry of Land, Infrastructure, Transport, and Tourism of Japan.
Author information
Authors and Affiliations
Contributions
S.N. and F.N. conceived and designed the study. S.N. and N.I. performed the field work. S.N. and A.M. performed the laboratory work. S.N., M.S., and S.K.H. analyzed the genetic data. S.N. wrote the manuscript, and M.S., S.K.H., N.I., A.M., Y.S., and N.F. edited and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Jane Hughes.
Supplementary information
Rights and permissions
About this article
Cite this article
Nakajima, S., Sueyoshi, M., Hirota, S.K. et al. A strategic sampling design revealed the local genetic structure of cold-water fluvial sculpin: a focus on groundwater-dependent water temperature heterogeneity. Heredity 127, 413–422 (2021). https://doi.org/10.1038/s41437-021-00468-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-021-00468-z
This article is cited by
-
The domestication and breeding history of Castanea crenata Siebold et Zucc. estimated by direction of gene flow and approximate Bayesian computation
Tree Genetics & Genomes (2023)
-
Population stability and asymmetric migration of caddisfly populations, Stenopsyche marmorata (Stenopsychidae), in a forest–agriculture landscape
Aquatic Sciences (2023)
-
Conservation genomics of an endangered floodplain dragonfly, Sympetrum pedemontanum elatum (Selys), in Japan
Conservation Genetics (2023)
-
Inferring future changes in gene flow under climate change in riverscapes: a pilot case study in fluvial sculpin
Landscape Ecology (2023)
-
Genetic structure and diversity of amphidromous sculpin in Shiretoko, a mountainous peninsula in Japan
Conservation Genetics (2022)