Abstract
Polyploidy, or whole-genome duplication, is a common speciation mechanism in plants. An important barrier to polyploid establishment is a lack of compatible mates. Because self-compatibility alleviates this problem, it has long been hypothesized that there should be an association between polyploidy and self-compatibility (SC), but empirical support for this prediction is mixed. Here, we investigate whether the molecular makeup of the Brassicaceae self-incompatibility (SI) system, and specifically dominance relationships among S-haplotypes mediated by small RNAs, could facilitate loss of SI in allopolyploid crucifers. We focus on the allotetraploid species Capsella bursa-pastoris, which formed ~300 kya by hybridization and whole-genome duplication involving progenitors from the lineages of Capsella orientalis and Capsella grandiflora. We conduct targeted long-read sequencing to assemble and analyze eight full-length S-locus haplotypes, representing both homeologous subgenomes of C. bursa-pastoris. We further analyze small RNA (sRNA) sequencing data from flower buds to identify candidate dominance modifiers. We find that C. orientalis-derived S-haplotypes of C. bursa-pastoris harbor truncated versions of the male SI specificity gene SCR and express a conserved sRNA-based candidate dominance modifier with a target in the C. grandiflora-derived S-haplotype. These results suggest that pollen-level dominance may have facilitated loss of SI in C. bursa-pastoris. Finally, we demonstrate that spontaneous somatic tetraploidization after a wide cross between C. orientalis and C. grandiflora can result in production of self-compatible tetraploid offspring. We discuss the implications of this finding on the mode of formation of this widespread weed.
Similar content being viewed by others
Introduction
Whole-genome duplication, or polyploidization, is a common speciation mechanism in plants (Grant 1981). It has been estimated that about 15% of flowering plant speciation occurs through polyploidization (Wood et al. 2009), and most angiosperms have undergone whole-genome duplication at some point in their history (Masterson 1994; Soltis et al. 2004). Polyploidy has therefore had a marked impact on plant biodiversity. Understanding the origin and evolution of polyploids can thus give insights into an important process that generates biodiversity in flowering plants.
Polyploids share the common feature of harboring more than two complete chromosome complements, but differ in their mode of origin. Autopolyploids originate by duplication of the genome of one species, whereas allopolyploids originate by hybridization between two biological species, accompanied by genome duplication (Ramsey and Schemske 1998). Allopolyploidy can thus restore the fertility of otherwise sterile hybrids (Winge 1917). In general, both auto- and allopolyploids can form either through (1) the merger of unreduced (2n) gametes, (2) through a triploid bridge which involves the fusion of reduced (n) and unreduced (2n) gametes and subsequent crosses among triploids or backcrosses to diploids, or (3) through spontaneous somatic polyploidization involving meristematic tissue (Ramsey and Schemske 1998). Studies of polyploid formation have long focused on the role of unreduced gamete production, although instances of polyploid formation through spontaneous somatic polyploidization have been described (Ramsey and Schemske 1998). For instance, one of the first cases of allopolyploid formation described in detail occurred through somatic polyploidization in Primula kewensis, a hybrid between Primula verticillata and Primula floribunda (Newton and Pellew 1929).
Polyploidy can contribute to the rapid evolution of reproductive barriers and thus speciation, but newly formed polyploids face several challenges to establishment and persistence. A particular barrier to polyploid establishment is the lack of mates of the same ploidy. If the new polyploid is rare and mostly mates with the more frequent diploid parental species, most of its matings will be ineffective. This leads to selection against the rarer ploidy, a process termed minority cytotype exclusion (Levin 1975). This barrier can be alleviated in polyploids that are self-compatible and capable of self-fertilization or vegetative reproduction, or that are perennial (Grant 1956; Oswald and Nuismer 2011; Fowler and Levin 2016). If polyploids differ from their diploid progenitors in their ecological tolerance or show wider niche breadth, polyploid establishment should also be facilitated (Levin 1975, 1983; Fowler and Levin 2016).
An association between ploidy and mating system is expected on theoretical grounds, both because self-fertilization should facilitate polyploid establishment (Stebbins 1950, 1974) and because polyploids may experience less inbreeding depression than diploids (Lande and Schemske 1985; Ronfort 1999). In line with these expectations, Grant (1956) described a higher rate of self-fertilization in polyploid lineages relative to their diploid relatives, and Barrington (2007) documented an association between ploidy and self-fertilization rates in angiosperms, using phylogenetically independent contrasts. A corollary of the prediction of an association between mating system and polyploidy is that polyploidy should be associated with the breakdown of genetic self-incompatibility (SI) systems that allow rejection of self pollen in many outcrossing angiosperms. Therefore, polyploidy should be associated with self-compatibility (SC). In a broad comparative analysis, Mable (2004) found no global support for such an association across all plant families studied. However, in particular plant families with gametophytic SI, such as the Solanaceae, there is evidence for an association between polyploidy and SC (Mable 2004; Robertson et al. 2011).
One reason for the association between SC and polyploidy in Solanaceae species with gametophytic SI is that in this system breakdown of SI can be an instant byproduct of polyploidization (Zenil-Ferguson et al. 2019). This depends crucially on the molecular details of the Solanaceae SI mechanism, which is based on collaborative non-self recognition (Fujii et al. 2016). The female SI determinant consists of a style-expressed S-RNase which prevents pollen tube growth unless it is degraded by a pollen-expressed F-box protein. Each S-haplotype harbors multiple S-linked F-box genes that can detoxify several different maternal S-RNases, but not their own. In this system, the presence of two S-alleles in unreduced pollen grains can allow detoxification of any maternal S-RNase and therefore immediately leads to self-compatibility in polyploids (Kubo et al. 2010). As a result of the particular molecular basis of the Solanaceae SI system, a general association between polyploidy and self-compatibility is expected, and indeed observed (Zenil-Ferguson et al. 2019).
In contrast, in plant families such as the Brassicaceae, which has a sporophytic SI system that depends on self-recognition and rejection of self pollen, whole-genome duplication is not necessarily expected to cause the loss of SI. Nevertheless, there is evidence that dominance relationships among S-haplotypes in the Brassicaceae sporophytic SI system may facilitate the breakdown of SI in allopolyploids. In the Brassicaceae, it is common for only one allele of the male specificity determinant SCR to be expressed in heterozygotes, due to pollen-level dominance relationships among alleles. Such dominance relationships are mediated by sRNAs expressed by dominant alleles, that target and transcriptionally silence SCR of recessive alleles (Tarutani et al. 2010; Durand et al. 2014). In the presence of pollen level dominance, decay of key S-locus genes in the dominant S-haplotype would be sufficient for breakdown of SI in a newly formed allopolyploid (discussed in Tsuchimatsu et al. 2012). Decay of S-locus genes could involve silencing or nonfunctionalization of the female SI specificity determinant SRK or the male SI specificity determinant SCR in the dominant S-haplotype. If the dominant S-haplotype was already nonfunctional when the polyploid formed, then the recently formed allopolyploid could be instantly SC (Novikova et al. 2017).
In the Brassicaceae, there are empirical examples of SC allopolyploids where dominance seems to have played a role in the breakdown of SI. For instance, in the allopolyploid Arabidopsis kamchatica, breakdown of SI has been linked to decay of the male SI specificity gene SCR in a relatively dominant S-haplotype (Tsuchimatsu et al. 2012). The allopolyploid Arabidopsis suecica was likely instantly SC because it inherited a dominant nonfunctional S-allele harboring a 213-bp inversion in the SCR gene from its A. thaliana parent (Novikova et al. 2017). The dominant A. thaliana-derived S-haplotype of A. suecica is further likely to have suppressed the expression of SCR from A. arenosa, the SI parent of A. suecica (Novikova et al. 2017). The sRNA-based epigenetic mechanisms that regulate dominance of S-alleles at the pollen level in the Brassicaceae (Tarutani et al. 2010; Durand et al. 2014) could thus also facilitate breakdown of SI in Brassicaceae allopolyploids.
The allotetraploid weed Shepherd’s Purse Capsella bursa-pastoris (Brassicaceae) constitutes a suitable system in which to address the role of S-locus dominance for the loss of SI. C. bursa-pastoris is a self-fertilizing weedy species with a nearly worldwide distribution that is in part anthropogenic (Hurka and Neuffer 1997). The Capsella genus harbors three diploid species that differ in their mating system. The SC C. orientalis occurs in central Asia, the SC and self-fertilizing C. rubella is mainly found in the Mediterranean region and central Europe, and the SI outcrosser C. grandiflora is found mainly in the north-western Balkans and sometimes in northern Italy (Hurka et al. 2012). We have previously shown that C. bursa-pastoris is an allotetraploid species that originated ~200–300 kya through hybridization and genome doubling between an ancestor of the extant SC species C. orientalis and an SI progenitor ancestral to the extant species C. grandiflora and C. rubella (Douglas et al. 2015). This means that the ranges of the progenitor lineages must previously have been overlapping although currently they are not (Douglas et al. 2015). The two subgenomes of C. bursa-pastoris from the C. grandiflora/rubella lineage and the C. orientalis lineage are independently inherited, and have been designated the A and B subgenomes, respectively (Slotte et al. 2006, 2008; 2009; Douglas et al. 2015). It is further known that the maternal progenitor of C. bursa-pastoris came from the C. orientalis lineage (Hurka et al. 2012).
While we have a broad understanding of the genome composition and origin of C. bursa-pastoris (Douglas et al. 2015), we still know little about its mode of formation and the breakdown of SI in this species. Our analyses of S-locus variation so far suggest that C. bursa-pastoris could have been SC when it formed (Bachmann et al. 2019). For instance, the SC species C. orientalis harbors a dominant and nonfunctional S-haplotype with a coding frameshift mutation in the SCR gene, which is shared with C. bursa-pastoris B (Bachmann et al. 2019). This suggests that C. orientalis was SC when it gave rise to C. bursa-pastoris. If the C. bursa-pastoris A subgenome S-haplotype from the C. grandiflora lineage is more recessive than the B-haplotype from the C. orientalis lineage, then it is possible that the newly formed allotetraploid C. bursa-pastoris was immediately SC. Such instant SC might have facilitated establishment of the new allopolyploid.
In order to improve our understanding of the role of dominance at the S-locus for the origin of SC allopolyploid crucifers, we have conducted targeted long-read sequencing, annotation and analysis of eight full-length S-haplotypes representing both A and B subgenomes of C. bursa-pastoris. Using short-read data, we document low worldwide S-allele diversity at both subgenomes in a worldwide sample of C. bura-pastori accessions. We further analyze small RNA (sRNA) sequencing data from flower buds to investigate whether the C. bursa-pastoris B S-haplotype harbors and expresses sRNA-based dominance modifiers, which might have facilitated the shift to self-compatibility. Finally, we report on the spontaneous somatic tetraploidization and production of SC allotetraploid Capsella offspring after a wide cross between C. orientalis and C. grandiflora, which demonstrates that SC can be instant upon allopolyploid formation in this system. We discuss the implications of this finding on the mode of formation of C. bursa-pastoris.
Materials and methods
Plant material for S-locus and sRNA sequencing
We grew seeds from five accessions of C. bursa-pastoris from west and east Eurasia (Table S1) for production of bacterial artificial chromosome (BAC) libraries to be used for S-locus sequencing. These accessions were chosen to be representative of major geographic and genetic clusters in this species (Slotte et al. 2009; Cornille et al. 2016). Seed germination, plant growth and sampling of material for BAC library production followed the procedure described in Bachmann et al. (2018). Our material was sufficient for a total of four BAC libraries, one of which was based on two accessions (Table S1). For small RNA sequencing, we grew and collected mixed-stage flower buds from one C. bursa-pastoris accession (Tables S1 and S3) as described in Steige et al. (2016).
S-locus sequencing and annotation
To obtain full-length S-haplotype sequences, we conducted targeted long-read sequencing of S-locus BACs, as described previously (Bachmann et al. 2018). Briefly, we extracted high molecular weight DNA from 10 g of flash-frozen young leaves per library to construct four C. bursa-pastoris BAC libraries (Table S1) at the French Plant Genomic Resource Centre CNRGV. We identified BACs with full-length S-haplotypes as in Goubet et al. (2012) by screening BAC libraries for S-locus flanking regions with DNA probes for U-box and ARK3, which flank the S-locus. We selected two different S-locus BACs per library, based on S-locus polymorphisms obtained by PCR and Sanger sequencing of ARK3 and U-box. S-locus BACs were sequenced with long-read SMRT sequencing (Pacific Biosciences of California, CA, USA) to a coverage of 179–399 and short-read sequencing (MiSeq, Illumina, Inc., San Diego, USA) to a coverage of 1967–4380 at the National Genomics Infrastructure (NGI) in Uppsala, Sweden (Table S2). We generated eight de novo S-haplotype long-read assemblies, corresponding to the subgenome A and B S-haplotypes of the four C. bursa-pastoris BAC libraries. Assemblies were indel error corrected using short reads as described previously (Bachmann et al. 2018).
We annotated the S-locus sequences as previously described (Bachmann et al. 2018). In short, we used Maker v2.31.9 (Holt and Yandell 2011) running RepeatMasker v4.0.7 (http://www.repeatmasker.org) and Augustus v3.2.3 (Stanke et al. 2004) with A. thaliana as prediction species and protein sequences of ARK3, U-box and SRK for gene prediction. The highly divergent SRK was often not identified by Maker, which is why we identified SRK based on sequence similarity to known SRK sequences, as described in Bachmann et al. (2018). SCR was annotated by sequence similarity to CoS12 SCR (Bachmann et al. 2019) (subgenome B) with BLAST v2.5.0+ (Altschul et al. 1990), or an approach based on conservation of 8 cysteine residues in SCR (subgenome A), described in (Bachmann et al. 2018). For annotation of SCR in C. bursa-pastoris subgenome A we further used BLAST v2.5.0+ (Altschul et al. 1990) to annotate sequence similarities to two similar S-haplotypes, A. lyrata SCR38 (Guo et al. 2011) and C. rubella SCR (Vekemans et al. 2014), using a minimum query (exon) coverage of 30 percent and e-value cutoff of 0.1.
To illustrate the phylogenetic relationship among C. bursa-pastoris A and B subgenome S-haplotypes and previously sequenced S-alleles, we constructed a phylogenetic tree based on an alignment of SRK exon 1 sequences from C. bursa-pastoris to a set of sequences downloaded from GenBank (Table S3) (Bachmann et al. 2018). Phylogenetic tree construction was done in RaXMl v8.2.3 (Stamatakis 2014) with the GTR + G model, and 100 bootstrap replicates. The tree was visualized using R v3.3 (R Core Team 2014). We further estimated nucleotide diversity and Watterson’s theta at SRK for both S-haplotypes based on our BAC sequences using Polymorphorama (Haddrill et al. 2008).
S-locus coverage analyses based on whole-genome resequencing data of C. bursa-pastoris
To check whether a larger sample of C. bursa-pastoris might uncover additional S-haplotypes, we analyzed whole-genome resequencing data from 39 samples of C. bursa-pastoris sampled across the species’ distribution in Europe, east Eurasia and the Middle East (Table S4). Three of these sequences were newly generated, whereas the remainder were generated by previous studies (24 from Huang et al. (2018), 12 from Kryvokhyzha et al. (2019)). We also included whole-genome resequencing data from five C. orientalis individuals as a control (Table S4). Raw reads were trimmed with fastp (Chen et al. 2018) and mapped with bwa-mem (Li 2013) against a modified version of C. rubella genome (Slotte et al. 2013). In the modified reference, we masked the the C. rubella S-locus region (scaffold_7 7523601:7562919), and added the A and B alleles of the S-locus region of C. bursa-pastoris accession CbpWEDE, that we assembled previously. We selected only properly paired reads, calculated read coverage with samtools (Li et al. 2009) and visualized coverage across the A and B S-haplotypes for each accession using R v3.3 (R Core Team 2014).
sRNA sequencing
To aid identification of expressed S-locus sRNA-based dominance modifiers, we extracted total RNA from mixed stage flower buds of C. bursa-pastoris accession CbpWEDE (Table S1) using the mirVana extraction kit (Thermo Fisher Scientific Inc., Waltham MA, USA) which isolates RNA from 10-mer to kilobase length. Libraries for sequencing were prepared with the Illumina TruSeq small RNA kit (Illumina, Inc., San Diego, USA) at NGI Stockholm, Sweden. Samples were sequenced with a 1 × 51 setup and HiSeq Rapid SBS Kit v2 chemistry on a HiSeq2500 machine (Illumina, Inc., San Diego, USA). This yielded a total of 14 million reads for this accession.
Identification of candidate dominance modifiers
We performed bioinformatic identification of sRNA precursor regions and targets in our S-locus BAC sequences, in combination with analyses of sRNA expression in flower buds to identify candidate dominance modifiers at the C. bursa-pastoris S-locus.
To identify putative sRNA precursor regions at the S-locus in our S-locus BACs, we followed the approach outlined in Durand et al. (2014) and Bachmann et al. (2019). First, we predicted inverted repeats in C. bursa-pastoris S-haplotypes using EMBOSS-einverted (Rice et al. 2000) with the following parameters: gap penalty = 8, match score = 4, mismatch score = 4, minimum score threshold = 50, maximum separation between start of repeat and end of inverted repeat = 350. We used an initial mapping of sRNA to screen for inverted repeats (Meyers et al. 2008; Durand et al. 2014), and checked the remaining inverted repeats for hairpin structure with rnafold (Lorenz et al. 2011) (Figs. S3 and S4) and kept hairpins with terminal loop smaller than 40 bp and a predicted hairpin structure >20 bp of high base-pairing probability with four or fewer mismatches and maximum two asymmetric bulges (Meyers et al. 2008; Durand et al. 2014).
To assess whether there was expression of sRNA in the precursor regions we identified, we analyzed our floral bud sRNA data. We used Trimmomatic v0.36 for removing sequencing adapters and for quality filtering of reads, before mapping small RNA of 18–27 nt length with STAR v2.5.3a (Dobin et al. 2013) to an adjusted genome of C. rubella (Slotte et al. 2013), where the C. rubella S-locus was replaced with a C. bursa-pastoris subgenome A or B S-locus, as in Bachmann et al. (2019), and the annotation of identified sRNA precursors predicted above. To identify targets of expressed candidate sRNA dominance modifiers, we used a modified Smith and Waterman algorithm (Smith and Waterman 1981) as in Durand et al. (2014) with the following scoring matrix: matches = +1; mismatches = −1; gaps = −2; G:U wobbles = −0.5 and a cutoff score for identifying targets of 18 (Burghgraeve et al. 2018).
Spontaneous polyploidization in a wide cross of C. orientalis and C. grandiflora
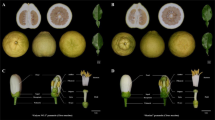
In a previous study, we crossed C. orientalis and C. grandiflora for the purpose of generating a mapping population to investigate the genetic basis of loss of SI (Bachmann et al. 2019). In Bachmann et al. (2019) we presented analyses of the sequences of the S-haplotypes segregating in that cross, and identify a potential dominance modifier at the C. orientalis S-locus. Here, we report on the finding of spontaneous polyploidization in interspecific C. orientalis × C. grandiflora F1s and their F2 offspring, as these results are relevant to an improved understanding of potential pathways to formation of the allopolyploid C. bursa-pastoris. For the mapping of loss of SI in C. orientalis, tetraploid F2 offspring were not considered further in Bachmann et al. (2019).
To generate interspecific F1 hybrids, we crossed the maternal parent C. orientalis Co2008-1 with the paternal parent C. grandiflora Cg88.15 and viable F1 individuals were obtained via embryo rescue (Bachmann et al. 2019). A total of eight mature F1 were obtained, of which all where self-compatible and autonomously self-pollinating (Bachmann et al. 2019). We collected mature seeds from F1 plants in bulk from different flowering shoots. We measured the ploidy of 589 F2 individuals by estimating DNA content of fresh leaf tissue in relation to control samples of known ploidy and to Carex acutiformis using flow cytometry (Plant Cytometry Services, Didam, The Netherlands). We further measured ploidy on three six month old C. orientalis x C. grandiflora F1 plants (Partec Cube 6, Sysmex, Kobe, Japan).
Self-compatibility of newly formed tetraploids
Due to the documented dominance of the C. orientalis S-haplotype over the C. grandiflora S-haplotype segregating in our F2s (Bachmann et al. 2019), we might expect instant self-compatibility in the tetraploid F2s. To assess whether this was the case, we scored 35 tetraploid F2s for silique elongation after autonomous self-pollination. Plants were scored as self-incompatible if they had no elongated siliques and otherwise as self-compatible. For a subset of four F2s we further scored pollen tube growth after manual self-pollination. To assess whether self-compatibility was stable across generations, we grew four F3 individuals that were offspring from a single self-compatible F2 plant and scored pollen tube growth after manual self-pollination. In these pollination assays we observed pollen tube elongation in the pistil after manual self-pollination of emasculated flowers, in six replicates per individual. We also used emasculated unpollinated pistils from one F2 and one F3 individual (in six replicates each) as negative controls. After 12 h flowers were fixed in a 9:1 solution of EtOH: glacial acetic acid for a minimum of 2 h. The flowers were then treated with 1 N NaOH at 60 °C for 20 min to soften the tissue and dipped into a staining solution with 0.01% decolorized aniline blue (in 2% K3PO4) for 2 h, before elongated pollen tubes in each pistil were observed under a Zeiss Axiovert epifluorescence microscope (Zeiss, Oberkochen, Germany). We tested for a difference in pollen tube counts between individuals and pollination treatments using a Kruskal–Wallis test in R 3.3.3.
Results
S-haplotype sequencing and annotation
We generated and annotated eight full-length S-haplotype sequences, four from the C. bursa-pastoris A (C. grandiflora-like) subgenome and four from the B (C. orientalis-like) subgenome. Final S-haplotype sequences (U-box – ARK3) were between 31,011 bp for subgenome A and 23,160–29,615 bp for subgenome B sequences (Table S2).
In the four S-haplotypes from the B subgenome, we identified the genes ARK3, U-box and SRK (Fig. 1). The male SI specificity gene SCR appears to be a pseudogene in all four C. bursa-pastoris B S-haplotypes. Two C. bursa-pastoris B S-haplotypes from western Eurasia shared a single-basepair coding frameshift deletion in SCR with C. orientalis, whereas two eastern Eurasian C. bursa-pastoris B S-haplotypes harbored a larger 31-bp frameshift coding deletion that overlapped with the single-basepair frameshift (Fig. 2). In the four C. bursa-pastoris A S-haplotypes, we identified the genes ARK3, U-box, and exon 1 of SRK. In addition we located short BLAST hits to the male SI specificity gene SCR (Fig. 1), but we were unable to identify the complete gene based on sequence similarity or using automated annotation procedures, including screening for a repetitive pattern of cysteine residues.
S-locus gene exons are shown as arrows in direction of transcription: U-box (red), mirS3 (green), SCR (yellow), SRK (blue), and ARK3 (gray). In C. bursa-pastoris subgenome A, we could not identify a complete SCR gene nor full-length SCR exons, and the bars for SCR here indicate short regions of similarity to C. rubella SCR and A. lyrata AlySCR38.
Evolutionary genetic patterns at SRK
SRK exon 1 sequences from C. bursa-pastoris B were phylogenetically close to those from C. orientalis, as expected, whereas those from C. bursa-pastoris A were closest to sequences S38 and S30 from A. lyrata, C. rubella sequences and C. grandiflora CgS37 (Fig. 3). All subgenome A S-haplotype sequences clustered together, separate from subgenome B S-haplotype sequences (Fig. 3). Both subgenomes harbored very low levels of diversity at SRK in our geographically broad sample (coding region diversity was π = 0.0009, θW = 0.0009 for SRK B and π = 0.0018, θW = 0.0014 for SRK A).
Maximum likelihood tree based on multiple sequence alignment of SRK exon 1 sequences. Large symbols indicate Capsella alleles. Arabidopsis alleles highly similar to C. bursa-pastoris subgenome B (AlS38 and AlS30) and A (AhS12) and SRK are annotated in the figure. Nodes with >95% bootstrap support are marked with an asterisk (*). The tree is rooted using ARK3 sequences. Labels in the outer circle indicate dominance classes according to Mable et al. (2018) (A1 < B < A2, A3 in order of increasing dominance).
Coverage analyses of 39 accessions support low S-allele diversity at the C. bursa-pastoris S-locus
To investigate whether a broader sample of C. bursa-pastoris accessions would support our inference of limited diversity of S-haplotypes at each of the two subgenomes, we analyzed publicly available whole-genome resequencing data from 39 C. bursa-pastoris accessions sampled worldwide. By mapping short reads to a modified C. rubella reference genome supplemented with BAC sequences of the A and B S-haplotypes of C. bursa-pastoris accession CbpWEDE, we identified accessions that shared these haplotypes as those that show broad and even coverage across each S-haplotype. We first demonstrated that mapping short reads of C. orientalis to such a reference, broad and even coverage was retreived only for the B S-haplotype (Figs. S1 and S2, Table S4). We then inspected coverage plots for C. bursa-pastoris. Overall, all 39 C. bursa-pastoris accessions showed evidence of harboring the same S-haplogroup at the A subgenome, and at the B subgenome all accessions except one accession from Kryvokhyzha et al. (2019) showed broad and even coverage, indicating that they harbor the same S-haplotype (Figs. S1 and S2, Table S4). These results thus support our conclusion that C. bursa-pastoris harbors limited S-allele diversity within each of its two subgenomes.
Candidate dominance modifiers and targets
We identified a total of 50 inverted repeats in C. bursa-pastoris subgenome B, out of which three were retained as putative dominance modifiers based on sRNA expression data (Table S5, Fig. S3). In C. bursa-pastoris B, one of the precursor regions, here termed CbpBmirS3 (Fig. 4a), is homologous to ComirS3. ComirS3 is a putative sRNA-based dominance modifier that we previously identified as associated with dominance of SC in C. orientalis (Bachmann et al. 2019) and that shows sequence conservation with AhMirS3 in A. halleri (Durand et al. 2014). Small RNAs are expressed in flower buds from CbpBmirS3 in C. bursa-pastoris accession CbpWEDE (Fig. 4b). We found one potential target region of CbpBmirS3 sRNA in the C. grandiflora derived subgenome A S-locus of C. bursa-pastoris with an affinity >18 (Durand et al. 2014; Burghgraeve et al. 2018), within 2 kb of the closest BLAST hit of SCR (Fig. 4d, e). One putative dominance modifier in C. bursa-pastoris subgenome A was retained from precursor prediction (Table S5, Fig. S4), with no target of expressed sRNA predicted in C. bursa-pastoris subgenome B (Durand et al. 2014; Burghgraeve et al. 2018). The combination of sequence conservation, small RNA expression and the existence of a target in the C. bursa-pastoris A S-haplotype together make CbpBmirS3 a promising candidate dominance modifier, although difficulties in annotating SCR in A subgenome sequences prevent us from directly assessing its impact on SCR expression.
a Predicted hairpin structure of CbpBmirS3 with rnafold. b Expression of CbpBmirS3 in C. bursa-pastoris subgenome B S-haplotype. The gray bar marks the length of the precursor, and the green bar shows the location of expressed 24 nt sRNA with a target predicted in subgenome A. c Predicted target of CbpBmirS3 expressed 24 nt sRNA in subgenome A. d Schematic of predicted dominance with C. bursa-pastoris subgenome B expressing CbpBmirS3 sRNA that target C. bursa-pastoris subgenome A.
Spontaneous polyploidization in a wide cross of C. orientalis and C. grandiflora
For the purpose of mapping the genetic basis of loss of SI in C. orientalis, we generated F2 offspring from a cross between C. orientalis and C. grandiflora. After a routine ploidy screen we unexpectedly found a substantial proportion of tetraploid F2 individuals (~18%, n = 105 tetraploids out of 589 screened F2s). We then screened the ploidy of three C. orientalis × C. grandiflora F1 hybrid individuals and found that one of them harbored both tetraploid and diploid flowering shoots. These results suggest a potential pathway to allopolyploid formation in Capsella through wide hybridization followed by spontaneous polyploidization.
Self-compatibility of newly formed tetraploids
Due to the documented dominance of the C. orientalis S-haplotype over the C. grandiflora S-haplotype segregating in our F2s (the diploid F1 was self-compatible; Bachmann et al. 2019), we expected that tetraploid F2s might be instantly self-compatible. In agreement with this expectation, all 35 tetraploid F2s had elongated siliques and were scored as self-compatible, although there was variation in the number of elongated siliques per plant. Our pollination assays revealed a significant difference in pollen tube counts among individuals and pollination treatments, with self-pollinated tetraploid F2s and F3s having higher pollen tube counts than negative controls (Fig. 5; Kruskal–Wallis rank-sum test statistic = 33.018, P = 0.0001, df = 9), as expected if assayed tetraploid F2s and F3s were self-compatible. This result therefore demonstrates that instant self-compatibility is a possible outcome upon allopolyploid formation in the Brassicaceae.
Discussion
Here, we have generated and analyzed eight full-length S-haplotypes representing both homeologous subgenomes of the allotetraploid crucifer C. bursa-pastoris, with the aim to investigate the role of dominant S-haplotypes for the transition to self-compatibility. More specifically, we were interested in examining whether the same sRNA-based dominance modifiers that are important for pollen-level dominance (Durand et al. 2014) might also facilitate transitions to self-compatibility in allopolyploid crucifers.
In line with a previous study (Bachmann et al. 2019) we found that all C. bursa-pastoris B S-haplotypes clustered together and harbored C. orientalis-derived mutations that are expected to result in the expression of a truncated and likely nonfunctional SCR protein. We further found that all our C. bursa-pastoris samples harbored very similar A S-haplogroup sequences, and polymorphism was very low at SRK of both subgenomes (θW = 0.0009 for SRK B; θW = 0.0014 for SRK A). S-locus coverage analyses based on whole-genome resequencing data from 39 C. bursa-pastoris accessions sampled worldwide further support our inference that the diversity of S-haplotypes is very low at both the C. bursa-pastoris A and B subgenomes. While low polymorphism at the B S-haplogroup might be expected given the limited diversity in the parental species C. orientalis (Douglas et al. 2015), limited polymorphism at the C. bursa-pastoris A S-haplogroup is more surprising given the high number of S-haplotypes currently segregating in C. grandiflora (Guo et al. 2009). While our results suggest that C. bursa-pastoris only inherited one S-haplotype from each of its parental species, a single polyploid origin of C. bursa-pastoris from two haploid gametes has previously been ruled out based on the presence of shared polymorphisms across the genome between C. bursa-pastoris and both of its parental species (Douglas et al. 2015). Other possible explanations for the low S-diversity at the A subgenome of C. bursa-pastoris include bottlenecks associated with the origin of C. bursa-pastoris or selection favoring a specific A subgenome S-haplotype after allopolyploid formation.
If the C. bursa-pastoris B S-haplotype was dominant over the A S-haplotype derived from the paternal C. grandiflora/C. rubella lineage, then the transition to SC might have occurred immediately upon allopolyploid formation. Therefore, we investigated whether the B S-haplotype harbored candidate sRNA-based dominance modifiers, which are known to confer pollen-level dominance in SI Brassicaceae (Tarutani et al. 2010; Durand et al. 2014). Using a combination of bioinformatic analyses and analyses of newly generated floral bud sRNA data, we identified an S-linked candidate dominance modifier termed CbpBmirS3 that is encoded by the C. orientalis-derived B subgenome, and that is expressed in flower buds. The CbpBmirS3 candidate dominance modifier has a predicted target in the A S-haplotype of C. bursa-pastoris. We further identified one potential dominance modifier in the C. grandiflora-derived A S-haplotype, however there were no targets of expressed sRNAs in the C. bursa-pastoris B S-haplotype. Taken together, these results suggest that dominant suppression of a recessive SCR allele derived from the C. grandiflora/rubella lineage by dominance modifiers expressed by the C. orientalis-derived dominant S-haplotype could have contributed to the transition to SC in the newly formed allotetraploid C. bursa-pastoris. These results therefore mirror those recently published regarding the transition to SC in the allotetraploid A. suecica, where a dominant loss-of-function allele at the A. thaliana-derived S-locus is thought to have conferred instant SC (Novikova et al. 2017).
A limitation of our study is that we were unable to empirically verify that SCR from the A S-haplotype is indeed repressed in C. bursa-pastoris, because we could not confidently annotate SCR in the A S-haplotype. We could further only annotate the first exon of the SRK gene in the A S-haplotype. The extremely polymorphic genes SCR and SRK are known to be notoriously difficult to annotate (e.g., Guo et al. 2009 and Vekemans et al. 2014). Therefore, it is possible that we were unable to fully annotate these genes due to bioinformatic limitations. Another possibility is that mutations have degraded C. bursa-pastoris A SCR and SRK to the point that they are no longer recognizable. At present we cannot distinguish between these scenarios. However, we note that CbpBmirS3 exhibits sequence conservation to the C. orientalis candidate dominance modifier ComirS3, which has been shown to be associated with dominant suppression of expression of SCR of more recessive S-haplotypes in diploid S-heterozygotes (Bachmann et al. 2019), although we have not directly assessed its effect in the tetraploid individuals studied here. Furthermore, pollination assays in spontaneously generated C. orientalis × C. grandiflora tetraploids support our hypothesis that instant self-compatibility is possible upon hybridization and polyploidization in the Brassicaceae. However, the C. grandiflora S-haplotype segregating in our F2 individuals likely belongs to a different S-haplogroup, with a different position in the dominance hierarchy among S-alleles than that of C. bursa-pastoris A (Fig. 3). Specifically, the most closely related SRK sequences to C. bursa-pastoris A belong to the dominance class A2, which is a more dominant allele class than the C. grandiflora allele segregating in our F2s, which likely belongs to the most recessive A1 class, according to the classification of Mable et al. (2018). This is important because whether instant self-compatibility upon allopolyploidization is expected depends on the dominance relationships among S-haplotypes inherited from the parental species of a newly formed allopolyploid.
In this study we report on the spontaneous generation of tetraploid offspring after a wide cross between C. orientalis and C. grandiflora, the two parental lineages of C. bursa-pastoris. Due to the direct observation of somatic tetraploidization in the F1 generation and the high proportion of tetraploid F2 offspring produced, we propose that tetraploid offspring likely formed through seed production by tetraploid flowering branches on F1 individuals. Based on this finding we propose that spontaneous somatic tetraploidization following wide hybridization might be a plausible pathway to formation of the widespread weed C. bursa-pastoris, as in the classic case of Primula kewensis (Newton and Pellew 1929). While unreduced gametes are currently considered to be the main route to auto- and allopolyploid formation (Ramsey and Schemske 1998; Mason and Pires 2015; Kreiner et al. 2017a; 2017b), our findings underscore the need for further systematic assessment of the contribution of somatic doubling.
Here, we have investigated the potential mode of formation and transition to SC of the allotetraploid species C. bursa-pastoris. Our results suggest that sRNA-based dominance modifiers may have been important, and we propose a potential mode of formation through spontaneous polyploidization after hybridization, possibly accompanied by instant self-compatibility. The high-quality full-length S-haplotype sequences presented here provide a basis for future detailed investigation of genetic variation and selection at the S-locus in this widespread allopolyploid species.
Data availability
S-locus sequences, sRNA sequences and whole-genome resequencing data have been uploaded to ENA (https://www.ebi.ac.uk/ena) under project accession numbers PRJEB38903, PRJEB32122 and PRJEB41157.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bachmann JA, Tedder A, Laenen B, Fracassetti M, Désamoré A, Lafon-Placette C, Steige KA, Callot C, Marande W, Neuffer B, Bergès H, Köhler C, Castric V, Slotte T (2019) Genetic basis and timing of a major mating system shift in Capsella. N. Phytol 224:505–517
Bachmann JA, Tedder A, Laenen B, Steige KA, Slotte T (2018) Targeted long-read sequencing of a locus under long-term balancing selection in Capsella. G3 8:1327–1333
Barrington BC (2007) Polyploidy and self-fertilization in flowering plants. Am J Bot 94:1527–1533
Burghgraeve N, Simon S, Barral S, Fobis-Loisy I, Holl AC, Poniztki C, Schmitt E, Vekemans X, Castric V (2018) Base-pairing requirements for small RNA-mediatedgene silencing of recessive self-incompatibility alleles in Arabidopsis halleri. Genetics 215:653–664
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890
Cornille A, Salcedo A, Kryvokhyzha D, Glémin S, Holm K, Wright SI, Lascoux M (2016) Genomic signature of successful colonization of Eurasia by the allopolyploid Shepherd’s Purse (Capsella bursa-pastoris). Mol Ecol 25:616–629
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Douglas GM, Gos G, Steige KA, Salcedo A, Holm K, Josephs EB, Arunkumar R, Agren JA, Hazzouri KM, Wang W, Platts AE, Williamson RJ, Neuffer B, Lascoux M, Slotte T, Wright SI (2015) Hybrid origins and the earliest stages of diploidization in the highly successful recent polyploid Capsella bursa-pastoris. Proc Natl Acad Sci USA 112:2806–2811
Durand E, Méheust R, Soucaze M, Goubet PM, Gallina S, Poux C, Fobis-Loisy I, Guillon E, Gaude T, Sarazin A, Figeac M, Prat E, Marande W, Bergès H, Vekemans X, Billiard S, Castric V (2014) Dominance hierarchy arising from the evolution of a complex small RNA regulatory network. Science 346:1200–1205
Fowler NL, Levin DA (2016) Critical factors in the establishment of allopolyploids. Am J Bot 103:1236–1251
Fujii S, Kubo K, Takayama S (2016) Non-self- and self-recognition models in plant self-incompatibility. Nat Plants 2:16130
Goubet P, Bergès H, Bellec A, Prat E, Helmstetter N, Mangenot S, Gallina S, Holl A-C, Fobis-Loisy I, Vekemans X, Castric V (2012) Contrasted patterns of molecular evolution in dominant and recessive self-incompatibility haplotypes in Arabidopsis. PLoS Genet 8:e1002495
Grant V (1956) The influence of breeding habit on the outcome of natural hybridization in plants. Am Nat 90:319–322
Grant V (1981) Plant speciation. Columbia University Press, New York, NY and London
Guo YL, Bechsgaard JS, Slotte T, Neuffer B, Lascoux M, Weigel D, Schierup MH (2009) Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci USA 106:5246–5251
Haddrill PR, Bachtrog D, Andolfatto P (2008) Positive and negative selection on noncoding DNA in Drosophila simulans. Mol Biol Evol 25:1825–1834
Holt C, Yandell M (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinforma 12:491
Huang H-R, Liu J-J, Xu Y, Lascoux M, Ge X-J, Wright SI (2018) Homeologue-specific expression divergence in the recently formed tetraploid Capsella bursa-pastoris (Brassicaceae). N. Phytol 220:624–635
Hurka H, Friesen N, German DA, Franzke A, Neuffer B (2012) ‘Missing link’ species Capsella orientalis and Capsella thracica elucidate evolution of model plant genus Capsella (Brassicaceae). Mol Ecol 21:1223–1238
Hurka H, Neuffer B (1997) Evolutionary processes in the genus Capsella (Brassicaceae). Plant Syst Evol 206:295–316
Kreiner JM, Kron P, Husband BC (2017a) Evolutionary dynamics of unreduced gametes. Trends Genet 33:583–593
Kreiner JM, Kron P, Husband BC (2017b) Frequency and maintenance of unreduced gametes in natural plant populations: associations with reproductive mode, life history and genome size. N. Phytol 214:879–889
Kryvokhyzha D, Milesi P, Duan T, Orsucci M, Wright SI, Glémin S, Lascoux M (2019) Towards the new normal: transcriptomic convergence and genomic legacy of the two subgenomes of an allopolyploid weed (Capsella bursa-pastoris). PLoS Genet 15:e1008131
Kubo K-i, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S-i, Ando T, Isogai A (2010) Collaborative non-self recognition system in S-RNase–based self-incompatibility. Science 330:796–799
Lande R, Schemske D (1985) The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution 39:24–40
Levin DA (1975) Minority cytotype exclusion in local plant populations. Taxon 24:35–43
Levin DA (1983) Polyploidy and novelty in flowering plants. Am Nat 122:1–25
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL (2011) ViennaRNA Package 2.0. Algorithm Mol Biol 6:26
Mable BK (2004) Polyploidy and self‐compatibility: is there an association. N Phytol 162:803–811
Mable BK, Brysting AK, Jørgensen MH, Carbonell AKZ, Kiefer C, Ruiz-Duarte P, Lagesen K, Koch MA (2018) Adding complexity to complexity: gene family evolution in polyploids. Front Ecol Evol 6:114
Mason AS, Pires JC (2015) Unreduced gametes: meiotic mishap or evolutionary mechanism. Trends Genet 31:5–10
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264:421–424
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhui JK (2008) Criteria for annotation for plant microRNAs. Plant Cell 20:3186–3190
Newton WCF, Pellew C (1929) Primula kewensis and its derivatives. J Genet 20:405–467
Novikova PY, Tsuchimatsu T, Simon S, Nizhynska V, Voronin V, Burns R, Fedorenko OM, Holm S, Säll T, Prat E (2017) Genome sequencing reveals the origin of the allotetraploid Arabidopsis suecica. Mol Biol Evol 34:957–968
Oswald BP, Nuismer SL (2011) A unified model of autopolyploid establishment and evolution. Am Nat 178:687–700
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org/
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Ann Rev Ecol Syst 29:467–501
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16:276–277
Robertson K, Goldberg EE, Igic B (2011) Comparative evidence for the correlated evolution of polyploidy and self-compatibility in Solanaceae. Evolution 65:139–155
Ronfort J (1999) The mutation load under tetrasomic inheritance and its consequences for the evolution of the selfing rate in autotetraploid species. Genet Res 74:31–42
Slotte T, Ceplitis A, Neuffer B, Hurka H, Lascoux M (2006) Intrageneric phylogeny of Capsella (Brassicaceae) and the origin of the tetraploid C. bursa-pastoris based on chloroplast and nuclear DNA sequences. Am J Bot 93:1714–1724
Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo YL, Steige K, Platts AE, Escobar JS, Newman LK, Wang W, Mandáková T, Vello E, Smith LM, Henz SR, Steffen J, Takuno S, Brandvain Y, Coop G, Andolfatto P, Hu TT, Blanchette M, Clark RM, Quesneville H, Nordborg M, Gaut BS, Lysak MA, Jenkins J, Grimwood J, Chapman J, Prochnik S, Shu S, Rokhsar D, Schmutz J, Weigel D, Wright SI (2013) The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet 45:831–835
Slotte T, Huang H-R, Holm K, Ceplitis A, Onge KS, Chen J, Lagercrantz U, Lascoux M (2009) Splicing variation at a FLOWERING LOCUS C homeolog is associated with flowering time variation in the tetraploid Capsella bursa-pastoris. Genetics 183:337–345
Slotte T, Huang H, Lascoux M, Ceplitis A (2008) Polyploid speciation did not confer instant reproductive isolation in Capsella (Brassicaceae). Mol Biol Evol 25:1472–1481
Smith TF, Waterman MS (1981) Identification of common molecular subsequences. J Mol Biol 147:195–197
Soltis DE, Soltis PS, Tate JA (2004) Advances in the study of polyploidy since plant speciation. N. Phytol 161:173–191
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Stanke M, Steinkamp R, Waack S, Morgenstern B (2004) AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32:W309–12
Stebbins GL (1950) Variation and Evolution in Plants. Columbia University Press, New York, NY
Stebbins GL (1974) Flowering plants: evolution above the species level. Belknap Press of Harvard University Press, Cambridge, Mass
Steige KA, Reimegård J, Rebernig CA, Köhler C, Scofield DG, Slotte T (2016) The role of transposable elements for gene expression in Capsella hybrids and allopolyploids. Preprint at https://www.biorxiv.org/content/10.1101/044016v1.full
Tarutani Y, Shiba H, Iwano M, Kakizaki T, Suzuki G, Watanabe M, Isogai A, Takayama S (2010) Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466:983–986
Tsuchimatsu T, Kaiser P, Yew C-L, Bachelier JB, Shimizu KK (2012) Recent loss of self-incompatibility by degradation of the male component in allotetraploid Arabidopsis kamchatica. Plos Genet 8:e1002838
Vekemans X, Poux C, Goubet PM, Castric V (2014) The evolution of selfing from outcrossing ancestors in Brassicaceae: what have we learned from variation at the S-locus? J Evol Biol 27:1372–1385
Winge Ø (1917) The chromosome. Their numbers and general importance. Compt Rend Trav Lab Carlsberg 13:131–175
Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH (2009) The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA 106:13875–13879
Zenil-Ferguson R, Burleigh JG, Freyman WA, Igić B, Mayrose I, Goldberg EE (2019) Interaction among ploidy, breeding system and lineage diversification. N Phytol 224:1252–1265
Acknowledgements
The authors thank Vincent Castric for advice on SCR annotation, Barbara Neuffer for kindly providing C. orientalis seeds, Olga Vinnere Pettersson for advice on long-read sequencing and Christian Tellgren-Roth for bioinformatic assistance with long-read assembly. Sequencing was performed by the Uppsala Genome Centre and the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The computations were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) partially funded by the Swedish Research Council through grant agreement no. 2016-07213, under Projects sllstore2017029, sllstore2017044, sllstore2017043, SNIC 2018/8-170, SNIC 2017/7-174, and SNIC 2017/7-175. The sampling and sequencing of the three C. bursa-pastoris samples was supported by grants from the Royal Swedish Academy of Sciences and by the Royal Physiographic Society of Lund to MF. This study was supported by a grant from the Swedish Research Council (grant #621-2013-4320) and by a grant from the Science for Life Laboratory, Swedish Biodiversity Program to TS. The Swedish Biodiversity Program has been made available by support from the Knut and Alice Wallenberg Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Olivier Hardy
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bachmann, J.A., Tedder, A., Fracassetti, M. et al. On the origin of the widespread self-compatible allotetraploid Capsella bursa-pastoris (Brassicaceae). Heredity 127, 124–134 (2021). https://doi.org/10.1038/s41437-021-00434-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-021-00434-9