Abstract
Muscle fiber diameter is an economically important trait because it affects meat yield and quality. However, the genetic basis underlying muscle fiber diameter has not been determined. In this study, we collected THREE muscular histological phenotypes in 479 ducks from an F2 segregating population generated by mallard × Pekin duck crosses. We performed genome-wide association studies (GWAS) and identified a quantitative trait locus (QTL) significantly associated with muscle fiber diameter on chromosome 3. Then, we discovered the selection signatures using the fixation index among 40 mallards and 30 Pekin ducks in this QTL region. Furthermore, we characterized the recombination event in this QTL region and identified a 6-kb block located on TASP1 that was significantly associated with muscle fiber diameter. Finally, five SNPs were screened as potential causative mutations within the 6-kb block. In conclusion, we demonstrated that TASP1 contributes to an increase in muscle fiber diameter, which helps to characterize muscle development and contributes to the genetic improvement of meat yield and quality in livestock.
Similar content being viewed by others
Introduction
Skeletal muscle is the most important component of the animal body, accounting for 40–50% of body weight, and muscle fiber is the main component of skeletal muscle (Janssen et al. 2000; Payne and Bearden 2006; Wang et al. 2001). The variation in muscle fiber is directly related to meat yield and quality (Chen et al. 2007; Joo et al. 2013). Muscle fiber diameter shows wide variations among different muscle tissues and individual muscles. This difference in muscle fiber diameter is regulated during the development stage, adapting the muscle fiber diameter according to the metabolic demands and neural activity of the body and promoting muscle growth or leading to muscle atrophy (Lee et al. 2017).
Generally, the number of muscle fibers is fixed during embryonic development, while the increase in muscle mass is attributed to the hypertrophy of skeletal muscle fiber after birth (Gonyea et al. 1977; Schiaffino et al. 2013). This phenomenon means that thickening the muscle fiber diameter can significantly affect muscle yield. In addition, meat tenderness is an important indicator for evaluating meat quality, and muscle fiber diameter has a high positive correlation with meat tenderness. Increases in muscle fiber diameter can promote meat tenderness and play an important role in meat flavor (An et al. 2010; Gondret et al. 2006; Hwang et al. 2010).
Duck (Anas platyrhynchos) is an economically important domestic fowl. Among the various phenotypically diverse indigenous breeds, Pekin duck is the most elite breed. Compared with the phenotypes of their wild ancestors (mallards) (Li et al. 2010), the phenotypes of Pekin ducks have diverged significantly due to intensive artificial selection. These phenotypes include white plumage, extraordinary body size, and large muscle fiber size. In our previous study, we identified the mechanisms leading to the white plumage and enlarged body size in Pekin ducks based on the F2 segregating population (Zhou et al. 2018). However, the genetic basis underlying fiber diameter increase is still poorly understood.
In this study, to dissect the genetic basis of the fiber diameter increase, we performed genome-wide association studies (GWAS) on the F2 segregating population generated by wild × domestic crosses. We identified that TASP1 was significantly associated with muscle fiber diameter and that TASP1 underwent artificial selection during domestication. We subsequently determined that five SNPs contributed to fiber diameter increase in breast muscle. Our results may help to elucidate muscle development and contribute to the genetic improvement of meat yield and quality in livestock.
Materials and methods
Animal populations
A total of 479 F2 ducks were used in this study from reciprocal crosses of mallards and Pekin ducks. The F1 generation was produced from reciprocal crosses of 10 ♂ Pekin ducks × 100 ♀ mallard ducks and 4 ♂ mallard ducks × 40 ♀ Pekin ducks. The size of the F1 generation reached more than 700 individuals, and the ratio of male to female ducks was relatively balanced. A total of 216 families were established using the F1 hybrids based on the following principles: (1) one male was mated to three females, (2) males and females in the same family were not full-siblings, and (3) female ducks within a family were not half-siblings. The F2 generation was composed of almost 2000 ducks, most of which were slaughtered for growth performance studies (Zhou et al. 2018). All experiments with ducks were performed under the guidance of ethics regulations from the Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing, China.
Genomic DNA extraction and sequencing
Genomic DNA was obtained from whole blood samples collected from the brachial veins of ducks and isolated using the standard phenol/chloroform extraction method. The quantity and quality of genomic DNA were assessed by Nanodrop and agarose gel electrophoresis. After the examinations, eligible DNA samples from the 479 F2 population individuals were generated to paired-end libraries using standard procedures. In addition, the average insert size was 500 bp, and the read length was 150 bp. All libraries were sequenced on an Illumina® Hiseq X-Ten platform to an average raw read sequence coverage of 5×.
Breast muscle histological data collection
The breast muscles were fixed in 4% paraformaldehyde and embedded in paraffin blocks. Hematoxylin and eosin were used to stain the muscle sections (3–5 μm). Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, USA) was used to measure the average area and minimum Feret’s diameter of at least 100 muscle fibers to estimate the fiber area and fiber diameter for each bird, and the muscle fiber density (fibers/mm2) was estimated by point-counting stereology using 300 points. In total, we collected the muscular histological traits of 10 Pekin ducks, 10 mallards, and 479 individuals of the F2 segregating population at 8 weeks.
Variant discovery and genotyping
The 150-bp paired-end raw reads were mapped to the Pekin duck reference genome (GCA_003850225.1) with Burrows-Wheeler alignment (BWA aln) (Li and Durbin 2009) using the default parameters. The paired reads that were mapped to the exact same position on the reference genome were identified with MarkDuplicates in Picard (Wysokar et al. 2014) to avoid any influence on variant detection. After mapping, SNP calling was performed using exclusively GATK HaplotypeCaller module (version 3.5) (DePristo et al. 2011), and the output was further filtered using VCFtools (version 0.1.15) (Danecek et al. 2011). SNPs were filtered based on the following criteria: (1) 3× < mean sequencing depth (overall included individuals) < 30×, (2) SNPs had to have a minor allele frequency >0.05, (3) the maximum missing rate was <0.1, and (4) SNPs had only two alleles. A total of 479 ducks from an F2 segregating population mated by Pekin duck and Mallard were genotyped, and 7307,089 SNPs were prepared for subsequent analysis.
Heritability estimation
The heritability of muscle fiber traits was estimated using the animal model implemented in the ASReml v3.0 software package (Gilmour et al 2009). The model is
where yij are vectors of phenotypic values of muscle fiber; the i refers to duck house and j refers to which individual duck in this duck house. X1, X2, and X3 are fixed effects of farm, sex, and mating strategies (10 ♂ Pekin ducks × 100 ♀ mallard ducks and 4 ♂ mallard ducks × 40 ♀ Pekin ducks); Aij represents additive genetic effects as random effects; and e is random residuals for muscle fiber data. The polygenic effects were treated as random and assumed to be mutually independent.
The heritability of each trait was estimated using
Variances of the random effects are defined as V(a) = A\({\upsigma}_{A}^2\) for the polygenes and V(e) = I\({\upsigma}_{e}^2\) for the residuals, where A was the additive kinship relationship matrix, I was the identity matrix. A matrix was inferred from the kinship where \({\upsigma}_{A}^2\) is the additive genetic variance and \({\upsigma}_{e}^2\) is the residual variance.
GWAS analysis
Population structure and cryptic relationships were considered to minimize false positives and increase statistical power. The Super model program GAPIT (Wang et al. 2014) was used for the association analysis. The model is
where y is the vector of phenotypic values of muscle fiber, W is the incidence matrix for v, v is marker effect, β is fixed effect including (farm, sex, mating strategies), u is random genetic effect, and e is random residuals for muscle fiber data.
PCA was performed based on all SNPs using EIGENSOFT software (version 4.2) (Patterson et al. 2006; Price et al. 2006). The first three PCA values (eigenvectors) derived from whole-genome SNPs, as well as the forward/backward cross, were set as fixed effects in the mixed model to correct for population stratification. The random effect was kinship matrix estimated based on the IBS algorithm among all individual whole-genome SNPs. We defined the whole-genome significance cutoff as the Bonferroni test threshold, and we set the association threshold as 0.05/total SNPs (total SNPs: 7307,089, −log10 (P) = 8.16).
Transcriptome
The breast muscle tissue was lyophilized in liquid nitrogen, dissolved in TRIzol reagent, and prepared for subsequent library construction. All extracted RNA quality and quantity were assessed by Nanodrop and agarose gel electrophoresis. RNA samples were reverse transcribed to cDNA with the use of PrimerScript™ RT Master Mix (RR036A, Takara, Dalian, China) following the manufacturer’s instructions. Twenty-four library preparations were sequenced on an Illumina X-Ten platform, and 150-bp single-end reads were generated. The average output was 6 Gb per library. Sequencing adaptors and low-complexity reads were removed by trimmomatic version 0.36 software in the initial data filtering step. Quality control and read statistics were estimated with FASTQC version 0.10.1 software. Then, clean data were mapped to the Pekin duck reference genome (GCA_003850225.1) using TopHat version 2.0.11 software (Trapnell et al. 2009). Read counts per gene were obtained by running HTSeq version 0.6.1 software (Anders et al. 2015). CPM (counts per million mapped sequence reads) for each gene were calculated by edgeR version 3.20.9 packages (Robinson et al. 2010), and genes with averaged CPM among replicates ≥1 at no less than one time point were considered expressed and retained for further analysis. Differential expression analysis of the two species (Mallard vs. Pekin duck) was performed using the edgeR version 3.20.9 packages. An adjusted P value < 0.05 and log2 fold change > |2| were set as the thresholds for significant differential expression.
Genome scanning for divergent regions
We calculated the value of FST for each SNP site on chr3 (11.00–12.00 Mbp) using VCFtools (version 0.1.15) among 30 Pekin ducks and 40 mallards. We then used extended haplotype homozygosity (EHH) for the core haplotype (chr3:11396601) to assess the value of each core haplotype by the decay of its association with alleles at various distances from the locus among Pekin ducks and mallards by selacan (version 1.20) (Szpiech and Hernandez 2014). PLINK (Purcell et al. 2007) was used to calculate the linkage disequilibrium (LD).
Results
Phenotypic and genetic parameters
We measured a total of three muscular histological traits (fiber area, fiber diameter, and fiber density) of ten Pekin ducks and ten mallards at 8 weeks and found significant differences in the breast muscle fiber, especially in muscle fiber diameter (P = 0.007, Fig. 1a, b and Table S1).
a The distribution of muscle fiber diameter is presented under the muscle fiber images. Blue and red refer to the mallard and Pekin duck at 8 weeks, respectively. b Pekin duck and mallard muscle fiber histomorphological data at 8 weeks, including muscle fiber area, muscle fiber diameter, and muscle fiber density. c F2 population muscle fiber histomorphological and heritability. d A heatmap depicting Pearson’s correlation coefficients between phenotype means (low triangle) and genetic correlations (upper triangle). Asterisks indicate significant correlations using a two-tailed t-test (**P < 0.001 and ***P < 0.0001), and the values between parenthesis indicate SE.
Then, we collected a total of five muscular histological traits (fiber area, fiber diameter, fiber density, perimysium, and endomysium) for 479 individuals of the F2 segregating population and estimates of heritability (h2). The coefficients of variation for fiber area, diameter, and density were 23.52%, 12.6%, 27.92%, respectively (Fig. 1c and Table S2). Our results showed that the estimated heritability did not vary noticeably for these muscular histological traits. We found that fiber diameter and fiber area displayed a moderate heritability at 0.32, and the estimated heritability for muscle fiber density was 0.20.
Phenotypic and genetic correlations among five muscle fiber traits mirrored one another. We found that high positive phenotypic and genetic correlations existed between fiber area and fiber diameter, while low positive correlations existed between fiber diameter and endomysium, fiber diameter and perimysium. In contrast, we found negative correlations between fiber density and fiber diameter, fiber density and fiber area (Fig. 1d).
GWAS identified a QTL associated with muscle fiber diameter
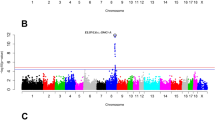
We performed a GWAS for three muscular histological traits (fiber diameter, fiber area, fiber density) on the Pekin ducks × mallards F2 segregating population, in which 461 phenotype F2 individuals were genotyped with 7307,089 SNPs. The GWAS analysis was performed based on the Pekin duck reference genome (GCA_003850225.1). The quantile–quantile (QQ) plot revealed that SNPs deviated from the distribution under the null hypothesis, which indicated an association between SNPs and phenotypes (Fig. S1). A single significant locus (−log10 P = 8.99) was detected at Chr3:11396601 bp of the Pekin duck reference genome in muscle fiber diameter and passed the Bonferroni significance threshold (−log10 P > 8.164; Fig. 2a and Table S3). However, a quantitative trait locus (QTL) range from Chr3:11.16 Mbp-11.40 Mbp was also associated with the muscle fiber area and muscle fiber density (Fig. S2). In addition, this QTL contained three annotated genes (NDUFAF5, ESF1, and TASP1).
a GWAS analyses of muscle fiber diameter. The gray horizontal dashed lines indicate the Bonferroni significance threshold of the GWAS (−log10 P > 8.164). b Regional plots for the loci ranging from 11.00 to 11.50 Mbp associated with muscle fiber diameter trait. All genotyped SNPs are color-coded according to their pairwise LD with the leader SNP (Chr3:11396601) calculated in the F2 population. SNPs are colored based on the strength of LD values (r2 values) considering the most strongly associated SNP and the other SNPs in the region. c Fixation index (Fst, blue dot) for chr3:10.00–12.50-Mbp SNP between Pekin duck and mallard. d Nucleotide diversity (π) for chr3:11.35–11.45-Mbp SNP between Pekin duck (red line) and mallard (blue line). e Extended haplotype homozygosity for core haplotype (chr3:11396601) to assess the age of each core haplotype by the decay of its association to alleles at various distances from the locus in Pekin duck (red line) and mallard (blue line).
Then, we examined the lead SNP (Chr3:11396601 bp) closely by calculating pairwise LD between the SNPs within the QTL (Chr3:9.39–13.39 Mbp) surrounding the lead SNP on chromosome 3. Thirty-five SNPs spanning a region from 11.36–11.40 Mbp were highly correlated (pairwise r2 > 0.4; Fig. 2b and Table S4).
Fine-mapping identified five SNPs as candidate causative mutations
To further determine the causative gene for muscle fiber diameter, we compared the mRNA expression of three candidate genes based on the transcriptome data in breast muscle tissues between Pekin ducks and mallards during duck growth and development (2, 4, 6, and 8 weeks), with three Pekin ducks and three mallards at each time point. Only TASP1 mRNA expression was higher in mallards than in Pekin ducks at 6 weeks (log2 FC = 0.85, FDR = 0.10). In addition, 6 weeks is the time of the fastest development stage for duck breast muscle, which suggests that TASP1 is a potential candidate gene. However, the candidate genes (NDUFAF5, ESF1, TASP1) were not differentially expressed at any developmental stages (Table S5). Finally, a comparison of the two breeds resulted in 2024 differentially expressed genes (DEGs), and the numbers of DEGs at 2, 4, 6, and 8 weeks were 13, 50, 1523, and 582, respectively (Fig. S3).
To validate and fine-map the QTL region (Chr3:11.16–11.40 Mbp) for muscle fiber diameter, we calculated the FST (fixation index) value of each SNP site between the Pekin duck population and mallard population and identified high fixation of 22 SNPs (FST > 0.7) (Fig. 2c and Table S6). The Pekin duck population showed lower nucleotide diversity (π = 9.3 × 10−4) than the mallard population (π = 2.6 × 10−3) in the QTL region (Fig. 2d and Table S7). The lead SNP as a core haplotype shows EHH, and we found that the homozygosity of Pekin duck haplotypes containing the lead SNP variants extends on average nearly 100 kb but only 2000 bp in mallard haplotypes (Fig. 2e and Table S8). Our results indicated that this region in Pekin duck had undergone strong artificial selection during domestication and breeding, which led to increased muscle fiber diameter.
To narrow the candidate region, we characterized the recombination events in the candidate region and identified three recombination breakpoints that divided the 440 F2 birds into thirteen haplotypes (Table S9). Only the haplotypes in block 2 (Chr3: 11,390,000–11,397,000 bp) located on the TASP1 gene could absolutely distinguish the individual phenotypic values of muscle fiber diameter (one-way ANOVA: muscle fiber diameter: P = 1.4 × 10−4). These results indicated that the segment of block 2 was the minimal candidate region responsible for muscle fiber diameter and that the effect of this block genotype could increase muscle fiber diameter by 5.21% (Fig. 3a).
a Recombination event analyses are shown in schematic form in this plot. Red bars refer to chromosomal segments originating from Pekin ducks, purple bars refer to segments originating from mallards, and orange bars refer to segments originating from heterozygotes. R1–10 refer to ten recombinant types. The left box plot refers to muscle fiber diameter. The numbers of individuals are given in brackets. The indicated P values are based on one-way ANOVA. Box plots denote median (centerline), 25–75th percentile (limits), the minimum and maximum values without outliers, and outliers (gray dots). The right breast muscle image shows F2 population muscle fiber diameter segregation of mallard, Pekin duck, and heterozygotic haplotypes of TASP1. b. The locations of all five candidate SNPs in the TASP1 gene and these contribute to muscle fiber diameter in block 2. Box plots denote median (centerline), 25–75th percentile (limits), minimum and maximum values without outliers, and outliers (gray dots).
However, we did not find high Fst values in this QTL with nonsynonymous substitutions or frameshift mutations. To ascertain the causative variations in block 2 (Chr3: 11,390,000–11,397,000 bp), we examined whether the genotypes of high Fst value loci (Fst > 0.6) are related to muscle fiber diameter. Ultimately, five SNPs were significantly associated with muscle fiber diameter (Fig. 3b). Therefore, these five SNPs located on the intron of TASP1 were considered to be candidate causative variations.
Discussion
Muscle fiber diameter is affected by TASP1
Animal breeders have long focused on meat production traits with the potential to increase yield and quality while decreasing input. More recently, improving meat yield and quality is of economic and social interest to help meet the needs of an increasing human population. Previous studies support that muscle fiber diameter is affected by factors such as breed (Ryu et al. 2008), selection (Larzul et al. 1999), and sex (Ozawa et al. 2000). However, it has not been reported that a single gene affects muscle fiber diameter in ducks. This study is the first to use a systems genetics approach to identify a gene and candidate loci for muscle fiber diameter in ducks.
Our study identified the TASP1 gene as a putative gene for muscle fiber diameter increase based on the following findings. (1) GWAS on the F2 populations enabled us to define the QTL for muscle fiber diameter within an interval of 240 kb on chromosome 3. This QTL contains only three genes, including TASP1. (2) The use of FST, π, and EHH analysis among the 30 Pekin ducks and 40 mallards identified a selection signal in this QTL. (3) Recombination events detected a 6-kb block located on the TASP1 gene.
Interestingly, we found a special alternative splicing phenomenon for TASP1 in 6-week Pekin ducks that did not exist in 6-week mallards (Fig. S4). Meanwhile, TASP1 mRNA expression was higher in mallards than in Pekin ducks at 6 weeks. In our previous studies, increasing muscle fiber diameter was greatest in Pekin ducks at 6 weeks (Fan et al. 2020). Evaluating whether alternative splicing in 6-week-old Pekin ducks directly causes an increase in muscle fiber diameter merits further research.
Furthermore, our study found that the effect of a favorable QTL genotype could increase muscle fiber diameter by 5.21% and significantly affect breast muscle yield (Fig. S5). This gene is economically important in fowl meat production.
F2 population GWAS signal underwent artificial selection during duck domestication
Compared with mallards, Pekin ducks show extraordinary body size and excellent meat production performance. The apparent difference in these traits is due to artificial selection, especially with regard to body weight (Zhou et al. 2018). In traditional duck breeding, breast muscle weight is one of the most economically important traits and is selected for during breeding. Pekin duck breast muscle weight is nearly three times greater than that of mallards (Gille and Salomon 1998). Furthermore, muscle fiber is the primary part of the muscle, and its differences are among the critical factors affecting muscle weight.
In this study, we first performed GWAS for muscle fiber diameter and showed a significant signal on chromosome 3 (Fig. 2a). Then, we found that this QTL also has a strong selective signal (Fig. 2b–e). This result supports the selection of this QTL during manual breeding. To explain this phenomenon, it is possible that the trait of breast muscle weight was strongly selected for during manual breeding, and the TASP1 gene, which appears to affect muscle fiber diameter, was selected simultaneously.
Hypothesis of the regulatory pathway by which TASP1 affects duck breast muscle fiber diameter
TASP1 encodes an endopeptidase that cleaves specific substrates following aspartate residues. This protein is required to cleave MLL, a protein required for the maintenance of HOX gene expression, and TFIIA (Khan et al. 2005; Lee et al. 2009; Takeda 2006). The Taspase1 proenzyme is intramolecularly proteolyzed, generating an active 28/22-kDa heterodimer. RNAi-mediated knockdown of Taspase1 results in the appearance of unprocessed MLL and the loss of proper HOX gene expression (Hsieh et al. 2003). Meanwhile, TASP1 plays a key role in cancer cell proliferation and apoptosis and can inhibit tumor growth after knockout (Chen et al. 2012). And TASP1 acting on cardiomyocytes in mouse to inhibit cardiomyocyte proliferation (Gan et al. 2015). These studies indicate that TASP1 may inhibit muscle cell proliferation to affect muscle development. The MLL directly affects HOXA9 function (Erfurth et al. 2008; Faber et al. 2009; Zeisig et al. 2004). Hoxa9 deletion increases the self-renewal and cell-autonomous nature of muscle fiber-associated satellite cells, and Hoxa9, in turn, activates several developmental pathways and represents a decisive factor that separates satellite cell gene expression (Schworer et al. 2016). The activated pathways include most of the currently known inhibitors of satellite cell function, including Wnt, TGFβ, JAK/STAT, and senescence signaling (Brack et al. 2007; Carlson et al. 2009; Sousa-Victor et al. 2014). Satellite cells act as major myogenic stem cells and undergo active proliferation and fusion during the formation of new muscle fibers (Ciciliot and Schiaffino 2010; Moss and Leblond 1971; Zammit 2008).
Considering the gene mRNA expression, HOXA9 was not significantly differentially expressed at 8 weeks, but mRNA expression in mallards (CPM = 25) was higher than in Pekin ducks (CPM =22). In addition, HOXA9 mRNA expression in Pekin duck had a declining trend during duck growth and development. Meanwhile, the expression levels of KAT2B (activating HOXA9 expression, log2 FC = 1.58, FDR = 0.10) were higher in mallards than in Pekin ducks at 8 weeks, but not significant differentially expressed between the Pekin ducks and mallards. The expression levels of the other genes (AFF1, KDM6A, KMT2A) were lower in mallards than in Pekin ducks at 8 weeks (Fig. 4 and Table S5). All of these data indicated that the increase in muscle fiber diameter may be related to this pathway.
The variation in TASP1 might influence its enzymatic activity, causing the appearance of unprocessed MLL and the loss of proper HOX gene expression, which in turn induces developmental pathways inhibiting SC function and muscle regeneration. Combined with the gene mRNA expression, the expression levels of both HOXA9 and KAT2B were higher in mallards than in Pekin ducks at 8 weeks. The expression levels of the other genes (AFF1, MLLT1, KDM6A, KMT2A) were lower in mallards than in Pekin ducks at 8 weeks. The 2- to 8-week CPM of Pekin ducks (orange line) and mallards (blue line).
Based on the findings of previous studies and our results, the regulatory process by which TASP1 affects muscle fiber diameter could be hypothesized as follows. TASP1 encodes an endopeptidase to cleave MLL. The variation in TASP1 might influence its enzymatic activity, causing the appearance of unprocessed MLL and the loss of proper HOX gene expression. Eventually, HOXA9 expression was repressed, resulting in activated pathways of satellite cell function and large-scale growth in muscle cells.
Conclusion
We have provided genetic evidence supporting that TASP1, located on chromosome 3, contributes to increased duck breast muscle fiber diameter, and this study is the first to demonstrate this association in poultry. We also found five SNPs as candidate mutations in TASP1. These results may provide a reference for studies on the muscle development of fowl and contribute to the genetic improvement of meat yield and quality in animals.
Data availability
All sequences have been deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with the accession codes PRJNA471401 and PRJNA450892. We deposited the genome assembly, all of the sequence data and SNP information in BIG Data Center (http://bigd.big.ac.cn/)65. The accession numbers are PRJCA000651, PRJCA000647, and GVM000015. The Illumina sequencing data used in this study can be available at BIG Data Center (http://bigd.big.ac.cn/) with the accession code PRJCA001307. Other data sets supporting the results of this article are included within the article and its supplementary information.
References
An JY, Zheng JX, Li JY, Zeng D, Qu LJ, Xu GY et al. (2010) Effect of myofiber characteristics and thickness of perimysium and endomysium on meat tenderness of chickens. Poult Sci 89(8):1750–1754
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169
Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C et al. (2007) Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317(5839):807–810
Carlson ME, Conboy MJ, Hsu M, Barchas L, Jeong J, Agrawal A et al. (2009) Relative roles of TGF‐β1 and Wnt in the systemic regulation and aging of satellite cell responses. Aging Cell 8(6):676–689
Chen DY, Lee Y, Van Tine BA, Searleman AC, Westergard TD, Liu H et al. (2012) A pharmacologic inhibitor of the protease Taspase1 effectively inhibits breast and brain tumor growth. Cancer Res 72(3):736–746
Chen XD, Ma QG, Tang MY, Ji C (2007) Development of breast muscle and meat quality in Arbor Acres broilers, Jingxing 100 crossbred chickens and Beijing fatty chickens. Meat Sci 77(2):220–227
Ciciliot S, Schiaffino S (2010) Regeneration of mammalian skeletal muscle: basic mechanisms and clinical implications. Curr Pharm Des 16(8):906–914
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011) The variant call format and VCFtools. Bioinformatics 27(15):2156–2158
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43(5):491
Erfurth FE, Popovic R, Grembecka J, Cierpicki T, Theisler C, Xia Z-B et al. (2008) MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc Natl Acad Sci 105(21):7517–7522
Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M et al. (2009) HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 113(11):2375–2385
Fan W, Liu W, Liu H, Meng Q, Xu Y, Guo Y et al. (2020) Dynamic accumulation of fatty acids in duck (Anas platyrhynchos) breast muscle and its correlations with gene expression. BMC Genom 21(1):58
Gan J, Sonntag HJ, Tang MK, Cai D, Lee KK (2015) Integrative analysis of the developing postnatal mouse heart transcriptome. PLoS ONE 10(7):e0133288
Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2009) ASReml user guide release 3.0. VSN International Ltd, Hemel Hempstead, UK. www.vsni.co.uk
Gondret F, Lefaucheur L, Juin H, Louveau I, Lebret B (2006) Low birth weight is associated with enlarged muscle fiber area and impaired meat tenderness of the longissimus muscle in pigs1,2. J Anim Sci 84(1):93–103
Gonyea W, Ericson GC, Bonde‐Petersen F (1977) Skeletal muscle fiber splitting induced by weight‐lifting exercise in cats. Acta Physiol Scand 99(1):105–109
Hsieh JJ, Cheng EH, Korsmeyer SJ (2003) Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell 115(3):293–303
Hwang YH, Kim GD, Jeong JY, Hur SJ, Joo ST (2010) The relationship between muscle fiber characteristics and meat quality traits of highly marbled Hanwoo (Korean native cattle) steers. Meat Sci 86(2):456–461
Gille U, Salomon FV (1998) Muscle growth in wild and domestic ducks. Br Poult Sci 39(4):500–505
Janssen I, Heymsfield SB, Wang Z, Ross R (2000) Skeletal muscle mass and distribution in 468 men and women aged 18–88 yr. J Appl Physiol 89(1):81–88
Joo ST, Kim GD, Hwang YH, Ryu YC (2013) Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat Sci 95(4):828–836
Khan JA, Dunn BM, Tong L (2005) Crystal structure of human Taspase1, a crucial protease regulating the function of MLL. Structure 13(10):1443–1452
Larzul C, Le Roy P, Gogué J, Talmant A, Jacquet B, Lefaucheur L et al. (1999) Selection for reduced muscle glycolytic potential in Large White pigs. II. Correlated responses in meat quality and muscle compositional traits. Genet Sel Evol 31(1):61
Lee JK, Hallock PT, Burden SJ (2017) Abelson tyrosine-protein kinase 2 regulates myoblast proliferation and controls muscle fiber length. eLife 6:e29905
Lee JT, Chen DY, Yang Z, Ramos AD, Hsieh JJD, Bogyo M (2009) Design, syntheses, and evaluation of Taspase1 inhibitors. Bioorg Medicinal Chem Lett 19(17):5086–5090
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
Li HF, Zhu WQ, Song WT, Shu JT, Han W, Chen KW (2010) Origin and genetic diversity of Chinese domestic ducks. Mol Phylogenet Evol 57(2):634–640
Moss FP, Leblond CP (1971) Satellite cells as the source of nuclei in muscles of growing rats. Anat Rec 170(4):421–435
Ozawa S, Mitsuhashi T, Mitsumoto M, Matsumoto S, Itoh N, Itagaki K et al. (2000) The characteristics of muscle fiber types of longissimus thoracis muscle and their influences on the quantity and quality of meat from Japanese Black steers. Meat Sci 54(1):65–70
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2(12):e190
Payne GW, Bearden SE (2006) The microcirculation of skeletal muscle in aging. Microcirculation 13(4):275–277
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38(8):904
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140
Ryu YC, Choi YM, Lee SH, Shin HG, Choe JH, Kim JM et al. (2008) Comparing the histochemical characteristics and meat quality traits of different pig breeds. Meat Sci 80(2):363–369
Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M (2013) Mechanisms regulating skeletal muscle growth and atrophy. FEBS J 280(17):4294–4314
Schworer S, Becker F, Feller C, Baig AH, Kober U, Henze H et al. (2016) Epigenetic stress responses induce muscle stem-cell aging by Hoxa9 developmental signals. Nature 540(7633):428–432
Sousa-Victor P, Gutarra S, García-Prat L, Rodriguez-Ubreva J, Ortet L, Ruiz-Bonilla V et al. (2014) Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506(7488):316
Suleiman J, Mundt M, Sampath S, El‐Hattab AW (2018) TASP1 is deleted in an infant with developmental delay, microcephaly, distinctive facial features, and multiple congenital anomalies. Clin Genet 94(1):170–173
Szpiech ZA, Hernandez RD (2014) selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol 31(10):2824–2827
Takeda S (2006) Proteolysis of MLL family proteins is essential for Taspase1-orchestrated cell cycle progression. Genes Dev 20(17):2397–2409
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111
Wang Q, Tian F, Pan Y, Buckler ES, Zhang Z (2014) A SUPER powerful method for genome wide association study. PLoS ONE 9(9):e107684
Wang Z, Heo M, Lee RC, Kotler DP, Withers RT, Heymsfield SB (2001) Muscularity in adult humans: proportion of adipose tissue‐free body mass as skeletal muscle. Am J Hum Biol: Off J Hum Biol Assoc 13(5):612–619
White RB, Biérinx A-S, Gnocchi VF, Zammit PS (2010) Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol 10(1):21
Wysokar A, Tibbetts K, McCown M, Homer N, Fennell T (2014) Picard: a set of tools for working with next generation sequencing data in BAM format. http://picard.sourceforge.net
Zammit PS (2008) The muscle satellite cell: the story of a cell on the edge! In: Skeletal muscle repair and regeneration. Springer, Dordrecht, p 45–64
Zeisig BB, Milne T, García-Cuéllar M-P, Schreiner S, Fuchs MartinM-E et al. (2004) Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol 24(2):617–628
Zhou Z, Li M, Cheng H, Fan W, Yuan Z, Gao Q et al. (2018) An intercross population study reveals genes associated with body size and plumage color in ducks. Nat Commun 9(1):1–10
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31972523), the Young Top-notch Talent Project of the National Ten Thousand Talent Program, the National Scientific Supporting Projects of China (2015BAD03B06), the China Agriculture Research System of Waterfowl (CARS-42), and the CAAS Innovation Team Project (ASTIP-2016-IAS-9 and CAAS-XTCX2016010-03).
Author information
Authors and Affiliations
Contributions
SH and ZZ conceived and coordinated the study. DL performed the study and wrote the manuscript. DL, WF, YX, SY, and WL carried out the bioinformatics and experimental analyses. SH, ZZ, ZG, and WH constructed the F2 population. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent to participate
All animals used in the study were treated following the guidelines for the experimental animals established by the Council of China Animal Welfare. Protocols of the experiments were approved by the Science Research Department of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (CAAS) (Beijing, China).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor Sara Knott
Rights and permissions
About this article
Cite this article
Liu, D., Fan, W., Xu, Y. et al. Genome-wide association studies demonstrate that TASP1 contributes to increased muscle fiber diameter. Heredity 126, 991–999 (2021). https://doi.org/10.1038/s41437-021-00425-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-021-00425-w
This article is cited by
-
Selection on the promoter regions plays an important role in complex traits during duck domestication
BMC Biology (2023)
-
Genome-wide mapping of signatures of selection using a high-density array identified candidate genes for growth traits and local adaptation in chickens
Genetics Selection Evolution (2023)