Abstract
Geographic isolation and reduced population sizes can lead to local extinction, low efficacy of selection and decreased speciation. However, population differentiation is an essential step of biological diversification. In allopatric speciation, geographically isolated populations differentiate and persist until the evolution of reproductive isolation and ecological divergence completes the speciation process. Pitcairnia flammea allows us to study the evolutionary consequences of habitat fragmentation on naturally disjoint rock-outcrop species from the Brazilian Atlantic Rainforest (BAF). Our main results showed low-to-moderate genetic diversity within populations, and deep population structuring caused by limited gene flow, low connectivity, genetic drift and inbreeding of long-term isolation and persistence of rock-outcrop populations throughout Quaternary climatic oscillations. Bayesian phylogenetic and model-based clustering analyses found no clear northern and southern phylogeographic structure commonly reported for many BAF organisms. Although we found two main lineages diverging by ~2 Mya during the early Pleistocene, species’ delimitation analysis assigned most of the populations as independent evolving entities, suggesting an important role of disjoint rock outcrops in promoting high endemism in this rich biome. Lastly, we detected limited gene flow in sympatric populations although some hybridization and introgression were observed, suggesting a continuous speciation process in this species complex. Our data not only inform us about the extensive differentiation and limited gene flow found among Pitcairnia flammea species complex, but they also contain information about the mechanisms that shape the genetic architecture of small and fragmented populations of isolated rock outcrop of recently radiated plants.
Similar content being viewed by others
Introduction
Species dispersal and colonization are determining ecological processes that affect the dynamics of populations and ecosystems in a changing world (Couvet 2002). In fact, spatial isolation of populations may restrict connectivity, leading to low levels of gene flow between fragments and subsequent higher genetic structure and lower genetic diversity in isolated populations (Honnay and Jacquemyn 2007). Furthermore, increased geographic isolation and reduced population sizes can lead to reduced fitness and survival rates (Leimu et al. 2006), promoting high levels of local extinction, low efficacy of selection and decreased speciation (Frankham et al. 2012). However, population genetic structure does not always lead to extinction. For instance, in allopatric speciation, a continuous process (Nosil 2012), geographically isolated populations differentiate and persist until the evolution of reproductive isolation and ecological divergence completes the speciation (Allmon 1992). As such, population differentiation is an essential step of diversification, implying that differentiation and diversification should be associated over a deep evolutionary time (Harvey et al. 2017). Surprisingly, proper estimations of the evolutionary consequences of habitat fragmentation and disturbance on naturally disjoint species on plant diversification still lack strong experimental and empirical support.
Deep genetic differentiation among populations due to demographic constraints of limited intraspecific gene exchange is a recurrent pattern observed in rock outcrops and sky islands all over the globe (Neotropics: Barbará et al. 2007; Pinheiro et al. 2014; Gonçalves-Oliveira et al. 2017; North America: Wiens et al. 2019; Australia: Byrne et al. 2019; Asia: He and Jiang 2014). The high levels of genetic differentiation found in some of these cases may suggest the existence of several endemic species occurring in different localities instead of only one broadly distributed species. Likewise, high levels of genetic differentiation in naturally fragmented populations suggest variable levels of reproductive isolation (Pinheiro and Cozzolino 2013). Thus, the reproductive barriers present among populations have great potential to clarify the mechanisms underlying the first step of speciation (Via 2009; Abbott et al. 2013), effectively connecting microevolutionary processes within species and the origin of species across macroevolutionary timescales. In fact, the first step of speciation is better understood when different populations from the same putative species are studied. In this context, studies of fragmented populations offer a great opportunity to illuminate the consequences of gene exchange restrictions in allopatric speciation and diversification.
The Brazilian Atlantic Rainforest (BAF) is the second largest tropical rainforest in South America, recognized as one of the most important biodiversity hotspots in the world, containing more than 60% of all terrestrial species on the planet (Myers et al. 2000; Mittermeier et al. 2005; Oliveira-Filho and Fontes 2000). The BAF consists of a complex of vegetation that shelters various types of plant communities (Scarano 2002, 2009). In addition to the predominant semi-deciduous and humid forest types, the biome has open-tree vegetation of sandy coastal plains, marsh forests and vegetation growing in low- and high-altitude rocky outcrops (Scarano 2002). Rocky outcrops are considered islands of terrestrial habitats due to their spatial and ecological isolation, which offers a barrier against dispersal and migration, thus allowing for the occurrence of species with disjoint populations (Porembski 2007). Furthermore, BAF rock outcrops are recognized as centres of species diversity and endemism, with a significant contribution to regional species diversity for the whole biome (Porembski 2007). Gradients in climatic and edaphic factors separate these rock outcrops from the neighbouring forest, which explains their highly specialized biota (Meirelles et al. 1999).
Questions of attention are how this region became so diverse, and whether its biota show similar patterns of speciation. Several phylogeographic studies attempted to reveal the complex evolutionary history of the BAF biota, considering different groups of continuous and connected forest-dwelling organisms as models to comprehend historical patterns of diversification (Cabanne et al. 2007; Carnaval and Moritz 2008; Palma-Silva et al. 2009; Colombi et al. 2010; Martins, de 2011; Leite et al. 2016; Menezes et al. 2016; Leal et al. 2016; Franco et al. 2017; Paz et al. 2019; Aguiar-Melo et al. 2019; Rivera et al. 2020). Nevertheless, phylogeographic patterns of naturally fragmented species from BAF rock outcrops remain largely unclear (but see Palma-Silva et al. 2011; Hmeljevski et al. 2017; de Paula et al. 2017). Furthermore, its high floristic diversity, species turnover, several ecological gradients and complex geological history make rock outcrops an excellent system for understanding evolutionary and ecological dynamics of BAF complex biome. In addition, it would be of interest to understand evolutionary processes promoting species diversity in such fragmented habitats because loss of the surrounding forest will certainly have an impact on rock outcrops endemic species (Scarano 2009).
Bromeliads adapted to rock outcrops, such as the recently radiated Pitcairnia genus (11.8 Mya, Schütz et al. 2016; Schubert 2017), are particularly suitable for addressing the potential role of intra- and interspecific gene flow during plant speciation and radiation, and establishing connections between macro- and microevolutionary processes (Palma-Silva et al. 2016, 2020; Mota et al. 2019). In fact, bromeliads are emerging as model systems for understanding evolutionary genetics and the ecology of species’ complex of radiations in naturally fragmented habitats (Barbará et al. 2007; Boisselier-Dubayle et al. 2010; Lavor et al. 2014; Palma-Silva et al. 2011, 2015; Lexer et al. 2016; Hmeljevski et al. 2017; Gonçalves-Oliveira et al. 2017; Hirsch et al. 2020; Pinange et al. 2017; Ruas et al. 2020; Wöhrmann et al. 2020).
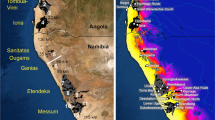
The P. flammea Lindl occupies a wide range of rocky outcrops and inselbergs along the Brazilian Atlantic Forest (latitudes 16°S–26°S—Fig. 2). This taxon has one of the largest geographical ranges and morphological variation among the species of the genus Pitcairnia (Smith and Downs 1974). According to Flora Neotropica (Smith and Downs 1974), seven varieties of P. flammea are recognized: var. flammea, var. roeltzii (E. Morren) L.B. Smith, var. corcovadensis (Wawra) L.B. Smith, var. glabrior L.B. Smith, var. pallida L.B. Smith, var. floccosa Lindl and var. macropoda L.B. Sm. & Reitz. P. corcovadensis was raised to the species category by Wendt et al. (2000). In a phylogenetic hypothesis, based on macromorphological and micromorphological characters, Saraiva et al. (2015) recognized the P. flammea group. More recently, Schubert (2017), based on a molecular phylogeny, recognized P. flammea as a monophyletic clade with unresolved branches. Furthermore, these species and varieties are found both in allopatry and sympatry throughout their distribution, and records of interspecific hybridization between sympatric populations emphasize the role of introgression and/or incomplete lineage sorting for the diversification of this radiating group (Palma-Silva et al. 2011; Mota et al. 2019). Here, we threaded the P. flammea group as proposed by Saraiva et al. (2015), and the varieties proposed by Smith and Downs (1974) as P. flammea species complex. Species complexes are ideal models to understand speciation due to the incomplete lineages’ divergence (Pinheiro et al. 2018). Thus, we believe that studying species’ delimitation in the P. flammea species complex will provide a framework for gaining insights on the first step of species formation.
This study intends to answer the following questions: (1) what phylogeographic and genetic structure patterns of the P. flammea complex tell us about the evolutionary mechanisms involved in speciation and diversification of rock-outcrop taxa? Are these patterns consistent with those previously reported for other dwelling-forest plants and animals in the Atlantic Forest? What is the extent of long-distance dispersal for this widely distributed species complex? (2) How many genetically differentiated populations exist throughout the distribution of the species, and can these be considered different evolutionary lineages? (3) Are the genetically differentiated populations corresponding to taxonomic varieties and species proposed for the P. flammea complex? (4) What interspecific gene flow tells us about diversification in sympatric populations of this group? To answer these questions, we reconstructed the evolutionary and demographic history of the P. flammea complex using phylogeographic approaches based on nuclear microsatellite loci and plastidial DNA analysis.
Materials and methods
Species studied and sampling
This study included a total of 501 samples from 21 localities, including 18 allopatric and three sympatric populations that cover most of the geographic range of P. flammea (Bromeliaceae) species complex. Different varieties collected in sympatry were considered different populations for the subsequent analyses, totalling 25 populations. Five varieties of the P. flammea complex were sampled, as well as three other morphologically similar species, namely P. carinata Mez, P. corcovadensis and P. curvidens L.B.Sm. & Read, which were included to test the degree of genetic differentiation among the populations of P. flammea (Table 1). The analysis of allopatric samples will provide estimates of genetic differentiation influenced directly by the fragmented nature of such populations. On the other hand, the analysis of sympatric populations will tell us if different varieties maintain species cohesion even in the absence of geographic barriers. Outgroup species were included from the early-diverging species in the genus: P. heterophyla and P. lanuginosa (GenBank accession numbers KJ188920/AF537904, MK423244, respectively). Information on each sampling locality and the number of individuals from each population used in DNA sequencing is in Table 1. Voucher specimens were deposited in Herbarium Rioclarense (HRCB), Rio Claro, Brazil.
DNA extraction, Sanger sequencing and genotyping of microsatellites
Fresh leaves were sampled from each individual plant, dried on silica gel and stored at –20 °C in a freezer until DNA extraction. Total genomic DNA was extracted using the DNeasy 96 Plant Kit (Qiagen) according to the instructions by the manufacturer.
Two plastid DNA regions (rpl32-trnL and rps16-trnK) were amplified and sequenced for a subset of 176 individuals from the populations sampled, using the primers described by Shaw et al. (2007). All polymerase chain reaction (PCR) amplifications were carried out in a total volume of 20 μL containing 10 ng of DNA template, 1× GoTaq buffer, 2.5 mM MgCl2, 0.25 mM dNTP mix, 5 pmol forward and reverse primers and 0.5 U GoTaq DNA polymerase (Promega), and run using the following parameters: initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, 54 °C (rpl32-trnL) or 58 °C (rps16-trnK) for 1 min and 72 °C for 1 min, and a final extension of 10 min at 72 °C. PCR products were sent for both forward and reverse sequencing to Macrogen (Seoul, Korea). The rpl32-trnL and rps16-trnK sequences were visualized, edited and manually verified using the software GENEIOUS version 10.2.3 (www.geneious.com, Kearse et al. 2012). Alignments were made with use of the MUSCLE algorithm with default parameters and checked by eyes for ambiguous alignments. Mononucleotide repeats were removed due to uncertain homology, and indels longer than 1 base pair were recorded as single characters. Sequence data of both plastid regions were then concatenated for subsequent analyses.
We amplified ten nuclear microsatellite loci that were previously developed for other bromeliad species (E6B: Boneh et al. 2003; VgA04: Palma-Silva et al. 2007; PaA05, PaA09, PaA10, PaC05, PaD07 and PaB12: Paggi et al. 2008; Acom12.12: Wöhrmann and Weising 2011; ngFos22: Wöhrmann et al. 2012) in a total of 501 individuals. For each microsatellite locus, the forward primer was synthesized with a 19-bp M13 tail (50-CACGACGTTGTAAAACGAC-30), following a method that uses three primers: a forward SSR-specific primer with the M13 tail added at its 5′-end, a reverse locus-specific primer and a universal M13 primer labelled with one of four fluorescent dyes: FAM, VIC, PET or NED (Applied Biosystems, Foster City, CA, USA). PCR amplifications of nuclear microsatellites were carried out in 10-µL reaction volumes comprising 1× GoTaqColorless Master Mix (Promega, Madison, WI, USA), 10 pmol of each forward and reverse microsatellite primer, 1 pmol of M13 universal primer and 10 ng of template DNA. We used a “touchdown” cycling programme and followed the protocol described by Palma-Silva et al. (2007). All reactions were performed using the Veriti 96-well thermal cycler (Applied Biosystems). Amplifications of nuclear microsatellites were conducted on an ABI 3500 sequencer (Applied Biosystems, Warrington, UK) and compared using GeneScan LIZ 500 molecular-size standard (Applied Biosystems). The loci were visually analyzed using GENEMARKER version 1.95 Demo (SoftGenetics, Pennsylvania, USA).
Molecular analyses on cpDNA sequence data
Genetic diversity and structure analyses
The number of haplotypes was identified using DnaSP (Rozas et al. 2017). We estimated the haplotype (Hd) and nucleotide diversities (π), and the number of polymorphic sites (S) using ARLEQUIN version 3.5 (Excoffier and Lischer 2010). This analysis was conducted for each population, as well as for the whole dataset. The evolutionary relationships between the haplotypes were estimated with Network version 4.6.1.1 (available at http://www.fluxus-engineering.com) using the median-joining (MJ) method (Bandelt et al. 1999). The “frequency > 1” criterion was used for MJ calculations to ignore singletons.
We examined the population genetic structure using “Clustering with linked loci” implemented in Bayesian clustering analysis (BAPS) 6.0 (Corander et al. 2013). To determine the most probable number of genetic groups (K), we performed ten algorithm repetitions for each K, from 1 to 24, using the default software parameters. We assessed the population structure with pairwise FST values calculated with ARLEQUIN version 3.5. We also performed an analysis of molecular variance (AMOVA) to test the structure among populations using the whole dataset and different hierarchical levels based on groups detected by the phylogenetic tree inferred in BEAST, and sorting by taxonomic varieties (Smith and Downs 1974).
Phylogenetic inference and divergence times
Phylogenetic trees were inferred based on Bayesian analyses, as implemented in BEAST version 1.8.4 (Drummond et al. 2012). Two phylogenetic trees were inferred: species tree for the entire dataset, and haplotype tree. The divergence times for individuals and haplotypes were estimated using P. lanuginosa and P. heterophyla as outgroups. The best-fit model of nucleotide substitution for each locus partition was estimated using jModelTest (Darriba et al. 2012) under default settings (NumModels = 88, +F, +I, +G, ML optimized, BEST) and following the corrected Akaike information criterion (AICM).
Analyses in BEAST consisted of three independent runs, each with a Markov chain Monte Carlo (MCMC) analysis performed for 10 × 107 steps, sampling every 10,000 steps. We estimated divergence times using a relaxed Bayesian molecular clock with uncorrelated lognormal rates (Drummond et al. 2012; Drummond and Rambaut 2007), a coalescent constant population-size prior. We applied a secondary calibration based on divergence time as inferred by Schubert (2017), set as a normal distribution. We verified the stationary posterior distributions of parameters (ESS > 200) and compared the likelihood of runs using the MCMC AICM in TRACER version 1.6.
Demographic analyses
To assess population demographic history, Tajima’s D and Fu’s Fs neutrality tests were carried out in Arlequin, based on 10,000 simulations. In addition, we estimated changes in population size over time with an Extended Bayesian Skyline Plot (EBSP) analysis. The analysis was run in the BEAST version 2.4.0 with the following priors: the HKY DNA substitution models (prior selected using jModelTest 2.1.10), and a strict clock prior using substitution rate intervals inferred by the BEAST tree (1.706E−3 ± 0.826). Two independent runs of 200,000,000 generations with a thinning interval of 20,000 were combined using LogCombiner to generate the final output after checking for convergence and stationarity in Tracer 1.7. To avoid bias in the demographic analysis interpretation due to the genetic structure found in P. flammea (see “Results”), we also performed neutrality tests and EBSP considering the two genetic groups recovered by BEAST using the same parameters described above.
Species delimitation
To determine genetic clusters representing possible independent evolving entities in the P. flammea complex, we applied the generalized mixed Yule coalescent method (GMYC). GMYC uses an ultrametric single-locus MCC tree to find a threshold in time between inter- and intraspecific branching rates, by maximizing the model likelihood. The analysis used the phylogenetic tree generated on BEAST, inferred for the entire dataset, and was implemented in software R using the package “splits” under the single-threshold version (https://r-forge.r-project.org/projects/splits/).
Molecular analyses on microsatellite data
Genetic diversity and structure analyses
The nuclear genetic diversity of microsatellite loci was characterized based on the number of alleles (A), allelic richness (AR), observed heterozygosity (Ho), expected heterozygosity (He) and inbreeding coefficient (FIS) calculated for each locus using MSAnalyzer version 4.05 (Dieringer and Schlötterer 2003). Deviations from Hardy–Weinberg equilibrium (HWE) were calculated in GENEPOP on the Web version 4.6 (Raymond and Rousset 2006).
To estimate the population genetic structure and admixture proportion of nuclear DNA, we performed a model-based BAPS using STRUCTURE version 2.3.2 (Pritchard et al. 2000). It was carried out under the admixture model assuming independent allele frequencies and using a burn-in period of 250,000 and run length of 1,000,000 with the MCMC and ten replicates per K, ranging from 1 to 30. No prior population information was used for analysis. To determine the most probable number of genetic groups (K), we used the method by Evanno et al. (2005) implemented in STRUCTURE-HARVESTER Web version 0.6.94 (Earl and vonHoldt 2012). We also assessed the population genetic differentiation with pairwise FST values and performed AMOVA to test the hierarchical structure within populations and among populations, using the whole dataset and sorting by varieties, all calculated on Arlequin version 3.5. To determine whether divergence among populations is an effect of isolation-by-distance, we tested the correlation between geographical and genetic distance matrices (FST/(1 − FST) with a standardized Mantel test using GENEPOP on the Web version 4.6 (Raymond and Rousset 2006). Significance was assessed with a randomized test using 10,000 Monte Carlo simulations. Estimates of gene flow were generated in BayesAss 3.0.3 (Wilson and Rannala 2003). The analysis consisted of 10 million iterations with the first 1 million iterations discarded as burn-in, and ΔA, ΔF and ΔM set to 0.3, 0.5 and 0.1, respectively.
Demographic analyses
In order to detect a recent and pronounced reduction in effective population size (Bottleneck effect), the two-phase mutation model (TPM) was implemented in BOTTLENECK version 1.2 using the recommendation for microsatellite data stated in the user manual (12% variance and 95% stepwise mutations) (Piry et al. 1999). Statistical significance was assessed in 10,000 replicates using a one-tailed “Wilcoxon signed-rank” test of heterozygosity excess for each population. In addition, we calculated the M ratio by Garza and Williamson (2001) for each population to detect recent decreases in effective population size using the software M_P_Val.exe. The M value represents the ratio between the total number of alleles and the range in allele size for each locus (Garza and Williamson 2001). When random alleles are lost due to a reduction in effective population size, the number of alleles tends to reduce faster than the overall allelic range (Garza and Williamson 2001). Significance for each population was assessed by comparisons of M ratios and critical values (MC values) obtained by simulating the M ratio under specific demographic and mutational conditions using the software Critical_M.exe (available at http://swfsc.noaa.gov/textblock.aspx?Division=FED&id=3298). M ratios below the critical M represent a strong sign of recent past bottleneck events.
Results
Sanger sequencing of cpDNA
Genetic diversity
The concatenated alignment with 176 P. flammea cpDNA sequences, measured 1480 base pairs (<5.00% missing data), with 74 polymorphic sites resulting in 26 haplotypes (Figs. 1 and 2). Extremely low intrapopulation diversity was observed in P. flammea, with few to no variations within populations, so the genetic diversity indexes were zero for most populations (Fig. 2; Table 2), with only three exceptions (CARMG, IBIMG-fla and JURSP, with two haplotypes each). Only two haplotypes were shared between different populations: TERJ-fla and GARJ shared haplotype H16, whereas PGSP-fla and BESP shared haplotype H21.The overall haplotype diversity for the whole dataset was h = 0.957 (SD = 0.0035) and nucleotide diversity was π = 0.0072 (SD = 0.0036) (Table 2).
Records were obtained from the GBIF (https://www.gbif.org/) and Species Link (http://splink.cria.org.br/) online databases and filtered according to each species’ checked distribution. Each haplotype assigned different colours, and circle sections represent the haplotype frequency of each sampled population. The codes indicate the sampling localities (for full names of populations, see Table 2).
cpDNA population structure
The haplotype network (Fig. 1) showed a star-like pattern, with H22 as the central haplotype. Most haplotypes are connected by one or two mutational steps. However, some haplotypes have more mutational steps from central H22. BAPS of the cpDNA sequences revealed an optimal structure of nine genetic clusters (K = 9) for P. flammea individuals (Fig. 3). The defined groups recovered the patterns observed in the haplotype network, where the most distant haplotypes were clustered together (Fig. 1).
a Indicates the results for STRUCTURE, with optimal K = 22, and b indicates the results for BAPS, with optimal partition of nine groups. Individuals are arranged by population. Distinct colours represent distinct genetic groups. See Table 1 for abbreviations of each population.
Pairwise FST values for cpDNA ranged from 0.745 to 1000, the latter being most of the values obtained between the populations (Fig. 4a; Table S1). All AMOVA tests (Table 3) indicated extremely high and significant genetic differentiation among populations and groups of P. flammea. For the whole dataset, the genetic variation was the highest among population (FST = 0.985, P < 0.001). AMOVA also showed significant genetic differentiation when contrasting genetic clusters indicated by BEAST (FCT = 0.434, P < 0.001). On the other hand, sorting by varieties was not significant among the groups (FCT = 0.065, P > 0.05), suggesting that taxonomy was not a good predictor of population genetic differentiation. Regardless of the clustering method utilized in the hierarchical AMOVA, differentiation among populations was always higher than among groups and within groups.
Divergence times and demographic analysis
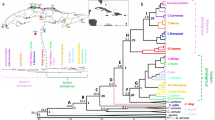
The Bayesian phylogenetic analysis of the cpDNA region, for both species/individuals and haplotype trees, clustered most of the populations into two strongly supported groups (posterior > 0.95), one comprising populations DESRJ, NFLRJ, NFRJ and SMARJ, distributed in the North region of Serra do Mar, in the state of Rio de Janeiro, and the second with the remaining populations, including populations in the northernmost region of the geographic distribution in the states of Minas Gerais and Espírito Santo (Fig. 5; Supplementary Fig. 1). The haplotypes of P. flammea diverged from a common ancestor with P. heterophylla at 3.866 Mya, during the late Pliocene. The time of the most recent common ancestor (TMRCA) of all P. flammea haplotypes was estimated at 2.008 Mya [95% highest posterior density (HPD)] in the early Pleistocene (Fig. 6), which corresponds to the divergence time between the two possible main haplotype lineages (Northern Rio de Janeiro with haplotypes H6, H7, H8 and H9, and all others). All the remaining divergence events are suggested to have occurred at less than 1 Mya. The phylogenetic tree constructed with all individuals recovered the same pattern of haplogroups, with 4.454 Mya for divergence with P. heterophylla and 2.089 for the TMRCA among P. flammea species complex (Figs. 5 and 6).
The results of Tajima’s D and Fu’s FS neutrality tests for cpDNA were not significant (P > 0.005). Hence, no demographic changes were detected for the P. flammea populations included in this study. The results were also not significant when tested for the two BEAST genetic groups. The EBSP analyses, with the present dataset, did not generate satisfactory results for any level of population clustering. The BEAST runs did not converge, presenting ESS < 200 in any cases, likely due to the high population structure observed among populations. Thus, we were unable to assess changes in population size over time.
The GMYC single-threshold results recovered 24 molecular operational taxonomic units (MOTUs) and identified a coalescence time threshold at 0.157 Mya. Thus, most populations were identified as a single independent MOTU, with few exceptions. CAPES was divided into two MOTUs: the geographic close populations of TERJ-fla and GARJ were grouped together into two MOTUs, similarly to IBIMG-fla/ceu/flo. The sympatric populations of PGSP-sp and PGSP-fla were each grouped into different MOTUs: PGSP-sp was grouped together with JURSP, and PGSP-fla together with BESP.
Microsatellite data of nDNA
Genetic diversity
Twenty-five populations of P. flammea were analyzed, totalling 501 individuals, with a mean of 21 individuals per population (Table 1). Allelic richness (AR) varied from 1.220 to 5.610, mean number of alleles (A) ranged from 1.5 to 9.1, observed heterozygosity (HO) ranged from 0.035 to 0.709 and expected heterozygosity (HE) ranged from 0.042 to 0.789 (Table 2). The population-level inbreeding coefficient (FIS) was high and significant for most populations, ranging from −0.090 to 0.414 (Table 2).
nDNA population structure
Bayesian admixture analysis revealed K = 22 genetic clusters. Most of the populations were grouped in independent genetic clusters, indicating extremely low gene flow among populations. Few geographically close populations of BESP, PISP and PGSP-fla, IBIMG-fla and TERJ-fla, IBIMG-flo and IBIMG-ceu and MPPR and MCPR were grouped together in the same genetic cluster. However, the Mantel test was not significant, showing that genetic differentiation was not correlated with geographic distance (P = 0.369, P > 0.05), thus suggesting that populations were not differentiated under the isolated-by-distance model. Among the three sympatric populations, only one (TERJ-fla and TERJ-co) showed genetic admixture (Fig. 6), suggesting hybridization and introgression between these two species (P. flammea and P. corcovadensis), as previously reported by Mota et al. (2019).
FST values were generally lower for nDNA than for cpDNA data, ranging from 0.116 to 0.750 (Fig. 4b; Table S2). AMOVA detected high and significant genetic differentiation, among all populations for nuclear microsatellite (FST = 0.374, P < 0.001—Table 3). Such high genetic differentiations are in agreement with limited gene flow among populations revealed by gene flow analysis in BayesAss (Table S3). For the hierarchical AMOVA, the genetic differentiation among varieties/species of the P. flammea complex was low and non-significant (FCT = –0.002, P > 0.05) (Table 2), in agreement with the results found using cpDNA data.
Demographic analysis
No excess of heterozygosity was detected in the bottleneck analysis for any of the 25 populations investigated with microsatellite loci, suggesting no recent changes in population sizes (Table 4). M-ratio values ranged from 0.632 to 1.117 across all populations. However, M-ratio analysis showed signs of recent past genetic bottleneck events in five populations (SMARJ, IBIMG-fla, SSSP, ATISP and MCPR) (Table 4).
Discussion
Extremely high genetic structure among rock outcrops
We found strong genetic structure at population level for both cpDNA and nDNA (Figs. 3 and 4), with the almost complete fixation of haplotypes within each population and a high number of clusters obtained in Structure (K = 22). Our results show low gene flow and genetic connectivity among P. flammea populations and the absence of isolation-by-distance, reinforcing the geographic isolation of populations restricted to naturally fragmented rock outcrops. The low gene flow for both nuclear and plastidial markers caused by limited pollen and seed dispersal demonstrates that genetic drift has a large influence on genetic diversity, randomly removing alleles from the gene pool. Assuming maternal inheritance of plastidial DNA as a rule for angiosperm (Ennos 1994) and biparental inheritance of nuclear DNA, population genetic differentiation results (see AMOVA—Table 3) indicated that gene flow via seed is comparatively less efficient than via pollen, despite the fact that both nuclear and plastidial genomes show high levels of population genetic differentiation. In fact, P. flammea has small and inconspicuous seeds without any flight appendages, characteristics of autochoric seed-dispersal syndrome, which might have limited their dispersal capability to maintain extensive gene flow among populations. However, infrequent but relatively long-distance dispersal of P. flammea seeds may also be sporadically expected during colonization process. For instance, the only bromeliad colonizing Africa has the same seed morphology of P. flammea and was an event of long-distance dispersal (P. feliciana: ~12 Mya, Givnish et al. 2004), demonstrating that long-distance dispersal events may be underestimated in the widely distributed genus Pitcairnia. Thus, it is plausible to accept that colonization of disjunct outcrops may be facilitated by the occasional long-distance dispersal (over several km), as shown for other rock-outcrop plants in Australia (Acacia woodmaniorum: Millar et al. 2013). Moreover, the P. flammea species is self-compatible (Wendt et al. 2002), suggesting that its populations can tolerate variable levels of inbreeding, as observed in our data (FIS up to 0.415), which might facilitate population colonization via rare events of long-distance dispersal.
Furthermore, the reduced nuclear gene flow among P. flammea populations (see Table S3) also suggests limited pollen dispersal and reduced ability of pollinating hummingbirds (Wendt et al. 2002) in maintaining gene exchange between rock outcrops, and ultimately promoting allopatric speciation, a key hypothesis raised for explaining diversification of this radiation family (Givnish et al. 2014; Kessler et al. 2020). In fact, reduced seed and/or pollen dispersal has likely promoted allopatric speciation in several Neotropical plants adapted to naturally fragmented habitats, such as inselberg (Alcantarea spp. and Encholirium ssp.: Barbará et al. 2007; Gonçalves-Oliveira et al. 2017; Hmeljevski et al. 2017), rock outcrops (Pinheiro et al. 2014), gallery forests (P. lanuginosa: Leal et al. 2019), Andean mountains (Fosterella rusbyi: Wöhrmann et al. 2020), iron outcrops (Dyckia excelsa: Ruas et al. 2020) and campos rupestres (Vellozia compacta: Lousada et al. 2013; Pilosocereus aurisetus: Bonatelli et al. 2014; Barres et al. 2019). In addition, differences in nuclear and plastidial levels of genetic differentiation are also likely reflecting their differences in mutation rates, recombination effects, effective population sizes and coalescent times.
Phylogeography of the endemic rock outcrop P. flammea species complex
We detected significant demographic changes in population sizes through time for five out of 25 populations, suggesting that most populations of P. flammea likely had a history of long-term persistence across time (Table 4). Moreover, the variable responses of demographic changes in the populations were observed all over the species’ range, suggesting that the effects might be due to uneven ecological and local adaptation responses in each population. High-altitude rock outcrops may have acted as refugia for xerophilic and/or cold-adapted species during glacial–interglacial periods (Scarano 2002; Speziale and Ezcurra 2012; Millar et al. 2013; Tapper et al. 2014; Maswanganye et al. 2017), implying that climatically stable microhabitats could have contributed to the maintenance of these rock-outcrop populations. Indeed, the complex topography and ecological peculiarity of BAF (Martinelli 2007), where rock outcrops provide habitat heterogeneity and a range of climatic and edaphic niches, with a mosaic of soil types, range of soil depth and water-retaining depressions (Benites et al. 2007), have likely allowed for the persistence of P. flammea populations during prolonged drought periods. Even though it is important to acknowledge high genetic structure and low gene flow, they might promote a bias in the inferences of demographic analysis. We believe that future high-throughput sequencing combined with a coalescent framework will increase our ability to infer demographic changes in highly fragmented populations.
The Bayesian phylogenetic reconstructions indicate the existence of two evolutionary lineages for P. flammea, which diverged c. 2 Mya during the early Pleistocene, revealing a distinct genetic grouping in the northeastern region of Serra do Mar. Thus, our results indicate a plausible role of the early Quaternary glaciations in shaping population divergence of this species complex, despite many studies pointing to climatic oscillations connected with the late Pleistocene and Last Glacial Maximum (LGM—about 21,000 ka) as major forces on promoting population divergence of South American species (Palma-Silva et al. 2009; Carnaval et al. 2014; Batalha-Filho and Miyaki 2016; Turchetto-Zolet et al. 2016; Vergara et al. 2017). In fact, several other studies have indicated the importance of older geological and climatic events as main drivers of the lineage diversification and speciation events on the plants and animals of the Atlantic Forest (Pennington et al. 2004; Menezes et al. 2016; Peres et al. 2019; Silva et al. 2018).
Despite the strong genetic structure and low levels of gene flow observed in P. flammea populations, a weak phylogeographic structure was evident in our results for both nDNA and cpDNA datasets. The BAF is a mosaic of different dynamics and patterns of diversification, where past climatic oscillations and geographic barriers probably influenced each organism in a different way (Cabanne et al. 2007; Carnaval and Moritz 2008; Colombi et al. 2010; Thomé et al. 2010; Ribeiro et al. 2011; Martins, de 2011; Menezes et al. 2016; Aguiar-Melo et al. 2019; Paz et al. 2019). The heterogeneous nature of the southern and northern regions of the BAF is a well-observed fact for both animals and plant groups (Costa 2003; Grazziotin et al. 2006; Cabanne et al. 2007; Tchaicka et al. 2007; Carnaval and Moritz 2008; Palma-Silva et al. 2009; Pinheiro and Cozzolino 2013; Paz et al. 2019). The northern portion of BAF was less affected by glacial/interglacial cycles (Carnaval and Moritz 2008), and shows floristic similarities with the eastern Amazonian forest (Oliveira‐Filho and Fontes 2000). On the other hand, several signs of fragmentation are observed during glacial cycles in the southern portion of the BAF (Carnaval and Moritz 2008), which is floristically related to southwestern portions of Amazonian forest (Oliveira‐Filho and Fontes 2000). Unlike most of the studies, we could not detect a clear sign of phylogeographic breaks between the northern and southern populations, demonstrating a much more complex history underlying current phylogeographic patterns in this species’ extremely rich biome. In fact, a lack of phylogeographic structure was reported for few other BAF animals and plants, such as Vriesea incurvata (plant: Aguiar-Melo et al. 2019), Schiffornis virescens (bird: Cabanne et al. 2013) and Mycetophylax simplex (ants: Cardoso et al. 2015).
Hybridization, introgression and species’ delimitation on recently radiated species complex
Species delimitation, which is assigning organisms to species, is a major challenging topic of phylogenetic and population genetics research (Coyne and Orr 2004), with important implications for modern systematics, evolutionary biology and conversation. There are two main issues that complicate species delimitation. The first is how to proceed with naturally fragmented populations, and the other is how to make decisions regarding nascent species that are hybridizing and introgressing (Wiens 2007). Our study allows us to explore these topics in this taxonomically challenging and recently radiated bromeliad species complex adapted to naturally disjoint habitats.
The Bayesian phylogenetic results supported the existence of two evolutionary lineages for the P. flammea species complex (Fig. 5). One comprising four populations distributed in the North region of Serra do Mar, situated in the state of Rio de Janeiro, and the second with the remaining populations. This lineage is composed of four distinct morphological taxa: P. flammea var. flammea, P. flammea var. roeltzii, P. flammea var. macropoda and P. carinata (Table 1). In the same way, AMOVA showed high genetic differentiation among populations of each lineage (FSC = 0.982, Table 3), indicating that the population continues to diversify as an independent unit within each main lineage. Consequently, the GMYC species’ delimitation analyses with cpDNA acknowledged most populations as independent evolving entities (24 MOTUs). Accordingly, nDNA in Structure revealed K = 22 independent clusters out of 25 populations. Sorting by varieties in AMOVA (for both nDNA and cpDNA) was not significant among the groups (here considered taxonomic varieties), but was high and significant among populations within varieties, suggesting that genetic clusters do not agree with the typological/morphological descriptions (Table 3). Thus, extremely high genetic differentiation among all populations may possibly be related to long-term population isolation, and the morphological variances found between the different varieties are likely the result of convergent evolution associated with local adaptations and/or phenotypic plasticity. Hence, the pattern observed here was an absence of congruence in genetic structure and the species or taxonomic varieties described in the literature (Smith and Downs 1974), which means our data do not support the classification in seven varieties. Naturally fragmented species’ groups are known for their challenges faced during discrimination between intraspecific differentiation and speciation (i.e. Folk and Freudenstein 2015; Hedin et al. 2015; Khan et al. 2018). Further studies investigating the origin of reproductive barriers in this group, as well as the role of environmental gradients in potential local adaptation events, would contribute to clarify how many species could be recognized in this morphological puzzling group (Sukumaran and Knowles 2017).
The current phylogeographic study offers some useful insight into possible species boundaries. Populations IBIMG-flo and IBIMG-ceu, although described as being of different taxa (P. flammea var. floccosa and P. curvidens, respectively) by Monteiro and Forzza (2008), were not delimited as distinct lineages in the haplotype and species’ phylogenetic trees. Moreover, these two populations were assigned to the same genetic cluster in the Structure and BAP analyses (Fig. 3). Altogether, the evidences indicate that these previously considered two different taxa effectively belong to the same population and species, although they are clearly different from the third taxa reported in the same population (P. flammea var. flammea—see Structure Fig. 3). P. carinata (DESRJ population) was the only population with a significant statistical support in the haplotype phylogenetic tree. In agreement with this, Structure classified it as a single genetic cluster, indicating that this species is an independent lineage.
Besides the high genetic differentiation and structure among populations, one sympatric population of Teresópolis (TERJ-fla and TERJ-co) showed intermediate admixture proportions in the Bayesian admixture analysis, suggesting interspecific gene flow, between P. flammea and P. corcovadensis, as previously reported by Mota et al. (2019). Strong but permeable reproductive isolation barriers for species occurring in sympatry were also reported for other Pitcairnia species (Wendt et al. 2001; Palma-Silva et al. 2011; Mota et al. 2019), supporting the hypothesis that rock outcrops of bromeliad species are maintained as discrete evolutionary units, despite the presence of low levels of intraspecific gene flow. On the other hand, sympatric populations of Ibitipoca (IBIMG) and Mogi das Cruzes (PGSP) showed no nuclear gene flow between the sympatric taxa. In addition, none of the taxa in sympatric populations shared haplotypes, suggesting the strength of reproductive isolation barriers among these taxa. Overall, those results indicated contrasting patterns of interspecific gene flow among the three sympatric populations, suggesting a continuous speciation process in this species complex, as reported for other inselberg bromeliad species (i.e. Alcantharea spp: Lexer et al. 2016). We believe that our results will pave the way to knowledge of the consequences of limited intraspecific and sporadic interspecific gene flow as crucial for assessing the process of speciation and species delimitation among recently radiated taxa occurring in naturally fragmented habitats (Lexer et al. 2016; Pinheiro et al. 2018). In the future, the use of wide genomic datasets (i.e. RAD-seq and whole-genome sequencing) together with a multispecies coalescent approach will greatly improve species delimitation, as well as providing a framework for getting insight into the speciation process and the first step of species formation on this complex taxonomic group.
Conclusion
Our study adds important information on how naturally fragmented species are maintained as discrete evolutionary units, despite their long-term isolation, low levels of genetic connection and low intraspecific gene flow. No phylogeographical break was observed between northern and southern populations of P. flammea as commonly reported for continuously distributed taxa from BAF. Our study has shown that the high genetic structure found in this species complex is the result of limited gene flow, random genetic drift and inbreeding, which is a consequence of a long-term isolation among populations, as well as demographic stability and persistence. Although most populations of P. flammea maintained constant population sizes during most Quaternary climatic oscillations, our study indicates an impact of early Quaternary glaciations (~2 Mya) on the distribution and population divergence of this species. Moreover, our results showed a high genetic structuring among populations, regardless of taxonomic variety, reinforcing the idea that these populations have been evolving and diversifying for a long time independently of each other, and the morphological discrepancies found among different varieties are likely the result of local adaptation, and/or a consequence of phenotypic plasticity.
Data availability
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.4tmpg4f73.
Change history
27 August 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Abbott R, Albach D, Ansell S et al. (2013) Hybridization and speciation. J Evol Biol 26:229–246
Aguiar-Melo C, Zanella CM, Goetze M et al. (2019) Ecological niche modeling and a lack of phylogeographic structure in Vriesea incurvata suggest historically stable areas in the southern Atlantic Forest. Am J Bot 106:971–983
Allmon WD (1992) A causal analysis of stages in allopatric speciation. Oxf Surv Evol Biol 8:219–257
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Barbará T, Martinelli G, Fay MF, Mayo SJ, Lexer C (2007) Population differentiation and species cohesion in two closely related plants adapted to Neotropical high-altitude ‘inselbergs’, Alcantarea imperialis and Alcantarea geniculata (Bromeliaceae). Mol Ecol 16:1981–1992
Barres L, Batalha-Filho H, Schnadelbach AS, Roque N (2019) Pleistocene climatic changes drove dispersal and isolation of Richterago discoidea (Asteraceae), an endemic plant of campos rupestres in the central and eastern Brazilian sky islands. Bot J Linn Soc 189:132–152
Batalha-Filho H, Miyaki CY (2016) Late Pleistocene divergence and postglacial expansion in the Brazilian Atlantic Forest: multilocus phylogeography of Rhopias gularis (Aves: Passeriformes). J Zool Syst Evol Res 54:137–147
Benites VM, Schaefer CEGR, Simas FNB, Santos HG (2007) Soils associated with rock outcrops in the Brazilian mountain ranges Mantiqueira and Espinhaço. Rev Bras Bot 30:569–577
Boisselier-Dubayle MC, Leblois R, Samadi S, Lambourdie’re J, Sarthou C (2010) Genetic structure of the xerophilous bromeliad Pitcairnia geyskesii on inselbergs in French guiana—a test of the forest refuge hypothesis. Ecography (Cop) 33:175–184
Bonatelli IAS, Perez MF, Peterson AT et al. (2014) Interglacial microrefugia and diversification of a cactus species complex: phylogeography and palaeodistributional reconstructions for Pilosocereus aurisetus and allies. Mol Ecol 23:3044–3063
Boneh L, Kuperus P, Van Tienderen PH (2003) Microsatellites in the bromeliads Tillandsia fasciculata and Guzmania monostachya. Mol Ecol Notes 3:302–303
Byrne M, Krauss SL, Millar MA et al. (2019) Persistence and stochasticity are key determinants of genetic diversity in plants associated with banded iron formation inselbergs Biol Rev 94:753–772
Cabanne GS, Santos FR, Miyaki CY (2007) Phylogeography of Xiphorhynchus fuscus (Passeriformes, Dendrocolaptidae): Vicariance and recent demographic expansion in southern Atlantic forest. Biol J Linn Soc 91:73–84
Cabanne GS, Sari EHR, Meyer D, Santos FR, Miyaki CY (2013) Matrilineal evidence for demographic expansion, low diversity and lack of phylogeographic structure in the Atlantic forest endemic Greenish Schiffornis Schiffornis virescens (Aves: Tityridae). J Ornithol 154:371–384
Cardoso DC, Cristiano MP, Tavares MG, Schubart CD, Heinze J (2015) Phylogeography of the sand dune ant Mycetophylax simplex along the Brazilian Atlantic Forest coast: Remarkably low mtDNA diversity and shallow population structure. BMC Evol Biol 15:1–13
Carnaval AC, Moritz C (2008) Historical climate modelling predicts patterns of current biodiversity in the Brazilian Atlantic forest. J Biogeogr 35:1187–1201
Carnaval AC, Waltari E, Rodrigues MT et al. (2014) Prediction of phylogeographic endemism in an environmentally complex biome. Proc R Soc B Biol Sci 281:20141461
Colombi VH, Lopes SR, Fagundes V (2010) Testing the Rio Doce as a riverine barrier in shaping the atlantic rainforest population divergence in the rodent Akodon cursor. Genet Mol Biol 33:785–789
Corander J, Cheng L, Marttinen P, Tang J (2013) BAPS: bayesian analysis of population structure. Manual v 6:0
Costa LP (2003) The historical bridge between the Amazon and the Atlantic Forest of Brazil: a study of molecular phylogeography with small mammals. J Biogeogr 30:71–86
Couvet D (2002) Deleterious effects of restricted gene flow in fragmented populations. Conserv Biol 16:369–376
Coyne JA, Orr HA (2004) Speciation. Sinauer Associates, Sunderland
Darriba D, Taboada GL, Doallo R, Posada D (2012) JModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772
de Paula LF, Leal BS, Rexroth J, Porembski S, Palma-Silva C (2017) Transferability of microsatellite loci to Vellozia plicata (Velloziaceae), a widespread species on Brazilian inselbergs. Rev Bras Bot 40:1071–1075
Dieringer D, Schlötterer C (2003) Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ennos R (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72:250–259
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Franco FF, Jojima CL, Perez MF, Zappi DC, Taylor N, Moraes EM (2017) The xeric side of the Brazilian Atlantic Forest: the forces shaping phylogeographic structure of cacti. Ecol Evol 7:9281–9293
Frankham R, Ballou JD, Dudash MR et al. (2012) Implications of different species concepts for conserving biodiversity. Biol Conserv 153:25–31
Folk RA, Freudenstein JV (2015) ‘Sky islands’ in the eastern U.S.A.?—Strong phylogenetic structure in the Heuchera parviflora group (Saxifragaceae). Taxon 64:254–271
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Givnish TJ, Barfuss MHJ, Van EeB et al. (2014) Adaptive radiation, correlated and contingent evolution, and net species diversification in Bromeliaceae. Mol Phylogenet Evol 71:55–78
Givnish TJ, Millam KC, Evans TM et al. (2004) Ancient vicariance or recent long-distance dispersal? inferences about phylogeny and South American–African disjunctions in Rapateaceae and Bromeliaceae based on ndh F sequence data. Int J Plant Sci 165:S35–S54
Gonçalves-Oliveira RC, Wöhrmann T, Benko-Iseppon AM et al. (2017) Population genetic structure of the rock outcrop species Encholirium spectabile (Bromeliaceae): the role of pollination vs. seed dispersal and evolutionary implications. Am J Bot 104:868–878
Grazziotin FG, Monzel M, Echeverrigaray S, Bonatto SL (2006) Phylogeography of the Bothrops jararaca complex (Serpentes: Viperidae): past fragmentation and island colonization in the Brazilian Atlantic Forest. Mol Ecol 15:3969–3982
Harvey MG, Seeholzer GF, Smith BT, Rabosky DL, Cuervo AM, Brumfield RT (2017) Positive association between population genetic differentiation and speciation rates in New World birds. Proc Natl Acad Sci USA 114:6328–6333
He K, Jiang X (2014) Sky islands of southwest China. I: an overview of phylogeographic patterns. Chin Sci Bull 59:585–597
Hedin M, Carlson D, Coyle F (2015) Sky island diversification meets the multispecies coalescent—Divergence in the spruce-fir moss spider (Microhexura montivaga, Araneae, Mygalomorphae) on the highest peaks of southern Appalachia. Mol Ecol 24:3467–3484
Hirsch LD, Zanella CM, Aguiar-Melo C, Costa LM, Bered F (2020) Interspecific gene flow and an intermediate molecular profile of Dyckiajulianae (Bromeliaceae), an endemic species from southern Brazil. Bot J Linn Soc 192:675–690
Hmeljevski KV, Nazareno AG, Leandro Bueno M, dos Reis MS, Forzza RC (2017) Do plant populations on distinct inselbergs talk to each other? A case study of genetic connectivity of a bromeliad species in an Ocbil landscape. Ecol Evol 7:4704–4716
Honnay O, Jacquemyn H (2007) Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv Biol 21:823–831
Kearse M, Moir R, Wilson A et al. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kessler M, Abrahamczyk S, Krömer T (2020) The role of hummingbirds in the evolution and diversification of Bromeliaceae: unsupported claims and untested hypotheses. Bot J Linn Soc 192:592–608
Khan G, Godoy MO, Franco FF, Perez MF, Taylor NP, Zappi DC, Machado MC, Moraes EM (2018) Extreme population subdivision or cryptic speciation in the cactus Pilosocereus jauruensis? A taxonomic challenge posed by a naturally fragmented system. Syst Biodivers 16:188–199
Lavor P, van den Berg C, Jacobi CM, Carmo FF, Versieux LM (2014) Population genetics of the endemic and endangered Vriesea minarum (Bromeliaceae) in the Iron Quadrangle, Espinhaço Range, Brazil. Am J Bot 101:1167–1175
Leal BSS, Graciano VA, Chaves CJN, Huacre LAP, Heuertz M, Palma-Silva C (2019) Dispersal and local persistence shape the genetic structure of a widespread Neotropical plant species with a patchy distribution. Ann Bot 124:499–512
Leal BSS, Palma da Silva C, Pinheiro F (2016) Phylogeographic studies depict the role of space and time scales of plant speciation in a highly diverse Neotropical region. CRC Crit Rev Plant Sci 35:215–230
Leimu R, Mutikainen P, Koricheva J, Fischer M (2006) How general are positive relationships between plant population size, fitness and genetic variation? J Ecol 94:942–952
Leite YLR, Costa LP, Loss AC et al. (2016) Neotropical forest expansion during the last glacial period challenges refuge hypothesis. Proc Natl Acad Sci 113:1008–1013
Lexer C, Marthaler F, Humbert S et al. (2016) Gene flow and diversification in a species complex of Alcantarea inselberg bromeliads. Bot J Linn Soc 181:505–520
Lousada JM, Lovato MB, Borba EL (2013) High genetic divergence and low genetic variability in disjunct populations of the endemic Vellozia compacta (Velloziaceae) occurring in two edaphic environments of Brazilian campos rupestres. Rev Bras Bot 36:45–53
Martinelli G (2007) Mountain biodiversity in Brazil. Rev Brasileira de Botanica 30:587–597
Martins F, de M (2011) Historical biogeography of the Brazilian Atlantic forest and the Carnaval-Moritz model of Pleistocene refugia: what do phylogeographical studies tell us? Biol J Linn Soc 104:499–509
Maswanganye KA, Cunningham MJ, Bennett NC, Chimimba CT, Bloomer P (2017) Life on the rocks: multilocus phylogeography of rock hyrax (Procavia capensis) from southern Africa. Mol Phylogenet Evol 114:49–62
Meirelles ST, Pivello VR, Joly CA (1999) The vegetation of granite rock outcrops in Rio de Janeiro, Brazil, and the need for its protection. Environ Conserv 26:10–20
Menezes L, Canedo C, Batalha-Filho H, Garda AA, Gehara M, Napoli MF (2016) Multilocus phylogeography of the treefrog Scinax eurydice (Anura, Hylidae) reveals a plio-pleistocene diversification in the atlantic forest. PLoS ONE 11:e0154626
Millar MA, Coates DJ, Byrne M (2013) Genetic connectivity and diversity in inselberg populations of Acacia woodmaniorum, a rare endemic of the Yilgarn Craton banded iron formations. Heredity 111:437–444
Mittermeier R, Gill P, Hoffmann M, Pilgrim J, Brooks J, Mittermeier C (2005) Hotspots revisited: earth’s biologically richest and most endangered terrestrial ecoregions. Chicago University: Cemex, Mexico City
Monteiro RF, Forzza RC (2008) A Família Bromeliaceae no Parque Estadual do Initipoca, Minas Gerais, Brasil. Bol Botânica da Univ São Paulo 26.
Mota MR, Pinheiro F, Leal BSS, Wendt T, Palma-Silva C (2019) The role of hybridization and introgression in maintaining species integrity and cohesion in naturally isolated inselberg bromeliad populations. Plant Biol 21:122–132
Myers N, Mittermeler RA, Mittermeler CG, Da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Nosil P (2012) Ecological speciation. Oxford University Press, Oxford; New York
Oliveira-Filho AT, Fontes MAL (2000) Patterns of floristic differentiation among Atlantic Forests in Southeastern Brazil and the influence of climate1. Biotropica 32:793–810
Paggi GM, Palma-Silva C, Bered F et al. (2008) Isolation and characterization of microsatellite loci in Pitcairnia albiflos (Bromeliaceae), an endemic bromeliad from the Atlantic Rainforest, and cross-amplification in other species. Mol Ecol Resour 8:980–982
Palma-Silva C, Fay MF (2020) Bromeliaceae as a model group in understanding the evolution of Neotropical biota. Bot J Linn Soc 569–586
Palma-Silva C, Cavallari MM, Barbará T et al. (2007) A set of polymorphic microsatellite loci for Vriesea gigantea and Alcantarea imperialis (Bromeliaceae) and cross-amplification in other bromeliad species. Mol Ecol Notes 7:654–657
Palma-Silva C, Cozzolino S, Paggi GM, Lexer C, Wendt T (2015) Mating system variation and assortative mating of sympatric bromeliads (Pitcairnia spp.) endemic to Neotropical inselbergs. Am J Bot 102:758–764
Palma-Silva C, Leal BS, Chaves CJ, Fay MF (2016) Advances in and perspectives on evolution in Bromeliaceae. Bot J Linn Soc 181:305–322
Palma-Silva C, Lexer C, Paggi GM, Barbará T, Bered F, Bodanese-Zanettini MH (2009) Range-wide patterns of nuclear and chloroplast DNA diversity in Vriesea gigantea (Bromeliaceae), a Neotropical forest species. Heredity 103:503–512
Palma-Silva C, Wendt T, Pinheiro F et al. (2011) Sympatric bromeliad species (Pitcairnia spp.) facilitate tests of mechanisms involved in species cohesion and reproductive isolation in Neotropical inselbergs. Mol Ecol 20:3185–3201
Paz A, Spanos Z, Brown JL et al. (2019) Phylogeography of Atlantic Forest glassfrogs (Vitreorana): when geography, climate dynamics and rivers matter. Heredity 122:545–557
Pennington RT, Lavin M, Prado DE, Pendry CA, Pell SK, Butterworth CA (2004) Historical climate change and speciation: Neotropical seasonally dry forest plants show patterns of both tertiary and quaternary diversification. Trans R Soc Lond B 359:515–537
Peres EA, Benedetti AR, Hiruma ST, Sobral-Souza T, Pinto-da-Rocha R (2019) Phylogeography of Sodreaninae harvestmen (Arachnida: Opiliones: Gonyleptidae): Insights into the biogeography of the southern Brazilian Atlantic Forest. Mol Phylogenet Evol 138:1–16
Pinangé DSB, Krapp F, Zizka G et al. (2017) Molecular phylogenetics, historical biogeography andcharacter evolution in Dyckia (Bromeliaceae, Pitcairnioideae). Bot J Linn Soc 183:39–56
Pinheiro F, Cozzolino S (2013) Epidendrum (Orchidaceae) as a model system for ecological and evolutionary studies in the Neotropics. Taxon 62:77–88
Pinheiro F, Cozzolino S, Draper D et al. (2014) Rock outcrop orchids reveal the genetic connectivity and diversity of inselbergs of northeastern Brazil. BMC Evol Biol 14:49
Pinheiro F, Dantas-Queiroz MV, Palma-Silva C (2018) Plant species complexes as models to understand speciation and evolution: a review of South American studies. CRC Crit Rev Plant Sci 37:54–80
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Porembski S (2007) Tropical inselbergs: habitat types, adaptive strategies and diversity patterns. Rev Bras Botânica 30:579–586
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (2006) An exact test for population differentiation. Evolution 49:1280–1283
Ribeiro RA, Lemos-Filho JP, Ramos ACS, Lovato MB (2011) Phylogeography of the endangered rosewood (Fabaceae): Insights into the evolutionary history and conservation of the Brazilian Atlantic Forest. Heredity 106:46–57
Rivera D, Prates I, Rodrigues MT, Carnaval AC (2020) Effects of climate and geography on spatial patterns of genetic structure in tropical skinks. Mol Phylogenet Evol 143:106661
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio J. et al. (2017) DnaSP v6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol 34:3299–3302
Ruas RDB, Paggi GM, Melo CA, Hirsch LD, Bered F (2020) Strong genetic structure in Dyckia excelsa (Bromeliaceae), an endangered species found on ironstone outcrops in Pantanal, Brazil. Bot J Linn Soc 192:691–705
Saraiva DP, Mantovani A, Campostrini Forzza R (2015) Insights into the evolution of Pitcairnia (Pitcairnioideae-Bromeliaceae), Based on Morphological Evidence. Syst Bot 40:726–736
Scarano FR (2002) Structure, function and floristic relationships of plant communities in stressful habitats marginal to the Brazilian Atlantic rainforest. Ann Bot 90:517–524
Scarano FR (2009) Plant communities at the periphery of the Atlantic rain forest: rare-species bias and its risks for conservation. Biol Conserv 142:1201–1208
Schubert K (2017) Systematik und Evolution der Gattung Pitcairnia L’Heritier (Bromeliaceae). Universität Kassel: Germany
Schütz N, Krapp F, Wagner N, Weising K (2016) Phylogenetics of Pitcairnioideae s.s. (Bromeliaceae): evidence from nuclear and plastid DNA sequence data. Bot J Linn Soc 181:323–342
Silva GAR, Antonelli A, Lendel A, Moraes E, de M, Manfrin MH (2018) The impact of early quaternary climate change on the diversification and population dynamics of a South American cactus species. J Biogeogr 45:76–88
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogeneticstudies in angiosperms: the tortoise and the hare III. Am J Bot 94:275–288
Smith LB, Downs RJ (1974) Flora Neotropica, Vol. 14, No. 1, Pitcairnioideae (Bromeliaceae). Hafner Press: New York, USA
Speziale KL, Ezcurra C (2012) The role of outcrops in the diversity of Patagonian vegetation: relicts of glacial palaeofloras? Flora Morphol Distrib Funct Ecol. Plants 207:141–149
Sukumaran J, Knowles LL (2017) Multispecies coalescent delimits structure, not species. Proc Natl Acad Sci 114:1607–1612
Tapper SL, Byrne M, Yates CJ et al. (2014) Prolonged isolation and persistence of a common endemic on granite outcrops in both mesic and semi-arid environments in south-western Australia. J Biogeogr 41:2032–2044
Tchaicka L, Eizirik E, De Oliveira TG, Cândido JF, Freitas TRO (2007) Phylogeography and population history of the crab-eating fox (Cerdocyon thous). Mol Ecol 16:819–838
Thomé MTC, Zamudio KR, Giovanelli JGR, Haddad CFB, Baldissera FA, Alexandrino J (2010) Phylogeography of endemic toads and post-Pliocene persistence of the Brazilian Atlantic Forest. Mol Phylogenet Evol 55:1018–1031
Turchetto-Zolet AC, Turchetto C, Cruz F et al. (2016) Phylogeography and ecological niche modelling in Eugenia uniflora (Myrtaceae) suggest distinct vegetational responses to climate change between the southern and the northern Atlantic Forest. Bot J Linn Soc 182:670–688
Vergara J, Acosta LE, González-Ittig RE, Vaschetto LM, Gardenal CN (2017) The disjunct pattern of the Neotropical harvestman Discocyrtus dilatatus (Gonyleptidae) explained by climate-driven range shifts in the Quaternary: paleodistributional and molecular evidence. PLoS ONE 12:1–32.
Via S (2009) Natural selection in action during speciation. Proc Natl Acad Sci USA 106:9939–9946
Wendt T, Canela MB, Gelli de Faria AP, Rios RI (2001) Reproductive biology and natural hybridization between two endemic species of Pitcairnia (Bromeliaceae). Am J Bot 88:1760–1767
Wendt T, Canela MBF, Klein DE, Rios RI (2002) Selfing facilitates reproductive isolation among three sympatric species of Pitcairnia (Bromeliaceae). Plant Syst Evol 232:201–212
Wendt T, Canela MBF, Morrey-Jones JE, Henriques AB, Rios RI (2000) Recognition of Pitcairnia corcovadensis (Bromeliaceae) at the species Level. Syst Bot 25:389–398
Wiens JJ (2007) Species delimitation: new approaches for discovering diversity. Syst Biol 56:875–878
Wiens JJ, Camacho A, Goldberg A et al. (2019) Climate change, extinction, and Sky Island biogeography in a montane lizard. Mol Ecol 28:2610–2624
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wöhrmann T, Michalak I, Zizka G, Weising K (2020) Strong genetic differentiation among populations of Fosterella rusbyi (Bromeliaceae) in Bolivia. Bot J Linn Soc 192:744–759
Wöhrmann T, Wagner N, Krapp F, Huettel B, Weising K (2012) Development of microsatellite markers in Fosterella rusbyi (bromeliaceae) using 454 pyrosequencing. Am J Bot 99:e160–e163
Wöhrmann T, Weising K (2011) In silico mining for simple sequence repeat loci in a pineapple expressed sequence tag database and cross-species amplification of EST-SSR markers across Bromeliaceae. Theor Appl Genet 123:635–647
Acknowledgements
We are thankful to Sérgio Luiz Nazareth for his assistance during fieldwork, to Juliana Ribeiro Martins Santin and Carolina Carvalho for their assistance in laboratory work. We also thank Karina Lucas da Silva-Brandão, Fabio Raposo do Amaral and Jordana Neri for comments in the early version of the paper. This study was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2009/52725-3, 2013/16440-0, 2013/12966-7, 2014/15588-6, 2014/02377-7, 2015/07685-4 and 2018/07596-0), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Edital Universal (475937/2013-9). We are thankful for the fellowship provided to CP (300819/2016-1) by Conselho Nacional de Desenvolvimento Científico e Tecnológico and to MMR by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). We also thank IAPT (International Association for Plant Taxonomy) for the IAPT research grants in plant systematics. The authors thank Espaço da Escrita – Pro-Reitoria de Pesquisa – UNICAMP – for the language services provided.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Thomas Meagher
Supplementary information
Rights and permissions
About this article
Cite this article
Mota, M.R., Pinheiro, F., Leal, B.S.d.S. et al. From micro- to macroevolution: insights from a Neotropical bromeliad with high population genetic structure adapted to rock outcrops. Heredity 125, 353–370 (2020). https://doi.org/10.1038/s41437-020-0342-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-020-0342-8
This article is cited by
-
Genetic diversity and population structure of Aechmea distichantha (Bromeliaceae), a widely geographically distributed species in South America
Plant Systematics and Evolution (2023)