Abstract
Understanding the effect of human-induced landscape fragmentation on gene flow and evolutionary potential of wild populations has become a major concern. Here, we investigated the effect of riverscape fragmentation on patterns of genetic diversity in the freshwater resident European brook lamprey (Lampetra planeri) that has a low ability to pass obstacles to migration. We tested the hypotheses of (i) asymmetric gene flow following water current and (ii) an effect of gene flow with the closely related anadromous river lamprey (L. fluviatilis) ecotype on L. planeri genetic diversity. We genotyped 2472 individuals, including 225 L. fluviatilis, sampled from 81 sites upstream and downstream barriers to migration, in 29 western European rivers. Linear modelling revealed a strong positive relationship between genetic diversity and the distance from the river source, consistent with expected patterns of decreased gene flow into upstream populations. However, the presence of anthropogenic barriers had a moderate effect on spatial genetic structure. Accordingly, we found evidence for downstream-directed gene flow, supporting the hypothesis that barriers do not limit dispersal mediated by water flow. Downstream L. planeri populations in sympatry with L. fluviatilis displayed consistently higher genetic diversity. We conclude that genetic drift and slight downstream gene flow drive the genetic make-up of upstream L. planeri populations whereas gene flow between ecotypes maintains higher levels of genetic diversity in L. planeri populations sympatric with L. fluviatilis. We discuss the implications of these results for the design of conservation strategies of lamprey, and other freshwater organisms with several ecotypes, in fragmented dendritic river networks.
Similar content being viewed by others
Introduction
Human activities strongly modify natural ecosystems (Vitousek et al. 1997) and impact evolutionary trajectories of wild species (Palumbi 2001) posing unprecedented threats to the maintenance of biodiversity (Ceballos et al. 2017). In particular, habitat fragmentation is a major threat to wild species (Fahrig 2003; Vitousek et al. 1997). Habitat fragmentation can reduce gene flow among subpopulations, which can ultimately decrease effective population size and genetic diversity (Blanchet et al. 2010; Couvet 2002; DiBattista 2008; Frankham 1998; Whiteley et al. 2013). Habitat fragmentation influences the genetic structure and diversity of various species including birds (Alonso et al. 2009), fishes (Blanchet et al. 2010; Hänfling and Weetman 2006; Raeymaekers et al. 2008; Torterotot et al. 2014) and plants (Young et al. 1996). Small isolated populations are expected to fix weakly deleterious alleles by random drift (Lynch et al. 1995; Wang et al. 1999; Glémin 2003) and can suffer higher extinction risks (Saccheri et al. 1998; Carlson et al. 2014; Smith et al. 2014). Maintaining high connectivity and genetic diversity levels is thus fundamental to preserve the evolutionary potential of populations (Frankham et al. 2014; Frankham 2015; Ralls et al. 2018).
Freshwater ecosystems have been particularly affected by fragmentation worldwide (Dynesius and Nilsson 1994; Nilsson et al. 2005) due to the construction of dams, weirs, and to artificial modifications of river channels. Such fragmentation alters the possibility of gene flow between populations of aquatic organisms, so that upstream isolated populations are particularly exposed to genetic drift and its consequences, namely reduced genetic diversity and ultimately increased inbreeding. In addition, river systems are naturally shaped as dendritic networks where migration preferentially occurs following downstream directed water flow, generating patterns of asymmetric gene flow (Hänfling and Weetman 2006; Pollux et al. 2008). As a result, populations are structured following a source–sink model (Hänfling and Weetman 2006; Kawecki and Holt 2002) in which the genetic diversity will be smaller in upstream source populations than in downstream sink populations. Three possible processes may explain this pattern of downstream increase in genetic diversity observed across taxa (Paz-Vinas et al. 2015): (i) downstream biased dispersal generating downstream gene flow (Paz-Vinas et al. 2013); (ii) increase in downstream habitat availability (e.g. Raeymaekers et al. 2008); and iii) upstream founding events with loss of genetic diversity, e.g. following postglacial colonization (Cyr and Angers 2011). However, it remains unclear how human mediated alterations of habitat connectivity in rivers may obscure or exacerbate this pattern.
To date, most studies focused on delineating the effect of barriers to migration in large species targeted by fisheries. This is particularly the case for salmonid fishes that display a strong migratory behaviour and a good ability to pass obstacles (Morita and Yamamoto 2002; Yamamoto et al. 2004). In contrast, few empirical studies have focused on species with modest dispersal abilities or weak capacities to pass obstacles (e.g. Hänfling and Weetman 2006; Raeymaekers et al. 2008; Blanchet et al. 2010), which are expected to be more impacted by the effect of river fragmentation. Even less work has been focussed on the effect of small barriers to migration (e.g. 0.5–5 m) despite the fact that they are more widespread than large dams. For instance, ~58,000 large dams (>15 m) are installed in the world (Mulligan et al. 2020) compared with more than 60,000 obstacles (<5 m) in France alone (sandre.eaufrance.fr). If these small dams affect a species’ ability to disperse or migrate then their effect should be widespread across the whole species’ range.
In addition, species can display various life history strategies. They may differ in their dispersal capacity and thus be differentially affected by changes in habitat connectivity. For instance, in certain fish species some individuals are freshwater-resident whereas others are anadromous (i.e. reproduce in freshwater and juveniles migrate to sea for growth) (Dodson et al. 2013; Jonsson and Jonsson 1993). Anadromous individuals can either display a homing behaviour as they return back to their natal river to spawn, or disperse into neighbouring rivers, which can enhance gene flow. Consequently, anadromous populations generally display lower levels of population genetic structure than resident populations (Hess et al. 2013; Hohenlohe et al. 2010; Quéméré et al. 2016; Rougemont et al. 2015; 2017; Spice et al. 2012). It has also been shown that anadromous salmonid populations usually display a higher level of genetic diversity than resident populations (e.g. Perrier et al. 2013) but it is not clear whether admixture between both forms may enhance genetic diversity of resident populations when both forms coexist (Tonteri et al. 2007; McPhee et al. 2014).
The European brook lamprey Lampetra planeri is a widespread freshwater resident species with a putatively low dispersal ability at the adult stage linked to its small size (15–22 cm) and its particular life cycle (Taverny and Elie 2010). Larvae spend between 4 and 6 years mostly buried in fine sediments, creating large opportunities for long distance passive downstream dispersal at low energetic expense. In contrast, adult upstream migration is limited to return to spawning grounds. However, this spawning migration up to several kilometers might be affected by migratory barriers (Malmqvist 1980; Moser et al. 2015). L. planeri is closely related to the river lamprey L. fluviatilis that is parasitic and anadromous at the juvenile stage (Eneqvist 1937). The two taxa share many similarities. They were originally recognized as ecotypes of the same species as a consequence of partial reproductive isolation (Eneqvist 1937). They can produce viable hybrids, display a low pre-zygotic and postzygotic isolation (Rougemont et al. 2015) and are best described as partially reproductively isolated ecotypes with patterns of extensive hybridization and genetic admixture in sympatry (Rougemont et al. 2017). Throughout the manuscript we will therefore refer to them as ecotypes, rather than species. L. fluviatilis populations from nearby watersheds remain connected and display low genetic differentiation in relation to dispersal abilities through the marine environment and an apparent absence of homing (Bracken et al. 2015; Rougemont et al. 2015). In contrast, L. planeri has a highly reduced migratory behaviour: it does not move outside its watershed and generally migrates over short upstream distances within the river for breeding purposes (Malmqvist 1980). Thus, the most isolated brook lamprey populations located in the upper reaches of rivers can be strongly genetically differentiated from other populations either downstream or in other rivers (Bracken et al. 2015; Mateus et al. 2011; Pereira et al. 2010; Dawson et al. 2015; Rougemont et al. 2015). These isolated populations often display a low genetic diversity at microsatellite loci (Rougemont et al. 2015). The dispersal ability of Lampetra larvae is largely unknown but downstream dispersal may be important at this stage. In particular, flood events induce the remobilization of fine sediments where larvae are burrowed and may favour passive drift of larvae. Such downstream dispersal should further enhance the natural tendency of increasing genetic diversity in downstream river networks. Therefore, we hypothesize that in brook lamprey gene flow should clearly be asymmetric. In addition, brook lamprey populations living in sympatry with river lampreys have been found to display a higher level of genetic diversity than populations located in upstream reaches where river lampreys are absent (Rougemont et al. 2015, 2016). Maintaining genetic diversity is key to maintain a species evolutionary potential (reviewed in Frankham 2015). Therefore, gene flow between the two ecotypes may act as a ‘reservoir’ of genetic diversity, which in turn may contribute to population adaption to an ever changing world (Ceballos et al. 2020). However, to disentangle the effects of gene flow between ecotypes and downstream biased dispersal, the genetic diversity of L. planeri populations should be compared between rivers where only L. planeri is present and watersheds where both species coexist, which has not yet been tested.
The main aims of this study were to understand: (i) the role of river fragmentation on population genetic diversity and structure of L. planeri in various river systems from North western Europe; (ii) the extent of asymmetry in gene flow among L. planeri populations from the same river; and (iii) the possible role of L. fluviatilis in increasing genetic diversity in sympatric L. planeri populations via introgression. We performed extensive sampling of L. planeri upstream and downstream of small barriers to migrations in 29 rivers from three hydrogeologic regions: Brittany, Normandy and Upper Rhône, in France. Moreover, two watersheds were sampled more extensively to further investigate the combined effects of multiple barriers to migration on patterns of genetic diversity. We were particularly interested in the effect of small obstacles since these are widespread and may therefore have a cumulatively higher impact on fish dispersal. To test the prediction that L. planeri populations found in sympatry with river lampreys may display greater levels of genetic diversity than populations where river lampreys are absent, we sampled sympatric and parapatric populations of L. planeri in Normandy and populations in Brittany where river lampreys are absent.
Materials and methods
Sampling design
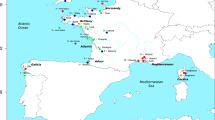
In 2013 and 2014 we sampled with electrofishing 2472 lamprey individuals distributed in 81 sites spread over 29 rivers (Fig. 1). We targeted L. planeri located upstream and downstream of a putative barrier to dispersal and, if possible, close to the barrier (<1 km upstream or downstream) to limit the effect of isolation by distance. We considered all kinds of barriers of moderate size (height between 0.50 and 5 m, described in Supplementary Table S1) that may restrict the dispersal of lampreys. The choice of obstacles was made by the description of these obstacles in the ROE data base of the Office Français de la Biodiversité (http://carmen.carmencarto.fr/66/ka_roe_current_metropole.map). None of these obstacles were equipped by fishways or spillover. Our goal was to explicitly test the effect of small migratory barriers, as these are the most widespread sites in France (>60,000; sandre.eaufrance.fr). Under experimental conditions, extremely low efficiency of river lamprey passage through fishways is frequently observed, even for small obstacles (Russon et al. 2011, Kemp et al. 2011). The restriction of passage under natural conditions may be even more pronounced depending on the presence of appropriate fishways and conditions of river flow (Foulds and Lucas 2013; Lucas et al. 2009; Tummers et al. 2018). While no data are available for brook lamprey, they display a smaller adult size. Therefore, we expected a negative relationship between barrier size and the probability of successfully passing the barrier. Although we initially planned to include the age of barriers in our model, such data were rarely available. In some cases, we were unable to capture lampreys immediately downstream or upstream of dams and some sampled points were separated by more than one obstacle. In addition, 12 pairs of sites from eight rivers without barriers to migration were included in the dataset.
Each points represents a sample site. Each site is numbered and the corresponding numbers are provided in Table S2 with details of river name and genetic diversity indices.
A total of 2247 L. planeri lampreys were collected from 73 sites in five distinct regions of France (Upper Rhône n = 575; Normandy n = 536; Atlantic coast n = 95;Brittany n = 969; and Upper Rhine n = 36), as well as two sites in United Kingdom (n = 83; from two sites above and below dam) and one in Ireland (n = 48; one single site). In Brittany, L. planeri were sampled from 32 sites. These were considered as allopatric since no observations of L. fluviatilis are currently or historically reported for these coastal rivers (Keith et al. 2011; Germis et al. 2018, Guirec et al. 2018). The same is true in the Upper Rhône and Upper Rhine area where L. fluviatilis has been extirpated (Renaud 1997). In Normandy, L. planeri were sampled from 17 sites (n = 536 individuals) and coexist in sympatry with L. fluviatilis at 8 sites (n = 225 individuals) or in parapatry for the 9 remaining sites. Here, we defined parapatry to be when populations from the same river were separated by obstacles to migration that are impassable. Sites were classified as sympatric or parapatric based on current expert knowledge (i.e. French Agency for Biodiversity, local angling association, and fisheries managers) and monitoring of these rivers throughout several years. In sympatric sites, populations of the two ecotypes were captured in the same nest, or in close vicinity without being separated by any barriers to gene flow. Among the sites in Brittany, two rivers were subjected to repeated sampling with n = 8 and 7 sites, respectively. Our goal was to dissect the joint effect of fragmentation and isolation by distance, independent of any confounding effect (e.g. presence of river lamprey) and in complement to our larger scale analysis. Adults individuals were collected in sympatric and parapatric sites since both ecotypes cannot be distinguished at early larval stage. In allopatric sites (Brittany, Upper Rhône) only brook lamprey larvae were collected during March–May of 2014. Individuals were collected by electric-fishing either on spawning sites of adults or on suitable habitat for larvae. Authorizations were obtained from the prefecture of each department in which a river was sampled.
A fin was clipped on each specimen and preserved in 95% EtOH. For adults, a small piece from the dorsal fin was taken, whereas for larvae we took a caudal fin clip. Explanatory variables of genetic parameters included the number of obstacles, their cumulative height, the geographic distances between each sample point, and the distance from the river source (i.e. distance from the headwaters). Data about obstacle height were gathered from the French “Referentiel des Obstacles à l’Ecoulement” (available at: http://carmen.carmencarto.fr/66/ka_roe_current_metropole.map). Geographic distances were computed using QGis 2.10.1.
Microsatellite genotyping
Genotyping was performed with 13 microsatellite markers specifically developed for L. planeri and L. fluviatilis after DNA extraction using a Chelex protocol modified from Estoup (1996) and strictly following protocols of Gaigher et al. (2013) and Rougemont et al. (2015).
Broad scale analysis
Genetic diversity within populations
We tested deviations from Hardy–Weinberg equilibrium using Genepop 4.1.0 (Rousset 2008) exact tests with Bonferroni corrections (Rice 1989, α = 0.05) and computed the inbreeding coefficient (Fis) for each population using Fstat 2.9.3 (Goudet 1995). Genetic diversity indices were computed and included the number of alleles (An), Allelic richness (Ar), observed heterozygosity (Hobs), and expected heterozygosity (He) using Fstat 2.9.3 (Goudet 1995) and Genetix 4.05.2 (Belkhir et al. 2004). We also measured relatedness (defined here as the probability that two allleles between two individuals are Identical By Descent) using the Loiselle coefficient (Loiselle et al. 1995). We tested for significant differences in levels of genetic diversity (Ar, He, and relatedness) as a function of “geographical connectivity” using Generalized Linear Models in R with Gaussian family. We considered five levels of connectivity: (1) downstream L. fluviatilis; (2) sympatry (i.e. the two species occur on the same spawning ground); (3) parapatry (where the two species co-occur on the same watershed but are geographically separated by impassable dams); (4) coastal allopatry (in coastal rivers where L. fluviatilis is absent); and (5) terrestrial allopatry (in the Upper Rhône, where L. fluviatilis is also absent).
Genetic differentation and structure among populations
To measure genetic differentation among sampling sites, we computed Weir & Cockerham’s estimator of FST (Weir and Cockerham 1984) between all pairs of populations and used permutations tests with Bonferroni corrections to test for significance in Fstat. We tested for global pairwise differences in FST between upstream and downstream sites and among the three major regions using permutations tested in Fstat (10,000–15,000 test permutations in each case) as well as pairwise t.test adjusted for multiple testing using FDR corrections in R (R Development Core Team 2015). However, populations are expected to deviate from migration drift equilibrium and to show a downstream increase in genetic diversity resulting in biased FST that may reflect this gradient effect rather than true differences. As a result, we also used indices of genetic differentiation that are independent from variations in genetic diversity among populations: the Jost D (Jost 2008) and Hedrick Gst (Hedrick 2005). We illustrated the distribution of pairwise FST values in R using the heatmap.2 function implemented in the gplots package (Warnes 2015).
To understand how the species were structured at a large geographic scale, a clustering analysis with the whole data was performed using the Bayesian clustering programme Structure 2.3.3 (Pritchard et al. 2000) and is provided as supplementary material. To evaluate the number of clusters (k), A total of 10 independent replicates per k value were performed. Markov Chain Monte Carlo simulations (MCMC) used 200,000 burn-in and 200,000 iterations under the admixture model with correlated allele frequencies (Falush et al. 2003). We used log likelihood Ln Pr(X | K) and the ΔK method (Evanno et al. 2005) to determine the most likely number of clusters in Structure harvester (Earl, vonHoldt 2011). Plots were drawn using Distruct 1.1 (Rosenberg 2003).
Isolation by distance
We were interested in testing if the samples follow isolation by distance (IBD) patterns as previously inferred in a smaller scale data set (Rougemont et al. 2015). To do so, we tested for IBD in each major geographic area separately. Given the complete disconnection between the Rhône drainage (in terms of waterway distance) and the rest of the sampled sites we did not compute IBD over the whole dataset. We computed Mantel tests using the linearized distance FST/(1−FST) against the waterway geographic distance between all sample sites using the R package Vegan, with the Mantel statistic being based on Spearman’s rank correlation rho (Oksanen et al. 2019).
Local scale analysis
Effect of river fragmentation on genetic diversity and differentiation
We aimed to test whether river fragmentation due to barriers to gene flow impacted genetic diversity and genetic differentiation. In particular, we predicted that in the absence of a barrier effect populations sampled upstream and downstream of a given obstacle should not differ in genetic diversity and not be differentiated. Conversely, significant differences should be indicative of a significant effect of river fragmentation. Here, we were interested in this effect on brook lamprey only. Therefore, we removed all river lamprey from the dataset as well as sites where we failed to capture individuals above or below any obstacles.
Effect on genetic diversity
To test this effect, the Allelic richness (Ar) differential was used as an estimator of difference in genetic diversity between upstream and downstream sites within each river. Independent variables included (i) the cumulative height of obstacles and (ii) geographic waterway distance to the source. We initially also included the number of obstacles as independent variable, but it was highly correlated with the cumulative height (r = 0.845) hence we only kept the cumulative height for analyses. All distances were computed manually in Qgis following water flow. Given that more than two upstream/downstream sites were sampled in a number of rivers, the river identity was fit as a random factor. Similarly, given the very different patterns observed in the different geographical areas (presence or absence of lampreys, reduced diversity in the Rhône), we fitted region as a random factor. Models were tested using AIC as implemented in lme4 (Bates et al. 2015) and car (Fox and Weisberg 2011) packages in the R software (R Development Core Team 2015). The significance of each variable was computed using type III sum-of-squares ANOVA and approximate F-tests. Pseudo-R² were then calculated using the function r.squaredGLMM implemented in the package MuMIn (Bartoń 2018).
Effect on genetic differentiation
Next, we tested the effect of barriers to gene flow on genetic differentiation. We used the linearized genetic distance FST/(1−FST) (Rousset 1997) between both sites in each river to test the effect of obstacles on genetic differentiation patterns. The exact same procedure as above with the exact same samples was performed implementing linear mixed models using distance and cumulative height as independent variables, and river as well as region as random variables.
Testing for downstream increase in genetic diversity
A common prediction across river networks or any network where dispersal is constrained (e.g. downstream biased) is that genetic diversity should increased downstream due to biased dispersal (Raeymaekers et al. 2008; Blanchet et al. 2010; Paz-Vinas et al. 2013). More importantly here, we predict that gene flow between ecotypes should further increase genetic diversity in areas of sympatry. To test this hypothesis, we used point estimates of Ar in linear mixed models and included all upstream and downstream sites from all rivers. The distance to the source was used as predictor variable (fixed effect) while the river and region were considered as random effects. The two other variables, namely number of obstacles and cumulated height to the source were all highly correlated with the distance to the source (r = 0.89 and r = 0.85) respectively and between each other (r = 0.956) and therefore not included in a single model but only tested separately. We also computed the pseudo-R2 using the r.squaredGLMM function. As above, we only included brook lamprey in our dataset since river lamprey were only sampled in downstream areas.
Testing for asymmetric gene flow
Another expectation of downstream directed dispersal is that gene flow should be asymetric and follow water currents. Thus, gene flow is expected to be higher from the upstream to downstream direction rather than the reverse. To measure the intensity and symmetry of recent migration between upstream and downstream populations, the software BayesAss 1.3 (Wilson and Rannala 2003) was used. We used a total of 10 millions iterations, discarding the first 1 million as burn-in and sampled the MCMC every 1000 intervals. Following the authors’ recommendations, we computed a rough 95% credible interval using the mean ± 1.96 std. We then considered a comparison to be informative only when the credible intervals of downstream and upstream-directed gene flow did not overlap. Then we assessed the symmetry of migration by normalizing the point estimates using (m1←2−m2←1)/max[m1←2,m2←1] so that this index varies between −1 and +1. Here m1←2 represents the fraction of individuals in population 1 (downstream) that are migrants from population 2 (upstream) each generation and m2←1 represents migration in the reverse direction. Therefore, positive values indicate higher downstream directed migration whereas negative values indicate stronger upstream migration.
Effect on local isolation by distance and population genetic structure
Finally, we measured IBD and tested the extent to which it was affected by the presence of obstacles in the Crano and Arz River (Brittany region) where more sites had been sampled (7 and 8 sites, respectively). We used Mantel and partial Mantel tests using the R package Vegan (Oksanen et al. 2019). We constructed matrices of linearized FST, computed as Fst(1−Fst), Ar and He differentials and matrices of pairwise waterway distances, number of obstacles and cumulative height for each river. Next, commonality analysis (Nimon et al. 2008) was applied in order to take into account collinearity between distance and cumulative barrier heights between sampling sites. This method enabled us to better assess the extent to which each predictor variable contributed to the variance in the response variable via a set of unique and shared effects (Prunier et al. 2015). The MBESS R package was used for this analysis (Kelly and Lai 2012).
Next, we evaluated whether population genetic structure increased along these two fragmented networks. We predicted that in the absence of an effect of barriers to gene flow, population admixture should follow a gradient of IBD and decrease as a function of distance separating sites (Meirmans 2012). To test this prediction, the Bayesian clustering programme Structure 2.3.3 (Pritchard et al. 2000) was used. The extact same settings as for the global analysis were used.
Results
Genetic diversity within populations
Fis was significantly >0 (Table S2, p < 0.01) in four populations: the downstream site on the Léguer River in Brittany (Fis = 0.259) and three downstream sites in the Rhône watershed (Oignin Fis = 0.598, Calonne Fis = 0.495, Neyrieux Fis = 0.395).
Levels of Allelic richness (Ar) based on a minimal sample size of 11 varied from 1.18 (Reyssouze River, Upper Rhône) to 3.79 (Béthune River, Normandy) and from 1.20 to 3.85 for the mean number of alleles per locus (Tables 1, S2, and S3). Levels of He averaged over all loci per population also varied substantially, ranging from 0.011 (Reyssouze River) to 0.563 (Aa River, Normandy). Populations of the Upper Rhine displayed similar levels of diversity to those of Brittany (Table 1). On average L. fluviatilis populations were significantly more diverse than L. planeri populations both in terms of allelic richness (Table 1, p < 0.0042, 15,000 permutations) and expected heterozygosity (Table 1, p < 0.0057, 15,000 permutations) (see also Fig. 2). L. fluviatilis populations in Normandy were not different from downstream L. planeri populations in Normandy in terms of expected heterozygosity or allelic richness (GLM, p > 0.05) (see Table S4 and Fig. 2). In contrast, the genetic diversity of L. fluviatilis populations was systematically higher than that of the upstream L. planeri populations of Normandy (i.e. parapatric) and of the neighbouring L. planeri populations of Brittany (all p < 0.05, see Table S4 and Fig. 2). Levels of genetic diversity of the Frome (UK) and Shannon (Ireland) populations (Table 1) were similar to those observed in Normandy. Comparisons among geographical areas revealed a significantly lower (GLM, p < 0.05) genetic diversity of L. planeri populations from the upper Rhône compared to Brittany and Normandy (Fig. 2, Table S4 for detailed p-values).
Genetic differentiation and structure among populations
Global FST was 0.377 (95% IC = 0.334–0.418) and reached 0.394 (95% IC = 0.353–0.437) when excluding L. fluviatilis (Table 1). Figure S1 illustrates two main groups of populations: the Upper Rhône vs. all other populations. L. fluviatilis populations were weakly differentiated (FST = 0.005). Populations of L. planeri were significantly more differentiated than L. fluviatilis populations (p < 0.00017, 6000 permutations with Hierfstat after pooling). The highest FST = 0.90, was observed between the Reyssouze (Rhône) and the Moulin du Rocher sites (Brittany). The lowest FST was 0 as observed in several cases (Fig. S1 and Table S5). Average pairwise FST between upstream and downstream sites within a river was 0.025 [min = 0−max = 0.095]. The maximal value of 0.095 was observed in the Crano between two sites located near the river source and in the absence of obstacles (Table S5). This difference was likely due to the fact that the uppermost site had the lowest genetic diversity as compared to the rest of the river (e.g. Arupstream = 2.13; Arall sites = 2.4). Pairwise FST between upstream and downstream sites were significant in 8 out of 43 pairwise comparisons with upstream–downstream sites from Normandy, with none of the sites where L. fluviatilis is present being significantly differentiated. Populations of the Upper Rhine, Frome and Shannon, were moderately differentiated from L. fluviatilis (Table S5). The Frome downstream site in particular displayed modest differentiation from L. fluviatilis as it was not significantly different from the Hem, Risle and Oir river (FST below 0.0125). Finally, genetic differentiation between L. planeri populations in sympatry with L. fluviatilis (i.e. in Normandy) was lower than the average FST between L. planeri population living in allopatry from L. fluviatilis (Brittany). Results from analyses using both Hedrick GST and Jost D were largely similar to those based on Weir and Cockerham FST with correlations of 0.989 and 0.973 between Fst and the two other indices, respectively. Details of population structure over the whole dataset are provided as supplementary materials (Figs. S3 and S4, Table S7). In short, we observed a large degree of admixture between L. fluviatilis and L. planeri in sympatry, whereas L. planeri from Brittany, Upper Rhône, and the Loire formed distinct clusters.
Landscape genetics
Global isolation by distance
Mantel tests, revealed contrasted patterns of isolation by distance, depending on the regions compared. First, we found a significant relationship between distance and linearized genetic differentiation in the upper Rhône area (Mantel r = 0.469, p = 2e−4). The pattern of IBD was less pronounced in Brittany, but still significant (Mantel r = 0.188, p = 0.016). In contrast, the pattern of isolation by distance was not significant in the Normandy area (Mantel r = 0.145, p = 0.143). This absence of relationship was largely driven by the lack of genetic differentiation between the upstream/downstream populations of the Oir River as compared to the remaining L. planeri Normandy populations. When this population was removed, the pattern of IBD appeared the strongest among Normandy populations (Mantel r = 0.55, p = 1e−4). To gain further insights about the evolutionary relationships among populations from coastal areas either connected to L. fluviatilis (Normandy) or disconnected (Brittany), we tested the pattern of IBD by keeping only the most downstream site per river. In this case the signal of IBD remained significant in Normandy (Mantel r = 0.43, p = 0.042) but not in Brittany (Mantel r = −0.0208, p = 0.53).
Effect of river fragmentation on genetic diversity and differentiation
To test the effect of river fragmentation on diversity and differentiation, a linear model was used, but the strong correlation between distance to the source, cumulative height or number of obstacles (all r > 0.5) precluded their joint analysis in a single model. Therefore, each variable was tested separately. The model selection procedure based on AIC indicated that the best model included only the pairwise geographic distance (Table 2) with a highly significant effect on AR differential between downstream and upstream sites. Even though the cumulative height had a significant effect (p = 0.002) this model had the highest AIC (Table 2). The amount of variance of the best model explained by fixed factors was R2m = 0.21 whereas the entire model explained a greater part of the variance (pseudo R2c = 0.59). Non-significant results were also observed when using expected heterozygosity differential instead of allelic richness (Table S8).
Regarding genetic differentiation, the linear modelling approach revealed no significant effect of the tested variables (cumulative height, distance) on brook lamprey differentiation within a given watershed (Table 2). Results obtained with the number of obstacles were similar and are provided in Table S9.
Interestingly, sites with no obstacle (n = 11 pairwise comparisons) displayed a slightly higher genetic diversity as compared to sites above and below obstacles (He = 0.22 vs. 0.20, Ar = 2.67 vs. 2.49 for sites with no obstacle versus sites with obstacles, respectively). Similarly, genetic differentiation between these pairs of sites was lower on average (Fst = 0.021) than the differentiation for sites separated by obstacles (Fst = 0.05). However, the small amount of pairs of sites with no obstacle prevented a robust comparison (e.g. using linear models) with sites fragmented by barriers to gene flow.
Effect of the distance from the source
As expected, we found a highly significant positive relationship between the distance from the source and the levels of genetic diversity (p = 1e−6, F = 29.7). Testing each dependant variable separately revealed a significant effect of the barrier count and cumulative height.
Downstream directed gene flow in brook lampreys
We expected that the particular life history of the brook lamprey, buried for several years in the sediments, should favour downstream directed gene-flow (Dawson et al. 2015). Analysis of recent migration rates in BayesAss indicated that confidence intervals do not overlap in 46% of the upstream–downstream comparisons (i.e. 18 out of 39 values were considered further for our analysis below (Fig. 3, Table S10)). In 100% of these informative cases, we found that migration was predominantly directed from the upstream to the downstream areas with the index of asymmetry reaching a median value of 0.90 (average = 0.88). Furthermore, there was no difference in the intensity of gene flow between L. planeri pairs located in sympatry areas (Normandy) and L. planeri pairs in allopatry (Brittany and upper Rhône) (Wilcoxon test W = 50, p = 0.117; GLM p > 0.1, Table S11), indicating that the highest genetic diversity observed in Normandy (Fig. 2) was not due to a higher downstream directed gene flow in this area.
Effect on local isolation by distance along two linear transects
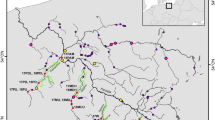
Mantel tests and partial Mantel tests on the two rivers where more than two sites had been sampled (Arz and Crano) indicated different influences of distance and obstacle-related variables. In the Arz River, all variables significantly influenced allelic richness (Table 3) whereas it was influenced solely by geographic distance in the Crano River. The extent of pairwise differentiation was also influenced by distance in the Crano River whereas this pattern was only revealed in the Arz when the influence of the number of obstacles was controlled for (Table 3). The commonality analysis (Table 4) also indicated a significant influence of the number of obstacles and geographic distance on genetic diversity (measured by heterozygosity) in the Arz with both contributions of unique and common effects, whereas only the number of obstacles influenced the pairwise differentiation. In the Crano River, commonality analysis indicated a strong influence (p < 0.001, Table 4) of the number of obstacles and geographic distance on pairwise differentiation whereas most of the variance in expected heterozygosity was explained uniquely by distance (Table 4).
Clustering analyses in the Crano (n = 7 sites) and Arz River (n = 8 sites) revealed similar patterns of admixture in these two systems. The Crano was composed of two distinct tributaries (Crano and St Sauveur River). The two most upstream sites from each of these tributaries formed distinct clusters with a lower degree of admixture than the downstream populations that displayed increased admixture values (Fig. S2). In the Arz, the source population formed a slightly distinct cluster with lower admixture than the downstream populations (Fig. S2).
Discussion
The goals of this study were threefold: testing the effect of anthropogenic river fragmentation on patterns of population genetic diversity, testing for asymmetric gene flow and exploring the potential influence of the presence of L. fluviatilis on genetic diversity levels in L. planeri. We used L. planeri as a model to test the effect of fragmentation as this species displays a reduced migratory behaviour (Malmqvist 1980). We found limited evidence for the effect of anthropogenic fragmentation on genetic diversity and differentiation of populations and the distance to the source was a more pertinent variable to explain patterns of genetic diversity within populations. Importantly, this lack of effect of river fragmentation could be explained by the strong downstream dispersal of L. planeri, which does not seem limited by obstacles of limited size considered in this study. The comparison of sympatric, parapatric and allopatric populations, located in downstream and isolated areas of different watersheds revealed a key role of L. fluviatilis in maintaining genetic diversity of L. planeri populations in the lower part of rivers where they co-occur.
Small impact of anthropogenic fragmentation on the distribution of genetic diversity
Several studies have reported strong impacts of barriers to migration on either genetic diversity and/or structure (Blanchet et al. 2010; Faulks et al. 2010; Gouskov et al. 2015; Hänfling and Weetman 2006; Leclerc et al. 2008; Raeymaekers et al. 2008; Torterotot et al. 2014). Here evidence for such effects was low and factors such as the distance to the source or the distance between sites, strong downstream directed gene flow, all contributed to erase genetic differentiation and homogenize diversity levels. Our results are therefore slightly different from those of Bracken et al. (2015) who suggested that barriers increased population differentiation. However, Bracken et al. (2015) analysed the effects of distance and barriers separately, complicating direct comparison with our results. Our results therefore suggest that small dams have only weak effects and that they might not constitue a significant obstacle under appropriate water flow conditions, as observed in the larger L. fluviatilis (Tummers et al. 2018) or that they do not restrict downstream dispersal.
Population genetic diversity was mostly affected by distance from the source, as upstream populations showed lower levels of allelic richness and heterozygosity (Fig. 2). This downstream increase in genetic diversity is expected in riverine habitat (Morrissey and de Kerckhove 2009; Paz-Vinas et al. 2015) and is frequently observed in empirical studies (e.g. Hänfling and Weetman 2006; Torterotot et al. 2014; Gouskov et al. 2015). Detailed investigations in the Arz River provided strong evidence for an increased downstream allelic richness and this pattern was also significantly influenced by all other physical variables. In the Crano, an increase in genetic diversity was not influenced by geographic variables other than distance. A recent simulation study investigated the underlying processes that can generate this pattern (Paz-Vinas et al. 2015), namely (i) downstream-biased dispersal, (ii) increase in habitat availability downstream, and (iii) upstream directed colonization. Among the three proposed processes, it appears likely that downstream dispersal plays a key role in L. planeri according to our analysis with BayesAss, which indicates higher upstream–downstream dispersal than downstream to upstream dispersal. Such dispersal is expected to occur mainly at the larval stage given the length of this phase that can reach 6 years in L. planeri (Hardisty and Potter 1971). Larvae that live mainly buried in the soft sediment may be passively transported downstream during flood events, whereas active downstream dispersal may also occur (Dawson et al. 2015). Accordingly, it has been observed in various lamprey species that older larvae are more frequent in downstream areas, compared to young larvae that are distributed closer to spawning grounds in upper reaches of river systems (Dawson et al. 2015). Admittedly, postglacial colonization history is also expected to shape the present distribution of genetic variation and its role is hard to separate from the above processes (Paz-Vinas et al. 2015). In a closely related lamprey species with similar lifestyle, Spice et al. (2019) found that both long term history and recent connectivity shape the distribution of genetic diversity. Similarly, Paz-Vinas et al. (2015) found that the observed downstream increase in genetic diversity was generally shaped by the interaction of different processes across species. Next, Bayesian clustering analysis (Fig. S2) in the St-Sauveur–Crano river system (the Crano is a small stream flowing into the St Sauveur) revealed another important pattern explaining the increase in downstream genetic diversity via admixture among individuals originating from different upstream sites. The two upstream populations of the St Sauveur and Crano form two genetically distinct clusters (FST = 0.265) and individuals located downstream of the Crano appear admixed, possibly having a shared ancestry stemming from these two source populations (and possibly from other unsampled populations). The second process that may have generated low upstream genetic diversity is the occurrence of bottlenecks through multiple serial founder effects, following usptream river colonization after glacial retreats (Hewitt 1996; Taberlet et al. 1998). It remains unclear so far whether L. planeri populations have recovered from ancestral bottlenecks and disentangling the three hypotheses will require further data. For instance, Spice et al. (2019) suggested that headwater areas tend to have higher stream gradients and less fine sediment than downstream habitat, potentially giving them a lower capacity to support lamprey larvae.
Overall, we hypothesized that our observations of strong downstream directed gene flow may explain our inability to detect the effect of river fragmentation globally. Moreover, even a small amount of upstream directed migration, as inferred here by Bayes Ass may contribute to reduce the effect of obstacles to migration. Alternatively, it is possible that subtle effects will be revealed later in time if most of the studied barriers are still relatively recent (Landguth et al. 2010). For instance, significant effects on genetic differentiation and genetic diversity were found in populations located upstream of a 90-year-old dam (Yamazaki et al. 2011) and of a 45–120 years old series of bigger dams (Coleman et al. 2018). Finer investigations in the Crano and the Arz revealed significant effects of distance and of the number of obstacles (according to the commonality analysis) on differentiation in the Crano River. In contrast, in the Arz River the effect of distance was only revealed when obstacles number was controlled for, in agreement with the commonality analysis. Finally, the impact of river fragmentation may be best revealed by studies focusing on a single catchment and with bigger obstacles to migration (e.g. Raeymaekers et al. 2008; Blanchet et al. 2010; Gouskov et al. 2015). We investigated the impact of obstacles of small to moderate size and it is possible that these obstacles do not influence the downstream passive drift of lamprey larvae, which may be sufficient to homogenize populations and obscure patterns of differentiation (Faubet et al. 2007).
River lamprey as a source of genetic diversity for resident lampreys
Understanding the evolutionary relationships between parasitic and nonparasitic lamprey ecotypes is a long-standing debate (Docker 2009). Recent studies (Bracken et al. 2015; Rougemont et al. 2015) have shown that gene flow is ongoing between the river lamprey and the brook lamprey, locally lowering their level of genetic differentiation. Here, our results also support ongoing introgression between the two ecotypes. Using an extensive SNP data set, Rougemont et al. (2017) inferred the occurrence of locally asymmetric introgression from anadromous to resident sympatric populations following secondary contacts. More specifically, we inferred that between 90 to 95% of the genome was freely introgressing between the two ecotypes, suggestive of partial reproductive isolation. Such results were due to both long term introgression and recent ongoing gene-flow as revealed by demographic analyses and structure analyses. Introgression from a large marine population toward freshwater populations is also known to occur in the stickleback Gasterosteus aculeatus (Hohenlohe et al. 2010, 2012). Here, genetic analyses of populations in sympatry (on the same nest), in parapatry (where the two species co-occur on the same watershed but are geographically separated by impassable dams) and in allopatry (in coastal rivers where L. fluviatilis is absent) revealed that allopatric L. planeri displayed a lower genetic diversity than sympatric and parapatric populations (Fig. 2). These results, with those from previous studies, further suggest that the current genetic makeup of L. planeri populations in Normandy is influenced by ongoing gene flow with L. fluviatilis. In addition, we found a much stronger pattern of IBD in the connected pairs of L. planeri (i.e. populations of downstream areas in Normandy) than in populations from Brittany. In the absence of inter-basin gene flow mediated by L. fluviatilis, populations of Brittany evolved independently from each other and do not seem globally at migration-drift equilibrium (which does not imply that sub-populations within rivers deviate from this equilibrium). In addition to this introgression, other factors can contribute to the increased genetic diversity and lower genetic differentiation in populations living in sympatry with L. fluviatilis. For instance, larger population sizes in sympatric areas are expected due to more habitat availability (Spice et al. 2019). Moreover, founder events and bottlenecks are expected following the upstream colonization of the rivers, resulting in lower Ne (and lower genetic diversity) upstream. Finally, it is possible that areas of allopatry (Britanny, Upper Rhone) were founded by more ancient colonization events so that L. planeri had more time to diverge from L. fluviatilis.
Populations from the Upper Rhône area displayed a highly reduced genetic diversity and were strongly differentiated from all other populations, which could be explained by different complementary hypotheses. First, there is evidence for at least three major evolutionary lineages existing in L. planeri (Espanhol et al. 2007). It is thus possible that colonization of the Mediterranean area (Upper Rhône region) following postglacial colonization originated from a different lineage than the one having colonized the Atlantic and Channel areas, as observed in various European fish species (Bernatchez and Wilson 1998). Similarly, some tributaries from the Iberian Peninsula have likely been colonized by different populations isolated for a long period of time (Mateus et al. 2011, 2016). In these conditions, it is possible that our microsatellite markers set (originally developed using L. planeri and L. fluviatilis samples from the Atlantic and Channel areas) is not the most appropriate to perform accurate population genetic inference of Rhône samples. Second, L. fluviatilis no longer colonizes this area and was already reported to be declining during the last century (Bernard 1909; Gensoul 1907). Consequently, it is possible that the history of divergence between Mediterranean and Atlantic populations was initiated a long time ago and that gene flow between neighbouring rivers of the Mediterranean area has been further reduced during the last century. The phylogeography of Iberian populations has been well studied (Espanhol et al. 2007; Mateus et al. 2016), but their relationships with the Upper Rhône and Northern populations still need to be explored. Decreasing costs of low coverage whole genome sequencing should provide insights into these questions.
Conservation implications
Fragmentation of rivers may impact lamprey populations, especially the most upstream populations that do not receive migrants from downstream sites. Whether the most isolated populations from headwaters suffer a mutation load and greater extinctions risks would require further investigation (Frankham 2005, 2015; Higgins and Lynch 2001; Lynch 1991; Spielman et al. 2004). On the other hand, it is not clear if maintaining a possibility for upstream migration by removing obstacles may help preventing the loss of genetic diversity in the most upstream populations of L. planeri through gene flow (Frankham 2015). This is of major importance as small isolated populations often undergo less efficient selection and can accumulate more deleterious mutations, threatening their persistence (Lynch 1991). Outbreeding depression is sometimes perceived as a greater risk (Edmands 2007), potentially also reducing local adaptation (Lenormand 2002), but it is rarely observed and meta-analyses revealed that maintaining opportunities for gene-flow between small and isolated populations is key for their maintenance (Ralls et al. 2018; Frankham 2015; Frankham 2016).
For instance, the upstream populations on the Crano, St Sauveur, and Tamoute river displayed increased genetic differentiation despite the absence of migratory barriers. These observations suggest a natural functioning where the most upstream populations are inevitably subject to a loss of genetic diversity (Hänfling and Weetman 2006; Barson et al. 2009; Dehais et al. 2010; Morrissey and de Kerckhove 2009).
Importantly, our study revealed positive impacts of the presence of L. fluviatilis on the maintenance of genetic diversity in sympatric populations. However, in Europe, L. fluviatilis abundance has strongly declined in some areas, due to habitat alteration and pollution (Maitland et al. 2015) and it is now considered as vulnerable in France on the IUCN red list (UICN Comité français MNHN SFI & AFB 2019). In addition, the low ability of the anadromous ecotype to pass migration barriers often restricts its distribution to downstream areas where L. planeri are less abundant. In terms of conservation priorities, it appears fundamental to first ensure that L. fluviatilis will have access to upstream reaches of rivers. This will benefit both the river and brook lampreys in sympatric and parapatric areas. In such areas, a joint regional management of the two ecotypes could be envisioned, whereas in allopatric areas, a management at the river scale may be more appropriate.
Conclusion
We have shown here that impacts of anthropogenic barriers to migration were modest on the extent of genetic differentiation, but we provided evidence that headwater populations of L. planeri displayed reduced genetic diversity and higher genetic differentiation when compared to downstream sites as a result of isolation by distance and biased downstream gene flow in a river network. Restoring the possibility for upstream active migration from downstream populations could increase genetic diversity and evolutionary potential in the most upstream populations (Brauer et al. 2016; Coleman et al. 2018; Pavlova et al. 2017), but it seems that such restoration practices could hardly counterbalance the strong downstream gene flow probably due to drift of larvae (Dawson et al. 2015).
In addition, our comparative analyses among sympatric, parapatric and allopatric areas support the hypothesis that sympatric populations display higher levels of genetic diversity due to introgression from L. fluviatilis (Rougemont et al. 2015, 2016, 2017). Potential strong gene flow or even introgression swamping from anadromous populations to resident populations thus plays a fundamental role in maintaining genetic diversity of L. planeri (Rougemont et al. 2017).
Data archiving
Raw genotype data used in this study will be available at the Dryad Digital Repository: https://doi.org/10.5061/dryad.3ffbg79gb.
References
Alonso H, Matias R, Granadeiro JP, Catry P (2009) Moult strategies of Cory’s Shearwaters Calonectris diomedea borealis the influence of colony location sex and individual breeding status. J Ornithol 150:329–337
Barson NJ, Cable J, Van Oosterhout C (2009) Population genetic analysis of microsatellite variation of guppies (Poecilia reticulata) in Trinidad and Tobago: evidence for a dynamic source-sink metapopulation structure, founder events and population bottlenecks. J Evol Biol 22:485–497
Bartoń K (2018) MuMIn: Multi-Model Inference. R package version 1.42.1. https://CRAN.R-project.org/package=MuMIn
Bates DM, Maechler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000 Université de Montpellier II, Montpellier, France
Bernard H (1909) Bulletin de la Société des Sciences Naturelle et d’Archeologie de l’Ain, vol 57 Imprimerie du Journal, Bourg-en-Bresse
Bernatchez L, Wilson CC (1998) Comparative phylogeography of Nearctic and Palearctic fishes. Mol Ecol 7:431–452
Blanchet S, Rey O, Etienne R, Lek S, Loot G (2010) Species-specific responses to landscape fragmentation: implications for management strategies. Evol Appl 3:291–304
Bracken FSA, Hoelzel AR, Hume JB, Lucas MC (2015) Contrasting population genetic structure among freshwater-resident and anadromous lampreys: the role of demographic history, differential dispersal, and anthropogenic barriers to movement. Mol Ecol 24:1188–1204
Brauer CJ, Hammer MP, Beheregaray LB (2016) Riverscape genomics of a threatened fish across a hydroclimatically heterogeneous river basin. Mol Ecol 25:5093–5113
Carlson SM, Cunningham CJ, Westley PAH (2014) Evolutionary rescue in a changing world. Trends Ecol Evol 29(9):521–530
Ceballos G, Ehrlich PR, Dirzo R (2017) Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc Natl Acad Sci USA 30:6089–6096
Ceballos G, Ehrlich PR, Raven PH (2020) Vertebrates on the brink as indicators of biological annihilation and the sixth mass extinction Proc Natl Acad Sci USA 117:13596–13602
Coleman RA, Gauffre B, Pavlova A, Beheregaray LB, Kearns, Lyon J et al. (2018) Artificial barriers prevent genetic recovery of small isolated populations of a low-mobility freshwater fish. Heredity 120:515–532
Couvet D (2002) Deleterious effects of restricted gene flow in fragmented populations. Conserv Biol 16:369–376
Cyr F, Angers B (2011) Historical process lead to false genetic signal of current connectivity among populations. Genetica 139:1417–1428
Dawson HA, Quintella BR, Almeida PR, Treble AJ, Jolley JC (2015) The ecology of larval and metamorphosing lampreys. In: Docker MF (ed.), Lampreys: biology, conservation and control. Springer, Netherlands, pp. 75–137
Dehais C, Eudeline R, Berrebi P, Argillier C (2010) Microgeographic genetic isolation in chub (Cyprinidae: Squalius cephalus) population of the Durance River: estimating fragmentation by dams. Ecol Freshw Fish 19:267–278
DiBattista JD (2008) Patterns of genetic variation in anthropogenically impacted populations. Conserv Genet 9:141–156
Docker MF (2009) A review of the evolution of non-parasitism in lampreys and an update of the paired species concept. In: Brown LR, Chase SD, Mesa MG, Beamish RJ, Moyle PB (eds) Biology, management and conservation of lampreys in North America. Am Fish Soc Symp 72, Bethesda, MD, pp. 71–114
Dodson JJ, Aubin-Horth N, Thériault V, Páez DJ (2013) The evolutionary ecology of alternative migratory tactics in salmonid fishes. Biol Rev Camb Philos Soc 88:602–625
Dynesius M, Nilsson C (1994) Fragmentation and flow regulation of river systems in the northern third of the world. Science 266:753–762
Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Edmands S (2007) Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol Ecol 16:463–475
Eneqvist P (1937) The brook lamprey as an ecological modification of the river lamprey. On the river and brook lampreys of Sweden. Ark Zool 29:1–22
Espanhol R, Almeida PR, Alves MJ (2007) Evolutionary history of lamprey paired species Lampetra fluviatilis (L.) and Lampetra planeri (Bloch) as inferred from mitochondrial DNA variation. Mol Ecol 16:1909–1924
Estoup A (1996) Rapid one-tube DNA extraction for reliable PCR detection of fish polymorphic markers and transgenes. Mol Mar Biol Biotechnol 5:295–298
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol, Evolution, Syst 34:487–515
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Faubet P, Waples RS, Gaggiotti OE (2007) Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol 16:1149–1166
Faulks LK, Gilligan DM, Beheregaray LB (2010) Islands of water in a sea of dry land: hydrological regime predicts genetic diversity and dispersal in a widespread fish from Australia’s arid zone, the golden perch (Macquaria ambigua). Mol Ecol 19:4723–4737
Fox J, Weisberg S (2011) An {R} companion to applied regression, 2nd edn. Sage, Thousand Oaks, CA
Foulds WL, Lucas MC (2013) Extreme inefficiency of two conventional, technical fishways used by European river lamprey (Lampetra fluviatilis). Ecol Eng 58:423–433
Frankham R (1998) Inbreeding and extinction: island populations. Conserv Biol 12:665–675
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618
Frankham R (2016) Genetic rescue benefits persist to at least the F3 generation, based on a meta-analysis. Biol Conserv 195:33–36
Frankham R, Bradshaw CJA, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Gaigher A, Launey S, Lasne E, Besnard AL, Evanno G (2013) Characterization of thirteen microsatellite markers in river and brook lampreys (Lampetra fluviatilis and L. planeri). Conserv Genet Resour 5:141–143
Gensoul J (1907) Monographie des poisons d’eau douce de Saone et Loire. Extrait du Bulletin de la société d’histoire naturelle d’Autun. 20è Bulletin
Germis G, Arago MA, Ampen N, Moulin C, Deleys, N (2018) Plan de Gestion des poissons migrateurs 2018–2023. available at http://www.bretagne.developpement-durable.gouv.fr/IMG/pdf/plagepomi_2018.pdf
Glémin S (2003) How are deleterious mutations purged? Drift versus nonrandom mating. Evolution 57:2678–2687
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Gouskov A, Reyes M, Wirthner‐Bitterlin L, Vorburger C (2015) Fish population genetic structure shaped by hydroelectric power plants in the upper Rhine catchment. Evol Appl 9:394–408
Guirec A Guillerme N, Sauvader C, Diouach O, Chapon PM, Beaulaton L (2018) Synthèse sur la répartition des lamproies et des aloses amphihalines enFrance. Rapport final. 2018.032 available at https://www.documentation.eauetbiodiversite.fr/notice/000000000168991aeaf427a64ba5487a
Hänfling B, Weetman D (2006) Concordant genetic estimators of migration reveal anthropogenically enhanced source–sink population structure in the River Sculpin, Cottus gobio. Genetics 173:1487–1501
Hardisty MW, Potter IC (1971) The biology of lampreys. Academic Press, London‐New York
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Hess JE, Campbell NR, Close DA, Docker MF, Narum SR (2013) Population genomics of Pacific lamprey: adaptive variation in a highly dispersive species. Mol Ecol 22:2898–2916
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Higgins K, Lynch M (2001) Metapopulation extinction caused by mutation accumulation. Proc Natl Acad Sci USA 98:2928–2933
Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA (2010) Population genomics of parallel adaptation in threespine stickleback using sequenced RAD Tags. PLoS Genet 6:e1000862
Hohenlohe PA, Catchen J, Cresko WA (2012) Population genomic analysis of model and nonmodel organisms using sequenced RAD tags. Methods Mol Biol 888:235–260
Jonsson B, Jonsson N (1993) Partial migration: niche shift versus sexual maturation in fishes. Rev Fish Biol Fish 3:348–365
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kawecki TJ, Holt RD (2002) Evolutionary consequences of asymmetric dispersal rates. Am Nat 160:333–347
Keith P, Persat H, Feuteun É. AllardiI J (2011) Les Poissons d’eau douce de France. Muséum national d’Histoire naturelle, Paris; Biotope, Mèze, 552 p. (Inventaires & biodiversité)
Kelly K, Lai K (2012) Package ‘MBESS.’ R Package version 3.3.3. http://www3.nd.edu/~kkelley/site/MBESS.html
Kemp PS, Russon IJ, Vowles AS, Lucas MC (2011) The influence of discharge and temperature on the ability of upstream migrant adult river lamprey (Lampetra fluviatilis) to pass experimental overshot and undershot weirs. River Res Appl 27:488–498
Landguth EL, Cushman SA, Schwarte MK, McKelvey KS, Murphy M, Luikart G (2010) Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol 19:4179–4191
Leclerc E, Mailhot Y, Mingelbier M, Bernatchez L (2008) The landscape genetics of yellow perch (Perca flavescens) in a large fluvial ecosystem. Mol Ecol 17:1702–1717
Lenormand T (2002) Gene flow and the limits to natural selection. Trends Ecol Evol 17:183–189
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Lucas MC, Bubb DH, Jang MH, Ha K, Masters JEG (2009) Availability of and access to critical habitats in regulated rivers: effects of low-head barriers on threatened lampreys. Freshw Biol 54:621–634
Lynch M (1991) The genetic interpretation of inbreeding depression and outbreeding depression. Evolution 45:622–629
Lynch M, Conery J, Burger R (1995) Mutation accumulation and the extinction of small populations. Am Nat 146:489–518
Maitland P, Renaud C, Quintella BA, Close D, Docker M (2015) Conservation of native lampreys. In: Docker M (ed.) Lampreys: Biology, Conservation and Control. Fish & Fisheries Series, vol 37. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-9306-3_8
Malmqvist B (1980) The spawning migration of the brook lamprey, Lampetra planeri Bloch, in a South Swedish stream. J Fish Biol 16:105–114
Mateus CS, Almeida PR, Quintella BR, Alves MJ (2011) MtDNA markers reveal the existence of allopatric evolutionary lineages in the threatened lampreys Lampetra fluviatilis (L.) and Lampetra planeri (Bloch) in the Iberian glacial refugium. Conserv Genet 12:1061–1074
Mateus CS, Almeida PR, Mesquita N, Quintella BR, Alves MJ (2016) European Lampreys: New Insights on Postglacial Colonization, Gene Flow and Speciation. PLoS ONE 11:e0148107
McPhee MV, Whited DC, Kuzishchin KV, Stanford JA (2014) The effects of riverine physical complexity on anadromy and genetic diversity in steelheador rainbow trout Oncorhynchus mykiss around the Pacific Rim. J Fish Biol 85:132–150
Meirmans PG (2012) The trouble with isolation by distance. Mol Ecol 21:2839–2846
Morita K, Yamamoto S (2002) Effects of habitat fragmentation by damming on the persistence of stream‐dwelling charr populations. Conserv Biol 16:1318–1323
Morrissey MB, de Kerckhove DT (2009) The maintenance of genetic variation due to asymmetric gene flow in dendritic metapopulations. Am Nat. 174:875–889
Moser ML, Almeida PR, Kemp PS, Sorensen PW (2015) Lamprey spawning migration. In: Docker MF (ed) Lampreys: biology, conservation and control. Springer, Netherlands, pp. 215–252
Mulligan M, van Soesbergen A, Sáenz L (2020) GOODD, a global dataset of more than 38,000 georeferenced dams. Sci Data 7:31
Nilsson C, Reidy CA, Dynesius M, Revenga C (2005) Fragmentation and flow regulation of the world’s large river systems. Science (New York, NY) 308:405–408
Nimon K, Lewis M, Kane R, Haynes RM (2008) An R package to compute commonality coefficients in the multiple regression case: an introduction to the package and a practical example. Behav Res Methods 40:457–466
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al. (2019) vegan: community ecology package. R package version 2.5-6
Palumbi SR (2001) Humans as the world’s greatest evolutionary force. Science 293:1786–1790
Pavlova A, Beheregaray LB, Coleman R, Gilligan D, Harrisson KA, Ingram BA et al. (2017) Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: a call for assisted gene flow. Evol Appl 10:531–550
Paz-Vinas I, Loot G, Stevens VM, Blanchet S (2015) Evolutionary processes driving spatial patterns of intraspecific genetic diversity in river ecosystems. Mol Ecol 24:4586–4604
Paz-Vinas I, Quéméré E, Chikhi L, Loot G, Blanchet S (2013) The demographic history of populations experiencing asymmetric gene flow: combining simulated and empirical data. Mol Ecol 22:3279–3291
Pereira AM, Robalo JI, Freyhof J, Maia C, Fonseca JP, Valente A et al. (2010) Phylogeographical analysis reveals multiple conservation units in brook lampreys Lampetra planeri of Portuguese streams. J Fish Biol 77:361–371
Perrier C, Bourret V, Kent MP, Bernatchez L (2013) Parallel and nonparallel genome-wide divergence among replicate population pairs of freshwater and anadromous Atlantic salmon. Mol Ecol 22:5577–5593
Pollux BJA, Luteijn A, Van Groenendael JM, Ouborg NJ (2008) Gene flow and genetic structure of the aquatic macrophyte Sparganium emersum in a linear unidirectional river. Freshw Biol 54:64–76
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Prunier JG, Colyn M, Legendre X, Nimon KF, Flamand MC (2015) Multicollinearity in spatial genetics: separating the wheat from the chaff using commonality analyses. Mol Ecol 24:263–283
Quéméré E, Baglinière JL, Roussel JM, Evanno G, McGinnity P, Launey S (2016) Seascape and its effect on migratory life‐history strategy influences gene flow among coastal brown trout (Salmo trutta) populations in the English Channel. J Biogeogr 43:498–509
R Development Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Raeymaekers JAM, Maes GE, Geldof S, Hontis I, Nackaerts K, Volckaert FAM (2008) Modeling genetic connectivity in sticklebacks as a guideline for river restoration. Evol Appl 1:475–488
Ralls K, Ballou JD, Dudash MR, Eldridge MDB, Fenster CB, Lacy RC et al. (2018) Call for a paradigm shift in the genetic management of fragmented populations. Conserv Lett 11:e12412
Renaud CB (1997) Conservation status of Northern Hemisphere lampreys (Petromyzontidae). J Appl Ichthyol 13:143–148
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223
Rosenberg NA (2003) DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes
Rougemont Q, Gagnaire PA, Perrier C, Genthon C, Besnard AL, Launey S, Evanno G (2017) Inferring the demographic history underlying parallel genomic divergence among pairs of parasitic and nonparasitic lamprey ecotypes. Mol Ecol 26:142–162
Rougemont Q, Gaigher A, Lasne E, Côte J, Coke M, Besnard AL et al. (2015) Low reproductive isolation and highly variable levels of gene flow reveal limited progress towards speciation between European river and brook lampreys. J Evol Biol 28:2248–2263
Rougemont Q, Roux C, Neuenschwander S, Goudet J, Launey S, Evanno G (2016) Reconstructing the demographic history of divergence between European river and brook lampreys using approximate Bayesian computations. PeerJ 4e1910
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (2008) genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Russon IJ, Kemp PS, Lucas MC (2011) Gauging weirs impede the upstream migration of adult river lamprey Lampetra fluviatilis: lamprey ability to pass gauging weirs. Fish Manag Ecol 18:201–210
Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I (1998) Inbreeding and extinction in a butterfly metapopulation. Nature 392:491–494
Smith TB, Kinnison MT, Strauss SY, Fuller TL, Carroll SP (2014) Prescriptive evolution to conserve and manage biodiversity. Ann Rev Ecol Evol Syst 45:1–22
Spice EK, Goodman DH, Reid SB, Docker MF (2012) Neither philopatric nor panmictic: microsatellite and mtDNA evidence suggests lack of natal homing but limits to dispersal in Pacific lamprey. Mol Ecol 21:2916–2930
Spice EK, Whitesel TA, Silver GS, Docker MF (2019) Contemporary and historical river connectivity influence population structure in western brooklamprey in the Columbia River Basin. Conserv Genet
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci USA 101:15261–15264
Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Taverny C, Elie P (2010) Les lamproies en Europe de l‟Ouest – Ecophases, espèces et habitats. Quae, Versailles, France
Tonteri A, Veselov AJ, Titov S, Lumme J, Primmer CR (2007) The effect of migratory behaviour on genetic diversity and population divergence: a comparison of anadromous and freshwater Atlantic salmon Salmo salar. J Fish Biol 70:381–398
Torterotot JB, Perrier C, Bergeron NE, Bernatchez L (2014) Influence of forest road culverts and waterfalls on the fine‐scale distribution of brook trout genetic diversity in a boreal watershed. Trans Am Fish Soc 143:1577–1591
Tummers JS, Kerr JR, O’Brien P, Kemp P, Lucas MC (2018) Enhancing the upstream passage of river lamprey at a microhydropower installation using horizontally-mounted studded tiles. Ecol Eng 125:87–97
UICN Comité français MNHN SFI & AFB (2019) La Liste rouge des espèces menacées en France—Chapitre Poissons d’eau douce de France métropolitaine. Paris, France
Vitousek PM, Mooney HA, Lubchenco J, Melillo JM (1997) Human domination of earth’s ecosystems. Science 277:494–499
Wang J, Hill WG, Charlesworth D, Charlesworth B (1999) Dynamics of inbreeding depression due to deleterious mutations in small populations: Mutation parameters and inbreeding rate. Genet Res 74(2):165–178. https://doi.org/10.1017/S0016672399003900
Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B (2015) gplots: Various R Programming Tools for Plotting Data. R package version 3.0.4. https://CRAN.R-project.org/package=gplots
Weir B, Cockerham C (1984) Estimating F-statistics for the analysis of population-structure. Evolution 38:1358–1370
Whiteley AR, Coombs JA, Hudy M, Robinson Z, Colton AR, Nislow KH, Letcher BH (2013) Fragmentation and patch size shape genetic structure of brook trout populations. Can J Fish Aquat Sci 70:678–688
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Yamamoto S, Morita K, Koizumi I, Maekawa K (2004) Genetic differentiation of white-spotted charr (Salvelinus leucomaenis) populations after habitat fragmentation: spatial–temporal changes in gene frequencies. Conserv Genet 5:529–538
Yamazaki Y, Yamano A, Oura K (2011) Recent microscale disturbance of gene flow in threatened fluvial lamprey, Lethenteron sp. N, living in a paddywater system. Conserv Genet 12:1373
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Acknowledgements
We thank B. Jacquot, B. Herodet, B. Bulle, B. Dufour, Y. Salaville, H. Catroux, G. Sanson V. Zunigas, R. Pélerin, V. Mouren, M. Lesimple, B. Rigault, G. Garot, P. Domalain, J.-L. Fagard, J.M. le Boucher, N. Jeanneau, A. Robbe, J. Tremblay (Experimental Unit of Aquatic Ecology and Ecotoxicology, U3E), Y. Perraud, P. McGinnity and B. Beaumont who helped us collecting samples. This study was funded by the Office Français de la Biodiversité.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Barbara Mable
Rights and permissions
About this article
Cite this article
Rougemont, Q., Dolo, V., Oger, A. et al. Riverscape genetics in brook lamprey: genetic diversity is less influenced by river fragmentation than by gene flow with the anadromous ecotype. Heredity 126, 235–250 (2021). https://doi.org/10.1038/s41437-020-00367-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-020-00367-9
This article is cited by
-
Population genetics of the endangered Clanwilliam sandfish Labeo seeberi: considerations for conservation management
Aquatic Sciences (2024)
-
A revised taxonomy and estimate of species diversity for western North American Lampetra
Environmental Biology of Fishes (2023)
-
A SNP marker to discriminate the european brook lamprey (Lampetra planeri), river lamprey (L. fluviatilis) and their hybrids
Molecular Biology Reports (2022)