Abstract
The human genome is not randomly organised, with respect to both the linear organisation of the DNA sequence along chromosomes and to the spatial organisation of chromosomes in the cell nucleus. Here I discuss how these patterns of sequence organisation were first discovered by molecular biologists and how they relate to the patterns revealed decades earlier by cytogeneticists and manifest as chromosome bands.
Similar content being viewed by others
Where are the genes?
As a postdoc in the late 1980s, I was trying to track down the gene responsible for a human genetic disease. It is hard to imagine now but, at that time, there was no human genome sequence, there was not even a map of the human genome. You had to make your own maps using the laborious methods of chromosome walking and jumping from genomic libraries cloned into cosmids and yeast artificial chromosomes, and constructing long-range restriction maps made using rare-cutter enzymes and pulsed field gel electrophoresis. These are methods now largely consigned to the dustbin of history—although pulsed field gels are still useful for sizing chromosomes that are up to a few Mb in size (e.g., from yeasts).
I was rather proud of my map—a total of about 7 Mb in size. So where were the genes located? The assumption was that the disease I was interested in was caused by a mutation in a protein-coding gene—almost no attention was paid to the non-coding genome in those days. One of the few ways to identify genes in long stretches of human DNA was to look for the unusual sequence content found at the 5ʹ end of a large proportion (70%) of human genes—regions where there is no depletion of CpG relative to GpC, and where CpGs are unmethylated. These are CpG islands or CGIs (Deaton and Bird 2011). I had noticed a strange pattern to my map—all the CGIs were squashed down one end of it and I had no idea why at the time. It would take a serendipitous encounter with cytogenetics to eventually figure that out.
I might have remained a disease gene cloner, and never explored the fascinating world of three-dimensional (3D) genome organisation, if it were not for institute building renovations. I never did manage to clone that gene I was after, but during the laboratory refurbishments I was relocated temporarily to the cytogenetics department of the institute I worked in—at that time called the MRC Clinical and Population Cytogenetics Unit (CAPCU; https://www.ed.ac.uk/mrc-human-genetics-unit/about/mrc-hgu-history). For a young gun-slinger of molecular biology who could clone, map and resolve DNA fragments of the human genome 100s of kilobases in length, looking down the light microscope at chromosomes seemed a deeply uncool and primitive activity. However, once I got the chance to see banded human metaphase chromosomes with my own eyes I was hooked by their beautiful and recognisable structures—each individual chromosome could be distinguished and different regions recognised from the chromosome banding pattern. Somewhere in my brain a connection was made between the pattern I had in my small genome map, and the transverse patterns of chromosome bands manifest in plain sight along the length of the human genome by chromosome banding techniques (Craig and Bickmore 1993).
At that time in the late 1980s, journal clubs were a very important activity in the institute. If it was your turn to do journal club you retired to the library for a week with a bunch of acetates, photocopies and bound volumes of old journals and researched the background to your chosen paper carefully. John Evans, the Director of CAPCU—who himself had contributed to the development of early chromosome banding techniques, suggested to me that I might be interested in presenting a paper by Julie Korenberg and Mary Rykowski (Korenberg and Rykowski 1988) that used the emerging technique of fluorescence in situ hybridisation (FISH). They had cleverly combined FISH with new developments in charge-coupled device (CCD) camera technology to investigate the genomic distribution of different classes of sequence across the human genome using metaphase chromosomes spread on glass slides as a visual readout of relative position in the genome. This suggestion from John Evans and reading the work of Korenberg and Rykowski proved to be a pivotal moment that opened up new research questions that I wanted to address and so has influenced my entire independent research career.
The Korenberg and Rykowski experiment
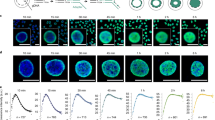
Approximately one-third of the human genome is composed of interspersed repeated sequences that fall into two main families—short- and long-interspersed repeat elements (abbreviated as SINEs and LINEs, respectively). The most abundant SINEs in the human genome are Alu elements—dimers of 7SL RNA-derived sequences (Kojima 2018). The retrotransposon L1 is the predominant LINE. By hybridising biotin-labelled probes detecting the consensus Alu and L1 repeats to human metaphase chromosomes stained with 4,6-diamidino-2-phenyl indole (DAPI), and detecting the hybridisation signals with streptavidin conjugated to Texas Red, Korenberg and Rykowski were able to see that Alus and L1s were non-uniformly distributed along the chromosomes, and hence along the genome sequence. Moreover, their hybridisation patterns corresponded with classical chromosome bands—Alus were concentrated in the so-called Reverse or R-bands, and L1s in the alternating Giemsa or G-bands (Korenberg and Rykowski 1988). Good examples include the high concentration of Alus at the distal tip of the short arm of chromosome 1 (1p34–p36) and the middle of the short arm of chromosome 6 (6p21) and the depletion of L1s from these same regions (Fig. 1). These regions are called T-bands, which are the most intensely stained and most GC-rich fraction of R-bands (Craig and Bickmore 1993).
Fluorescence in situ hybridisation (FISH) reveals the distribution of Alu and L1 repeats on human chromosomes 1 and 6. a Left: Alu hybridisation signal (white) for human chromosome 1 (top) and 6 (bottom). Right: Ideogram of chromosomes with T-bands (the most extreme R-bands) in red, R-bands in white and G-bands in black. b Left: 4,6-Diamidino-2-phenyl indole (DAPI)-stained chromosomes (R-bands are pale). Right: L1 hybridisation signal (white). Adapted with permission from Korenberg and Rykowski (1988)
The Korenberg and Rykowski paper revealed that there is a non-random distribution of DNA sequence (in this case interspersed repeats) along the human genome and that this relates to the visible compartmentalisation of chromosomes in the form of metaphase chromosome bands. It also exemplified the compelling nature of the visual image, and the way in which banded metaphase chromosomes could be used as a visually ordered representation of the human genome sequence. It also demonstrated that, through the use of CCD cameras, which provide a linear measure of signal intensity over a large dynamic range, imaging with fluorescent light could be quantitative.
Patterns in the distribution of genes along the human genome
Inspired by the Korenberg and Rykowski experiment, and given that the genome map I had been making had shown an apparently uneven distribution of CGIs, myself and my first PhD student—Jeff Craig—decided to see if we could take a similar approach to ask how genes are distributed across the human genome. As a probe to detect human genes, we used the small restriction fragments liberated from the human genome by the CpG methylation sensitive enzyme HpaII (CCGG). These HpaII tiny fragments (HTFs) originate mainly from CGIs (Bickmore and Bird 1992) at the promoter of approximately 70% of human genes. Hybridising this fraction of the genome to metaphase chromosomes together with a probe (late replicating DNA) for the inactive gene-poor portion of the genome dramatically revealed the concentration of human genes—or at least those associated with CGIs—in specific chromosomes bands, particularly in T-bands (Craig and Bickmore 1994) (Fig. 2). As with the Alu hybridisation pattern (Fig. 1a), a high density of CGIs is seen on the distal tip of the short arm of chromosome 1 and the middle of the short arm of chromosome 6. Other striking features of the hybridisation patterns that we saw is the high density of CGIs on human chromosome 19 and the paucity of them on human chromosome 18. This pronounced difference in the sequence composition of chromosomes 18 and 19 eventually led us to investigate the organisation of these two chromosomes in the nucleus. Using FISH with probes for chromosomes 18 and 19, or probes for different genomic regions, we discovered that mammalian chromosomes are non-randomly organised in the nucleus and have a radial organisation with the gene-poor chromatin localised toward the nuclear periphery (Croft et al. 1999; Boyle et al. 2011).
Fluorescence in situ hybridisation (FISH) reveals the distribution of CpG islands across the human genome. For each metaphase chromosome, the hybridisation signal from CpG islands (red) is shown on the left of each pair. 4,6-Diamidino-2-phenyl indole (DAPI)-stained chromosomes are on the left. Late replicating G-bands are shown in green. Modified from Craig and Bickmore (1994)
By digesting the human genome with restriction enzymes that cleave, on average, once per CGI we also used pulsed field gel electrophoresis to isolated fractions of the human genome with progressively decreasing CGI density, i.e., fractions where inter-island distances were 15–100, 100–500, 500–1000, 1000–6000 kb. The hybridisation pattern to metaphase chromosomes from the fraction with the shortest inter-CGI distances (15–100 kb), and therefore the highest CGI density, was very similar to that of the HTFs—i.e., concentrated in T-bands. Fractions with inter-island CGI distances of 100–500 kb—a slightly lower CGI density, highlighted the remaining (non-T) R-bands. G-bands were lit up by fractions of the human genome with extremely low CGI density ( > 1 Mb between CGIs). We found a similar organisation on rodent chromosomes (Cross et al. 1997). We therefore concluded that mammalian genomes have a non-random organisation, with genes concentrated together into specific regions of chromosomes that are manifest as T- and R-bands, and that—as Kornberg and Rykowsi had shown—are also enriched in Alu repeats. This organisation was later borne out when the sequencing of the human genome was finally completed (Lander et al. 2001).
The function of this level of linear genome organisation remains unclear. Although there may be a functional advantage to keep broadly and highly expressed genes clustered together in chromosomal domains (R-bands) (Sproul et al. 2005), gene ‘deserts’ may be the location of complex regulatory landscapes for genes with intricate roles and patterns of expression during development, and populated by multiple enhancers—non-coding regulatory elements (Salzburger et al. 2009).
Patterns in interphase chromosome structure and function
The striking hybridisation patterns of repeated sequences and genes along chromosomes demonstrated that human mitotic chromosomes spread on a glass slide could be an ordered visual readout of primary genome sequence organisation. Mitotic chromosome spreads can also report on functional aspects of genome organisation that occur during the preceding interphase. The most striking example of this is replication banding, revealed by the incorporation of thymidine analogues—most typically 5-bromo-2’-deoxyuridine (BrdU)—into the genome during defined periods of S-phase (Dutrillaux et al. 1976; Vogel et al. 1989). The period of S-phase is normally controlled using drugs that interfere with S-phase progression. Detection of the thymidine analogue on the metaphase chromosomes formed after S-phase showed that the human genome sequence is organised into domains (bands) that replicate at defined periods of S-phase. Domains that correspond to genomic regions with analogue incorporation during the early period of S-phase (early replicating) were shown to correspond to R-bands and later replicating bands correspond to G-bands (Fig. 2). Moreover, T-bands replicate on average earlier than ordinary R-bands (Drouin et al. 1994).
Immunofluorescence with antibodies detecting histone post-translational modifications has also been used to explore the distribution of different epigenetic states along metaphase chromosomes revealing, perhaps unexpectedly given their high gene density, that R-bands are more enriched in acetylated histones than G-bands (Jeppesen and Turner 1993) and that other specific modifications of histone H3 found at the promoters of active genes—acetylation of histone H3 lysine 9 (H3K9ac) or 27 (H3K27ac) or trimethylation of lysine 4 (H3K4me3) in the interphase genome are also quantitatively enriched at R-bands during mitosis (Terrenoire et al. 2010). Thus, despite the general cessation of mRNA transcription during mitosis, aspects of the epigenome associated with active genes are inherited to daughter nuclei through mitotic chromosomes. Contemporary research is now extending this concept of mitotic inheritance to binding of transcription factors (Festuccia et al. 2019) and the core transcriptional machinery itself (Teves et al. 2018).
We went on to use hybridisation to metaphase chromosome spreads to begin investigating different aspects of the structural organisation of the interphase genome. We first tried to analyse how different parts of the human genome may attach to various substructures within the nucleus. Such structures are defined by different biochemical extraction methods and termed the nuclear matrix, nuclear scaffold and nuclear skeleton. Less than 10% of genomic DNA remained attached to these substructures after extraction (Craig et al. 1997). Regions of the genome corresponding to G-bands appeared to have more frequent attachments to the nuclear matrix (remaining after high salt extraction) and the nuclear scaffold—a residual structure after extraction of nuclei with the anionic detergent lithium diiodosalicylate. This result is consistent with the suggestion that sites of attachment to a nuclear and mitotic chromosome scaffold would be most frequent in G-bands (Saitoh and Laemmli 1994). In contrast, genomic regions corresponding to R-bands had more attachments to the nuclear skeleton, the substructure remaining inside nuclei after electroelution of unattached sequences (Craig et al. 1997). Nuclear skeleton preparation methods had been designed to preserve more functional aspects of interphase function (transcription and replication) by avoiding the use of high salt or detergents that might cause protein aggregation (Jackson et al. 1988). Therefore, our finding of nucleoskeleton associations being more frequent in the gene-rich R-band fraction of the genome is consistent with a link between the aspect of nuclear organisation probed by this method and gene regulation.
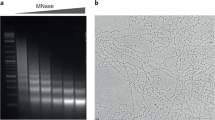
A lot of the controversy that surrounded the field of nuclear organisation in the 1980s and 1990s arose from the fact that no one really knew what the various experimental methods used did to native chromatin structure and nuclear architecture, nor which method—if any—might give the most physiologically relevant result. In an attempt to apply a more biophysical approach to higher-order interphase chromatin organisation, and to examine chromatin fibres per se, rather than their association with nuclear substructures, we used micrococcal nuclease (MNase) digestion and analytical sucrose gradient ultracentrifugation to separate fractions of the human genome with more open or more compact chromatin fibres. Sedimentation coefficient depends on the ratio between mass and fractional coefficient. Therefore, chromatin sedimenting fast for its mass has a compact hydrodynamic shape, and when we hybridised this chromatin fraction to metaphase chromosomes we could show that it originated from the gene-poor (G-band) fraction of the genome (Gilbert et al. 2004). Conversely, the slow sedimenting most ‘open’ chromatin fractions with high frictional coefficients originated from the genomic regions corresponding to the most gene-rich T-bands—see chromosome 1 in Fig. 3. These experiments provided the first map of higher-order chromatin structure in the human genome.
Fluorescence in situ hybridisation (FISH) reveals the distribution of the open chromatin fibres on human chromosome 1. Left: Hybridisation signal (green) from the open chromatin fraction on a 4,6-diamidino-2-phenyl indole (DAPI)-stained human chromosome 1. Right: Ideogram of chromosome 1 with T-bands in red, R-bands in white and G-bands in black. Adapted from Gilbert et al. (2004)
From chromosomes to microarrays
The experiments described above give a consistent picture of the human genome as organised into distinctive compartments with genes most concentrated together into domains that replicate early in S-phase and packaged into chromatin fibres with an open disordered structure. These are manifest on metaphase chromosomes as T- and R-bands. The other euchromatic fraction of the genome (G-bands) has a low gene density, replicates in the second half of S-phase and is packaged into more compact chromatin fibres. However, this broad compartmentalisation is very coarse-grained due to the limited resolution (5–10 Mb) of metaphase chromosomes.
Progress toward higher-resolution analysis came with the advent of genomic microarrays. These were initially often composed of arrayed bacterial or P1 artificial chromosomes tiled across the genome and had limited resolution (Mb) (Ishkanian et al. 2004; Lodén and van Steensel 2005; Woodfine et al. 2005), but the spotting of oligonucleotides at high density allows for higher-resolution interrogation of specific genomic regions. Early applications of this approach were the determination of replication timing domains (Repli-chip), allowing for the borders between early and later replicating domains to be demarcated in a way that was not possible by hybridisation to metaphase chromosomes (Ryba et al. 2011), and study of the distribution of specific histone modifications across the genome purified by chromatin immunoprecipitation (Kondo et al. 2004). We used hybridisation to microarrays to improve the resolution of our mapping of open and closed chromatin fibres in the human genome (Gilbert et al. 2004). A related approach (Weil et al. 2004) used the differential solubility of MNase digested chromatin to MgCl2 and KCl to assess the genomic distribution of chromatin compaction states.
Bas van Steensel and colleagues devised a new method based on targeted adenine methylation of DNA—DAMID—to determine domains of the genome that come into close contact with an intermediate filament protein—Lamin B1—that is a major component of the nuclear lamina that underlies the inner nuclear membrane. Originally applied to study of the Drosophila genome, they went on to perform DAMID for regions of the human genome associated with Lamin B1, hybridising the resulting DNA adenine methylated by a Lamin B1–Dam fusion protein compared with that methylated by free Dam, to high-density microarrays across the entire human genome (Guelen et al. 2008). This revealed 1000–1500 large (on average 0.5 Mb) domains of the genome that preferentially associate with lamin B1 (lamin-associated domains/LADs) and that are often seen by FISH to locate at the nuclear periphery. LADs are characterised by being gene-poor, CGI-poor, and containing inactive, late replicating chromatin.
Where are we today
Fast forward to the modern day and we are, of course, in the world of high-throughput sequencing, which can be applied to any genome-wide assay of chromosome organisation and function whose output is a sequenceable library of DNA fragments. This has opened the door to a plethora of high-throughput genome-wide assays of genome organisation, whose resolution is potentially very high, dependent on the details of the assay and the depth of the sequencing. Many of these assays—e.g., DAMID—were developments of techniques originally designed to be readout on microarrays, but are now being assayed by high-throughput sequencing and are even being applied to study genome organisation in single cells (Kind et al. 2015). High-resolution analysis of replication timing by sequencing (Repli-seq) (Marchal et al. 2018) has allowed identification of domains of the genome replicating at defined periods of S-phase—of approximately the same size as LADs. Moreover, as for DAMID-seq, high sequencing depths have facilitated a precise mapping of the transition regions between domains.
The most notable new technology that has opened our eyes to the compartmentalisation of the mammalian genome is Hi-C, a proximity ligation method that can inform on 3D chromatin conformation genome-wide. Two levels of compartmentalisation are generally recognised in Hi-C data—the first of these is the division of the genome into A and B compartments, which tend to cluster with in the nucleus—A with A and B with B. These categories seem to generally correspond to early replicating T/R-bands (A compartment) and late replicating G-bands (B compartment) (Lieberman-Aiden et al. 2009; Ryba et al. 2010; Kalhor et al. 2011). Very high-resolution Hi-C also reveals the organisation of the mammalian and Drosophila genomes into self-interacting domains—topologically associating domains (TADs) (Dixon et al. 2012; Sexton et al. 2012; Eagen et al. 2015). In mammals, very elegant mechanistic experiments and modelling have shown that TADs are formed by a process of loop extrusion by the cohesin complex, interrupted by particular orientations of CTCF sites (Nuebler et al. 2018). In Drosophila, the formation of TAD boundaries is less well understood (Ramírez et al. 2018). There are at least 2000 TADs in the mammalian genome, though it is hard to give a precise number because of the presence of sub-TADs within TADs. However, there is a very good correspondence between TADs, LADs and replication domains, with the transition points between replication domains aligning well with TAD boundaries (Pope et al. 2014). Therefore, a set of specific structural and functional properties (gene density, replication timing, lamin-association and intra- and inter-TAD interactions) seem to co-segregate with each other across the genome and with the properties of different metaphase chromosome bands. So are they one and the same thing, are we—using the powerful tool box of modern molecular biology—just rediscovering chromosome bands? Without very high-resolution mapping by FISH to metaphase chromosomes it is hard to be sure, but certainly the overall number of TADs/replication domains/LADs (one to two thousand) in the human genome is very close to the number of chromosome bands that have been seen on early prometaphase chromosomes (Yunis 1981; Drouin and Richer 1989) (Fig. 4).
As well as remarkable advances in genomic methods based on high-throughput sequencing assays, there has also been a revolution in fluorescence imaging. Technological advances have made available to the cell biologist a number of imaging modalities whose resolution is not diffraction limited. Of these ‘super-resolution’ imaging methods, structured illumination microscopy and stochastic optical reconstruction microscopy (STORM), coupled to FISH, have been most widely applied to investigate genome organisation. FISH probe technology has also advanced. Massively parallel oligonucleotide synthesis allows flexibly labelled oligonucleotide probe pools to be made to any chosen set of sequences (Boyle et al. 2011). In OLIGOPAINT, the introduction of non-genomic sequences to the oligonucleotides, allows for multiple different probe sets, labelled with different fluorophores, to be made from the same oligonucleotide library (Beliveau et al. 2015). A modification of OLIGOPAINT, called OLIGO STORM, makes probe sets that are particularly suited to very high-resolution imaging by STORM (Beliveau et al. 2017).
There is a remarkable correspondence between TADs and TAD boundaries and the bands and inter-bands seen on Drosophila polytene chromosomes (Eagen et al. 2015), and indeed a correspondence to chromomeres—locally coiled chromatin domains that had been seen decades ago by microscopy on mitotic and meiotic prophase chromosomes (Macgregor 2012; Eagen 2018). Chromomeres have now been revealed in the interphase nucleus of diploid cells using FISH and the latest super-resolution optical imaging methods (Szabo et al. 2018).
So maybe there is nothing really new in biology and we are often just rediscovering principles and phenomena that were first revealed many decades before the advent of the latest molecular and cellular technologies. However, we now have the ability to reveal and resolve these structures at unprecedented levels of resolution, in different cells at different stages of development or in disease states. Moreover, we have the tools to begin manipulating these levels of genome organisation. That surely must remain the grand challenge for genome organisation—what do all of these domains, compartments and structures mean for genome functions.
References
Beliveau BJ, Boettiger AN, Avendaño MS, Jungmann R, McCole RB, Joyce EF, Kim-Kiselak C, Bantignies F, Fonseka CY, Erceg J et al. (2015) Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun 6:7147
Beliveau BJ, Boettiger AN, Nir G, Bintu B, Yin P, Zhuang X, Wu CT (2017) In situ super-resolution imaging of genomic DNA with OligoSTORM and OligoDNA-PAINT. Methods Mol Biol 1663:231–252
Bickmore WA, Bird AP (1992) Use of restriction endonucleases to detect and isolate genes from mammalian cells. Methods Enzymol 216:224–244
Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA (2011) Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res 19:901–909
Craig JM, Bickmore WA (1993) Chromosome bands–flavours to savour. Bioessays 15:349–354
Craig JM, Bickmore WA (1994) The distribution of CpG islands in mammalian chromosomes. Nat Genet 7:376–382
Craig JM, Boyle S, Perry P, Bickmore WA (1997) Scaffold attachments within the human genome. J Cell Sci 110:2673–2682
Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145:1119–1131
Cross SH, Lee M, Clark VH, Craig JM, Bird AP, Bickmore WA (1997) The chromosomal distribution of CpG islands in the mouse: evidence for genome scrambling in the rodent lineage. Genomics 40:454–461
Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25:1010–1022
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380
Drouin R, Richer CL (1989) High-resolution R-banding at the 1250-band level. II. Schematic representation and nomenclature of human RBG-banded chromosomes. Genome 32:425–439
Drouin R, Holmquist GP, Richer CL (1994) High-resolution replication bands compared with morphologic G- and R-bands. Adv Hum Genet 22:47–115
Dutrillaux B, Couturier J, Richer CL, Viegas-Péquignot E (1976) Sequence of DNA replication in 277 R- and Q-bands of human chromosomes using a BrdU treatment. Chromosoma 58:51–61
Eagen KP, Hartl TA, Kornberg RD (2015) Stable chromosome condensation revealed by chromosome conformation capture. Cell 163:934–946
Eagen KP (2018) Principles of chromosome architecture revealed by Hi-C. Trends Biochem Sci 43:469–478
Festuccia N, Owens N, Papadopoulou T, Gonzalez I, Tachtsidi A, Vandoermel-Pournin S et al. (2019) Transcription factor activity and nucleosome organization in mitosis. Genome Res 29:250–260. https://doi.org/10.1101/gr.243048.118
Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA (2004) Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118:555–566
Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W et al. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453:948–951
Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP et al. (2004) A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet 36:299–303
Jackson DA, Yuan J, Cook PR (1988) A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J Cell Sci 90:365–378
Jeppesen P, Turner BM (1993) The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74:281–289
Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L (2011) Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol 30:90–98
Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M et al. (2015) Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163:134–147
Kojima KK (2018) Human transposable elements in Repbase: genomic footprints from fish to humans. Mob DNA 9:2
Kondo Y, Shen L, Yan PS, Huang TH, Issa JP (2004) Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc Natl Acad Sci USA 101:7398–7403
Korenberg JR, Rykowski MC (1988) Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell 53:391–400
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J et al. (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293
Lodén M, van Steensel B (2005) Whole-genome views of chromatin structure. Chromosome Res 13:289–298
Macgregor HC (2012) Chromomeres revisited. Chromosome Res 20:911–924
Marchal C, Sasaki T, Vera D, Wilson K, Sima J, Rivera-Mulia JC et al. (2018) Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat Protoc 13:819–839
Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA (2018) Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci USA 115:E6697–E6706
Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O et al. (2014) Topologically associating domains are stable units of replication-timing regulation. Nature 515:402–405
Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J et al. (2018) High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9:189
Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J et al. (2010) Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20:761–770
Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM (2011) Genome-scale analysis of replication timing: from bench to bioinformatics. Nat Protoc 6:870–895
Saitoh Y, Laemmli UK (1994) Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell 76:609–622
Salzburger W, Steinke D, Braasch I, Meyer A (2009) Genome desertification in eutherians: can gene deserts explain the uneven distribution of genes in placental mammalian genomes? J Mol Evol 69:207–216
Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M et al. (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148:458–472
Sproul D, Gilbert N, Bickmore WA (2005) The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet 6:775–781
Szabo Q, Jost D, Chang JM, Cattoni DI, Papadopoulos GL, Bonev B et al. (2018) TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci Adv 4:eaar8082
Terrenoire E, McRonald F, Halsall JA, Page P, Illingworth RS, Taylor AM et al. (2010) Immunostaining of modified histones defines high-level features of the human metaphase epigenome. Genome Biol 11:R110
Teves SS, An L, Bhargava-Shah A, Xie L, Darzacq X, Tjian R (2018) A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. Elife 7:pii: e35621
Vogel W, Autenrieth M, Mehnert K (1989) Analysis of chromosome replication by a BrdU antibody technique. Chromosoma 98:335–341
Weil MR, Widlak P, Minna JD, Garner HR (2004) Global survey of chromatin accessibility using DNA microarrays. Genome Res 14:1374–1381
Woodfine K, Carter NP, Dunham I, Fiegler H (2005) Investigating chromosome organization with genomic microarrays. Chromosome Res 13:249–257
Yunis JJ (1981) Mid-prophase human chromosomes. The attainment of 2000 bands. Hum Genet 56:293–298
Acknowledgements
WAB is funded by a Medical Research Council University Unit programme grant MC_ UU_00007/2.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that she has no conflict of interest.
Additional information
This article is dedicated to the memory of Herbert Macgregor who sadly died in 2018. Herbert truly appreciated the beauty of chromosome structure and he tirelessly dedicated his time and effort into promoting the field of Chromosome Biology. He founded the journal Chromosome Research in 1992, he edited it for 20 years and I had the pleasure of serving alongside him on the Editorial board of that journal.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bickmore, W.A. Patterns in the genome. Heredity 123, 50–57 (2019). https://doi.org/10.1038/s41437-019-0220-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-019-0220-4