Abstract
Conserving tree populations safeguards forests since they represent key elements of the ecosystem. The genetic characteristics underlying the evolutionary success of the tree growth form: high genetic diversity, extensive gene flow and strong species integrity, contribute to their survival in terms of adaptability. However, different biological and landscape contexts challenge these characteristics. This study employs 63 de novo developed microsatellite or SSR (Single Sequence Repeat) markers in different datasets of nine Neotropical Magnolia species. The genetic patterns of these protogynous, insect-pollinated tree species occurring in fragmented, highly-disturbed landscapes were investigated. Datasets containing a total of 340 individuals were tested for their genetic structure and degree of inbreeding. Analyses for genetic structure depicted structuring between species, i.e. strong species integrity. Within the species, all but one population pair were considered moderate to highly differentiated, i.e. no indication of extensive gene flow between populations. No overall correlation was observed between genetic and geographic distance of the pairwise species’ populations. In contrast to the pronounced genetic structure, there was no evidence of inbreeding within the populations, suggesting mechanisms favouring cross pollination and/or selection for more genetically diverse, heterozygous offspring. In conclusion, the data illustrate that the Neotropical Magnolias in the context of a fragmented landscape still have ample gene flow within populations, yet little gene flow between populations.
Similar content being viewed by others
Introduction
Conservation genetics utilises a representative sample of DNA and organisms to quantify and study genetic diversity to preserve species as dynamic entities capable of coping with environmental change (Frankham et al. 2010). A collection of DNA fragments representing the genome is realised by employing molecular markers: fragments of DNA associated with a certain location within the genome, providing information about the allelic variation at the given locus (Schlötterer 2004). Microsatellite or SSR (Simple Sequence Repeat) markers are often the preferred type of molecular marker in conservation genetics because they are codominant, highly polymorphic, ubiquitous, reproducible and neutral; and they have a high mutation rate, as well as an easy sample preparation (Selkoe and Toonen 2006). Although it is labour and cost intensive to develop and test SSR primer pairs, these can often be employed across species, with success decreasing proportionally to relatedness (Kalia et al. 2011). A representative sampling of organisms can be interpreted at different levels: individuals for populations, populations for species, and species for ecosystems. The latter strategy makes use of the umbrella species concept (Roberge and Angelstam 2004).
An exemplar group of umbrella species are trees: they maintain the structure and function of forest ecosystems, and create resource niches and patches for other organisms (Pautasso 2009). Trees also provide various ecosystem services and resources for human use (Neale and Kremer 2011) and their genetics and evolution have paradoxical features (Petit and Hampe 2006). Trees were found to maintain high levels of genetic diversity (Hamrick et al. 1992), but experience low nucleotide substitution rates and low speciation rates when compared to annual plant lineages (e.g. Bousquet et al. 1992; Petit and Hampe 2006; Whittle and Johnston 2003). They combine high local differentiation for adaptive traits (Aitken et al. 2008) with extensive gene flow (Austerlitz et al. 2000; Kremer and Le Corre 2012). Furthermore, they maintain species integrity, while expressing abundant interspecific gene flow (Ellstrand et al. 1996). The abovementioned features provide an expected capacity for tree survival, as they create resilience against threats such as climate change or habitat fragmentation (Aitken et al. 2008; Hamrick 2004). However, the interplay of the biological and landscape context challenges these generalised characteristics and creates the need for context-oriented tree conservation genetic studies and subsequent management guidelines (Aparicio et al. 2012; Dick et al. 2008).
To investigate the general patterns of tree genetics in an empirical setting, and to contribute to the conservation of the species and forests under study, we focus on New World representatives of the tree genus Magnolia (Magnoliaceae) occurring at tropical latitudes, hereafter named Neotropical Magnolias. Magnolia trees provide an interesting case-study with bisexual, protogynous flowers, specialised beetle pollination with tepal movement, variable flowering phenology and seed dispersal by animals (Thien 1974). The Red List of Magnoliaceae (Rivers et al. 2016) states that 76% of the Neotropical Magnolias are threatened, with an additional 16% listed as data deficient. Neotropical Magnolia populations have not been studied from a molecular point of view (Cires et al. 2013) and their species are delineated based on morphological and distributional argumentation (e.g. Howard 1948; Palmarola et al. 2016; Vázquez-García et al. 2013b). Many of the Magnolia species and populations occur in fragmented, highly-disturbed, relict primary forest landscapes, such as the cloud forests of the Caribbean islands and the cloud and rain forests of Mexico (Rivers et al. 2016).
This study aims to (1) provide de novo developed SSR markers for Neotropical Magnolia species; (2) employ the SSR markers for genetic species delimitation between Caribbean Magnolia species; (3) search for patterns of extensive gene flow between Caribbean Magnolia (sub)species and populations; and (4) test for signs of inbreeding within the Neotropical Magnolia populations.
Material and methods
Sampling and DNA extraction
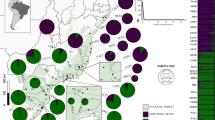
Sample information of the 17 different taxa (i.e. 16 species, of which one species consists of two subspecies) and 17 populations included in this study are given in Table 1. A map, showing the location information of the wild collected accessions of Neotropical Magnolia from the Caribbean and Mexico, is given in Fig. 1. The wild collected samples comprise 346 samples, of which 340 represent the 17 populations. The additional six wild collected samples represent single collections of different species. One further sample is from an ex situ collection of M. dealbata.
Location map of 16 Magnolia taxa (i.e. 15 Magnolia species, of which one species consists of two subspecies) from the Caribbean and Mexico, collected in the wild. Circles represent the species of the section Talauma subsection Splendentes. Squares represent species of the Talauma subsection Talauma. Triangles represent species of the section Magnolia. Classification is according to Figlar and Nooteboom (2004)
For the 17 populations included in the full genetic analyses, Average Pairwise Distance between individuals (APD), Maximum distance between consecutive individuals (Max), Spatial extent of the populations (SpE) and number of sampled individuals per populations (NS) are given in Table 2. Pairwise distances were calculated using the fossil package (Vavrek 2011) in R v.3.4.3 (R Core Team 2016).
All 347 leaf samples were dried in silica gel and their DNA was isolated using a modified cetyltrimethylammonium bromide (CTAB) (Doyle and Doyle 1987) extraction protocol, with MagAttract Suspension G solution (Qiagen, Germantown, USA) (Xin and Chen 2012) mediated cleaning (Larridon et al. 2015). DNA quantity and quality control was executed using a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Massachusetts, USA) and Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific), respectively.
SSR markers: development and testing
Primer pairs were developed to amplify sequences containing SSR repeats based on four Neotropical Magnolia species: Magnolia lacandonica (MA39), M. mayae (MA40), M. dealbata (MA41), and M. cubensis subsp. acunae (MA42). The development of the enriched microsatellite library was outsourced to Allgenetics® (A Coruña, Spain) where enrichment was performed using the Nextera XT DNA kit probes (Illumina, California, USA) with the following motifs: AGG, ACG, AAG, AAC, ACAC and ATCT. The library was sequenced on an Illumina MiSeq® platform.
From the 4 × 500 predetermined SSR primer pairs provided by Allgenetics®, 176 were selected for further testing: 49 developed from MA39-reads, 20 developed from MA40-reads, 20 developed from MA41-reads and 87 developed from MA42-reads. Selection of the 176 SSR markers was carried out randomly, respecting the characteristics specified in Guichoux et al. (2011). The forward primers were linked with a universal tail to accomplish multiplex pooling in a three-primer PCR (Vartia et al. 2014). The following universal tags were used: T3: 5′ AATTAACCCTCACTAAAGGG 3′, M13(-20): 5′ GTAAAACGACGGCCAGT 3’, Hill: 5’ TGACCGGCAGCAAAATTG 3′ (Tozaki et al. 2001) and Neomycin reverse: 5′ AGGTGAGATGACAGGAGATC 3’. The reverse primers had a PIG-tail (Brownstein et al. 1996).
All 176 markers were screened for amplification success on the 17 taxa, each represented by one randomly selected sample. PCRs were performed on a total volume of 13 µL under the following conditions: 2 min at 95 °C; 35 cycles of 95 °C for 30 s, 52 °C for 30 s, 72 °C for 90 s; 72 °C for 6 min. The Master Mix contained 0.2 µM forward primer, 0.2 µM reverse primer, 5 ng/ml DNA (suspended in 1 × TE buffer), 1 × TrueStart Taq Buffer (Thermo Fisher Scientific), 1.5 µM MgCl2 (Thermo Fisher Scientific), 0.125 µM dNTP, 5U of TrueStart Hot Start DNA polymerase (Thermo Fisher Scientific), and 0.4 mg/ml BSA (bovine serum albumin) per reaction. PCR products were run on a 1% agarose gel, stained with ethidium bromide and visualised under UV-light. Every (sub)species × primer combination was scored. Amplification scores of the 63 published SSR markers are given in the Supplementary Table S1. The (sub)species × primer combinations which were scored to have a single band were submitted to polymorphism testing.
Polymorphism tests were executed on eight individuals per Magnolia species, comprising four individuals per predefined population. The individuals for the test-multiplexes were selected to be spatially spread throughout the populations and have 260/230 and 260/280 OD (Optical Density) ratios approximating 2. The (sub)species × primer combinations were scored: 63 were considered polymorphic and unambiguous SSR markers in at least one of the ten tested taxa (Supplementary Table S2). These 63 SSR markers were used for species-specific multiplex design and final genotyping. Their primer information can be found in Supplementary Table S3.
Genotyping of individuals was executed by a multiplex pooling with a three-primer PCR (Vartia et al. 2014). The fluorescent labels FAM, NED, PET and VIC were linked to the tails T3, Hill, Neo and M13, respectively. The multiplex pools were designed using Multiplex Manager (Holleley and Geerts 2009). Multiplex PCRs were performed on a total volume of 5 µL, under the following conditions: 15 min at 95 °C; 35 cycles of 94 °C for 30 s, 57 °C for 90 s, 72 °C for 90 s; 72 °C for 10 min. Each multiplex reaction contained 2 × QIA Multiplex PCR Master Mix (Qiagen), 5 ng/µL DNA, 0.025 µM for each forward primer, 0.1 µM for each reverse primer and 0.1 µM for each specified dye, carrying the same universal tail as the selected forward primer of the chosen primer pairs. Fragment analyses were executed by Macrogen Inc. (Seoul, South Korea) on an ABI 3730XL fragment analyser (Thermo Fisher Scientific) with a GeneScanTM 500 LIZTM ladder (Thermo Fisher Scientific). The results were analysed in Geneious v.8.1.9 (http://www.geneious.com, Kearse et al. 2012) using the microsatellite plugin. When the test on the subset of individuals appeared promising (i.e. one set of clear peaks, good amplification and more than one allele), 20 individuals per population were genotyped for that marker. The ten taxa were genotyped for 21–36 polymorphic markers, delivering ten separate taxon-datasets (Supplementary Table S2: one taxon-dataset = one column with the markers coded “A”).
Error rates (Selkoe and Toonen 2006) for the markers (Supplementary Table S3) across all ten taxon-datasets were calculated, but were not actively and consistently tested for: duplicate genotyping was produced as a side-product during testing for polymorphism, optimizing multiplexes, re-genotyping a complete multiplex for (a) low/unclear peak(s), or as positive control between PCR batches.
The ten taxon-datasets were submitted to MICRO-CHECKER v.2.2.3 (Van Oosterhout et al. 2004) and ML-NullFreq (Kalinowski and Taper 2006) to test for null alleles. MICRO-CHECKER was run with 1000, and ML-NullFreq was run with 100 000 repetitions. Based on the results, markers with a high probability of representing null alleles were discarded from all downstream analyses.
To ensure that all amplified genetic regions were independent samples of the genome, allelic associations (Lewontin and Kojima 1960) (synonym: Linkage Disequilibrium = LD) per population were analysed in each of the ten taxon-datasets using the software program GENEPOP v.4.3 (Rousset 2008) with the dememorization number set to 10 000, batches set to 1000 and 50 000 iterations per batch. Evaluation of allelic associations was executed by examining both the uncorrected (Waples 2015) and (sequential Bonferroni) corrected p-values (Holm 1979) with nominal p-values of 0.05 per species and per population.
Genetic structure
To assess the utility of the SSR markers for genetic species delimitation between closely located Caribbean Magnolia species and to search for patterns of extensive gene flow between Caribbean Magnolia (sub)species, five different supraspecific (i.e. above species level) datasets were instated. Dataset 1 comprises 340 individuals representing 17 populations, genotyped for all their polymorphic and monomorphic loci (see Supplementary Table S2: all marker × taxon combinations coded A, B and C). Hence, for this dataset it was assumed that the loci that tested to be monomorphic for four or eight individuals were monomorphic for all 20 individuals. Dataset 2 comprises 340 individuals representing 17 populations, genotyped for all the polymorphic and monomorphic loci, but not the assumed monomorphic loci (See Supplementary Table S2: all marker × taxon combinations coded A and B). Dataset 3, or the Splendentes-normalized-dataset, comprises ten loci (see Supplementary Table S2: SSR markers labelled with an asterisk) that were genotyped for 260 individuals representing 13 populations and eight taxa of section Talauma subsection Splendentes (Table 1: Class. = TAS). Added to datasets 1, 2 and 3, two smaller supraspecific datasets were instated, representing the apparently closely related species i.e. the two species of Puerto Rico: the PR-dataset; and the three species of the Dominican Republic: the DR-dataset. To search for patterns of extensive gene flow between Caribbean Magnolia population pairs within the defined species, the 17 populations were studied on the infraspecific (i.e. below species) level using nine species-datasets (i.e. the taxon-datasets of the two M. cubensis subspecies were joined) and 17 population-datasets.
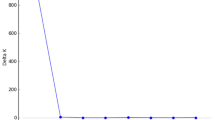
A first batch of analyses was conducted in STRUCTURE v.2.3.4 (Pritchard et al. 2000) on datasets 1, 2 and 3, the PR- and DR-datasets, the nine species-datasets and the 17 population-datasets. STRUCTURE analyses were run with a burn-in of 100 000, 100 000 MCMC steps after the burn-in and the admixture model as ancestry model. Datasets 1, 2 and 3 were run with the allele frequency model set to independent allele frequencies. They were expected to consist of 13 (dataset 3) or 17 (dataset 1 and 2) populations and were run with K set from 1 to 25. The PR- and DR-datasets were run both with the independent allele frequency model and the correlated allele frequency model and their results were compared. They were expected to have between 2 and 6 populations and K was set from 1 to 15. The nine species-datasets and 17 population-datasets were run with the allele frequency model set to correlated allele frequencies. They were run with K set from 1 to 10. For all datasets, each value of K was run 10 times. The results were visualized with Structure Harvester Web v.0.6.94 (Earl and vonHoldt 2012). The best K-value was selected using the ΔK statistic (Evanno et al. 2005) and the results for mean maximum likelihood (Mean LnK). The latter was taken into consideration because the ΔK statistic appointed K-values with unstable replicate results for datasets 1, 2 and 3 and because the ΔK statistic cannot detect single clusters: an outcome expected at the infraspecific level (i.e. population-datasets and possibly the species-datasets). Barplots were visualised using DISTRUCT v.1.1 (Rosenberg 2004).
DAPC analyses (Discriminant Analysis of Principal Components) on datasets 1, 2 and 3 were executed in R using the package adegenet (Jombart 2008). In the find.clusters function we retained 300 PCs for dataset 1 and 2, and 140 PCs for dataset 3. The number of PCs to retain for the PCA eigenvalues was determined using cross-validation. All discriminant functions (DA eigenvalues) were kept.
Pairwise FST values (Weir and Cockerham 1984) and their confidence intervals were calculated in R using the package diveRsity (Keenan et al. 2013). To visualize the genetic distances for dataset 1, 2 and 3, an unrooted network applying the Neighbour-joining (NJ) method based on Nei’s genetic distance: DA (Nei et al. 1983), was constructed using Populations v.1.2.32 (http://bioinformatics.org/populations/) using 1000 bootstrap replicates as a confidence measure.
Mantel tests on the supraspecific level were performed in GenAlEx v.6.5 (Peakall and Smouse 2006; Peakall and Smouse 2012) on the pairwise log-transformed geographic distance and pairwise FST values using 9999 permutations. Coordinates of one individual were taken as a representative of its population. Species geographic distance was averaged over the populations of the species.
Inbreeding and population statistics
To test for inbreeding within the Caribbean Magnolia populations, the inbreeding coefficient (FIS) for each locus and population was calculated in FSTAT. Tests to detect significant deviations from Hardy-Weinberg proportions (HWP) were calculated in GENEPOP, performing 2-tailed exact tests for each locus in each population. Complete enumeration was performed whenever possible (Louis and Dempster 1987), otherwise MCMC chains were run with 200 batches and 50 000 iterations (Guo and Thompson 1992). Deviations of both the uncorrected and sequential Bonferroni corrected p-values were used to evaluate if populations were truly deviating from HWP (Waples 2015). To frame and discuss the results, different statistical parameters were calculated for each locus and population within the ten taxon-datasets using GenAlEx, i.e. the percentage of polymorphic loci (P), the number of genotyped individuals (N), mean number of alleles (A), expected heterozygosity (He), and observed heterozygosity (Ho).
Results
SSR markers
Overall, 82–92% of the primer pairs amplified, of which 53–67% were scored to be a single amplification product (Supplementary Table S1). The polymorphism tests of the markers giving a single amplification product classified 16–37% of the primer pairs unambiguous and polymorphic (Supplementary Table S2). The reported SSR primers all have heterozygote states in at least one individual and a perfect motif (Weber 1990). For 56 SSR markers, the duplicate runs rendered the same genotypes (Supplementary Table S3: error rate: 0%). For one SSR marker no genotypes were duplicated. The error rates of the other six SSR markers ranged from 1–3.85%.
Results of detection and frequency of null alleles per marker × population combination are given in Supplementary Table S4. Twelve marker × species combinations were considered to have a high probability of showing null alleles: M. cubensis (MA42_028), M. domingensis (MA39_199), M. ekmanii (MA39_023, MA42_087), M. hamorii (MA40_223, MA42_413), M. lacandonica (MA39_182), M. pallescens (MA39_023, MA42_472), M. portoricensis (MA42_481) and M. splendens (MA39_023, MA42_481).
Associated alleles per marker × species combination are given in Supplementary Table S4. Magnolia domingensis and M. lacandonica showed a number of SSR markers with associated alleles that were higher than expected for the number of pairwise tests executed. The other eight taxa fell within their confidence intervals of false positives, whereby one significantly associated pair of SSR markers was detected in M. pallescens (MA40_045 × MA42_472).
Genetic structure: supraspecific level
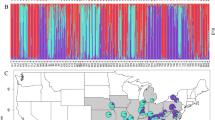
Supraspecific ΔK and Mean LnK plots are depicted in Supplementary Figure S5A–E and their interpretation is summarized in Table 3. Barplots of the STRUCTURE analyses on the three full supraspecific datasets are depicted in Fig. 2a–d. The DR-dataset and PR-dataset structured according to the species given both criteria and correlation frequency models. In the DAPC analysis, the “true” K in the replicate runs of the find.clusters algorithm was not univocal, and ranged between 9–13 for dataset 1, 9–15 for dataset 2 and 8–11 for dataset 3. For each dataset, a representative DAPC analysis is visualised in Fig. 3. Supraspecific pairwise FST values range from 0.216 to 0.618 for dataset 1, 0.166 to 0.472 for dataset 2 and 0.130 to 0.308 for dataset 3 (See Table 4). Their confidence intervals are visualized in Supplementary Figure S6. The unrooted NJ trees based on DA are depicted in Fig. 4. The Mantel tests for all three datasets including all population-pairs were significant (p = 0.000–0.003). Mantel tests on the supraspecific pairwise distances were significant for dataset 1 (p = 0.000), but not for dataset 2 (p = 0.080) and dataset 3 (p = 0.256). See Supplementary Figure S7 for visualisation of the relationship between geographic and genetic distance and Table 4 for the Pairwise Geographic Distance (PGD) between the population pairs.
STRUCTURE barplots of Magnolias from the Caribbean and Mexico. The replicate with the highest likelihood score is given. a STRUCTURE barplot of dataset 1 and dataset 2, K = 2. b STRUCTURE barplot of dataset 1: K = 9. c STRUCTURE barplot of dataset 3, K = 3. d STRUCTURE barplot of dataset 3, K = 8. e STRUCTURE barplot of the Guadeloupe population of Magnolia dodecapetala. f STRUCTURE barplot of the Toro Negro population of Magnolia portoricensis. Dataset 1 comprises 340 individuals representing 17 populations, genotyped for all 63 microsatellite markers where possible, including the assumed monomorphic data (See Supplementary Table S2: categories A, B and C). Dataset 2 comprises 340 individuals representing 17 populations, genotyped for all 63 microsatellite markers where possible, excluding the assumed monomorphic data (See Supplementary Table S2: categories A and B). Dataset 3 comprises 260 individuals representing 13 populations of the 8 taxa of the section Talauma subsection Splendentes (See Table 1: Class. = TAS), genotyped for 10 microsatellite markers (See Supplementary Table S2: marker names indicated with an asterisk)
DAPC plots of Magnolias from the Caribbean and Mexico. DAPC: Discriminant Analysis of Principal Components. Populations and (sub)species are abbreviated cf. Table 1 and CU: Magnolia cubensis. a DAPC plot of dataset 1 which comprises 340 individuals representing 17 populations, genotyped for all 63 microsatellite markers where possible, including the assumed monomorphic data (See Supplementary Table S2: categories A, B and C). Nine clusters are visualised following the nine species: CU, DOD, DOM, EKM, HAM, LAC, PAL, POR, SPL b DAPC plot of dataset 2 which comprises 340 individuals representing 17 populations, genotyped for all 63 microsatellite markers where possible, excluding the assumed monomorphic data (See Supplementary Table S2: categories A and B). Eleven clusters are visualised: CU (behind SPL), DOD, DOM, HAM, GRA, LAC (population), MAN, PAL (behind POR), POR (behind DOM), SPL, YAJ. C DAPC plot of dataset 3 which comprises 260 individuals representing 13 populations of the 8 taxa of the section Talauma subsection Splendentes (See Table 1: Class. = TAS), genotyped for 10 microsatellite markers (See Supplementary Table S2: marker names indicated with an asterisk). mix1: all 40 individuals of DOM and 3 individuals of SAL. mix2: all 40 individuals of PAL and 1 individual of PIC. Nine clusters are visualised: GRA, MAN, mix1 (behind PAL), mix2, PAL, PIC, POR, SPL, TOP
NJ trees of the Magnolias from the Caribbean and Mexico. Unrooted networks are constructed by the Neighbour-joining (NJ) method based on Nei’s genetic distance: DA (Nei et al. 1983). Bootstrap values above 70 are depicted. a NJ-tree of dataset 1 which comprises 340 individuals representing 17 populations, genotyped for all 63 microsatellite markers where possible, including the assumed monomorphic data (See Supplementary Table S2: categories A, B and C). b NJ-tree of dataset 2 which comprises 340 individuals representing 17 populations, genotyped for all 63 microsatellite markers where possible, excluding the assumed monomorphic data (See Supplementary Table S2: categories A and B). c NJ-tree of dataset 3 which comprises 260 individuals representing 13 populations of the 8 taxa of the section Talauma subsection Splendentes (See Table 1: Class. = TAS), genotyped for 10 microsatellite markers (See Supplementary Table S2: marker names indicated with an asterisk).
Genetic structure: infraspecific level
Infraspecific ΔK and Mean LnK plots are depicted in Supplementary Figure S5F–V2 and their interpretation is summarized in Table 3. Barplots of the two infraspecific STRUCTURE analyses exceeding the predefined clusters: GUA and TOR are given in Figs. 2e, f, respectively. Infraspecific pairwise FST values can be found in Table 4 and range from 0.044 to 0.222 for the species-datasets and 0.035 to 0.226 when standardized cf. dataset 3. Confidence intervals of the infraspecific pairwise FST values are depicted in Supplementary Figure S6. Mantel tests at the infraspecific level were not significant (dataset 1 and dataset 2: p = 0.084, dataset 3: p = 0.080): see Supplementary Figure S7.
Inbreeding: infraspecific level
Detailed results on the population statistics calculated on the ten taxon-datasets are listed per marker, population and subset in Supplementary Table S4. Population statistics of the most representative subset are listed in Table 2. Three populations: GUA, MART and TOR showed significant departure from HWP. GUA and MART presented significant deviation from HWP for 5/21 and 4/21 loci (1.45 [0, 3] expected to test false positive when p = 0.05). TOR showed significant deviation from HWP for 7/29 loci (1.45 [0, 3] expected to test false positive when p = 0.05).
Discussion
SSR markers
The data on marker development show an attrition of usable SSR primer pairs during post-sequencing marker development, which is a general issue in SSR development (Hodel et al. 2016). Patterns in success of the polymorphism tests should be treated with caution because (1) multiplexing SSR markers can lead to marker interaction; (2) testing with more individuals or populations can show more markers to be polymorphic; (3) massive parallel testing was executed, for which some SSR marker-species combinations were not replicated; (4) random isolated PCR artefacts have been observed. Because all 63 reported SSR markers had a heterozygous state in at least one individual and contain di- or tri-repeats, they are labelled nuclear SSR loci (Wheeler et al. 2014).
Sampling design
The sampling design renders a dataset which is standardised yet limited in the number of individuals per population and populations per species (Hoban et al. 2013; Ward and Jasieniuk 2009). It is possible that the limited number of samples invoked false positives or false negatives due to random sampling error (Waples 2015), hence, we recommend including SSR markers that reported to have null alleles when genotyping more individuals and populations in further analyses, except for the markers that have very strong evidence i.e. MA42_028 for M. cubensis, MA39_182 for M. lacandonica and MA42_481 for M. portoricensis.
Genetic structure: supraspecific level
In general, results of all supraspecific analyses (Tables 3–4, Figs. 2, 3, 4, Supplementary Figure S5A–E2, S6, S7) are influenced by the datasets used. Firstly, due to the resolution: inclusion of more differentiated species/populations conceals the signal of the lower genetic structural levels (e.g. Fig. 2a vs. Fig. 2b). Secondly, due to inclusion or exclusion of the assumed monomorphic SSR loci or fixed alleles (e.g. Fig. 3a vs. Fig. 3b). On the one hand, fixed alleles determined a higher differentiation among species. This is apparent in the NJ-tree when comparing branch lengths and bootstrap values in Figs. 4a, b and in the DAPC plots when comparing Fig. 3a with Fig. 3b. On the other hand, the monomorphic loci strengthen genetically similar species groups, illustrated by the three species of the Dominican Republic to be clustered together in Fig. 4a, while when omitting the assumed monomorphic data (Fig. 4b), M. hamorii is differentiated from the other two Dominican Magnolias.
Currently, a molecular phylogenetic analysis including a representative sampling of section Talauma and its four subsections (Figlar and Nooteboom 2004; Pérez et al. 2016) is not available. On the basis of the SSR results, it can be stated that the species delineations of the studied seven species of subsection Splendentes are genetically confirmed. Clustering methods placed individuals and populations in their respective species genetic cluster (Figs. 2b, d, 3a, b and 4). However, the likelihood of clustering according to the species was not significant enough for the ΔK method to recognize the K corresponding to the number of species (Figs. 2a, c) and species-clusters often overlap in the two-dimensional visualization of the DAPC analysis (Fig. 3) or even consistently cluster with another species (Fig. 3c: mix1, mix2).
Although the SSR data is able to deliver evidence for species boundaries, there can be little conclusions drawn on their evolutionary relationships (Fig. 3, Table 4, Fig. 4). The data illustrates that the set of three Dominican Magnolias and the set of two Puerto Rican Magnolias are the least genetically differentiated (Table 4, Figs. 3a, 4a), which is also visible as a gap in pairwise FST values (Supplementary Figure S6) and the significant results of the Mantel tests (Supplementary Figure S7). The pairwise FST values (Table 4, Supplementary Figure S6A, S6B) suggest (M. domingensis + M. hamorii) + M. pallescens; however, in Figs. 3, 4b, c (M. domingensis + M. pallescens) + M. hamorii is put forward. Although native to the same island as the three Dominican Magnolias, M. ekmanii is conspicuously differentiated from them, as well as from all other species. There is a hint that EKM is most closely related to the Cuban Magnolias: their pairwise FST calculated on dataset 1 is significantly lower compared to the other EKM pairwise comparisons (Table 4, Supplementary Figure S6A), the DAPC analyses (Fig. 3) place them more closely together according to the two most explanatory axes in the ordination space, and the NJ-tree of dataset 1 and 2 display shared ancestry, albeit unsupported (Figs. 4a, b). For EKM and species relationships across the different Caribbean islands, the SSR loci have accumulated too many (homologous) mutations for supported relationships to be deducted (Calonje et al. 2009). Therefore, studying more conservative DNA fragments by phylogenetic studies (e.g. on chloroplast DNA or single copy nuclear genes) would be valuable.
Genetic structure: infraspecific level
GUA, MART and TOR are suspected to suffer from the Wahlund effect given the larger spatial distances (Table 2: SpE, Max, APD), significantly high number of null alleles (Supplementary Table S4), significant FIS value (Table 2), high number of alleles (Table 2: A) and their population STRUCTURE (Figs. 2e, f). The absence of genetic HWP-based structure in the MART population could be due to unequal mixture fractions (Waples 2015) combined with a small sample size. For more in-depth study of these populations, it is recommended to invoke more substructure in future sampling design and analyses.
The range of pairwise infraspecific FST values (Table 4) is large (0.035–0.226) and the genetic differentiation can be labelled: little (HAM), moderate (DOM, PAL: dataset 2, POR), great (CUB, DOD, EKM, LAC, PAL: dataset 1) (Hartl and Clark 1997) or significant (CUB, DOD, EKM, LAC, PAL: dataset 1) (Frankham et al. 2010). The large range of pairwise, infraspecific FST values reminds us of the conflict between the continuity of lineage separation and the discrete entity of a species (de Queiroz 1998). Theoretically, infraspecific genetic differentiation was expected to be counteracted by extensive gene flow between populations: either by long-distance pollen dispersal (Petit and Hampe 2006) or seed dispersal by natural disturbances (Lugo et al. 1981).
The Wahlund effect and moderate to great genetic differentiation indicate that the population dynamics of the studied Neotropical Magnolias occur at a fine spatial scale; in this sampling design suggested to be limited in the spatial extent of 4 km (Table 4: PGD of HAM) to 6 km (Table 2: SpE of TOR). The Mantel tests on the infraspecific level (Supplementary Figure S7) and comparisons with Magnolia SSR literature (Kikuchi and Isagi 2002; Setsuko et al. 2007; Zhao et al. 2012) show no correlations or trends between pairwise geographic and infraspecific genetic distance. For this result, the biological context (i.e. different animal vectors), different evolutionary histories (i.e. recent long-distance dispersal), and different landscape context (i.e. less fragmented landscapes vs. highly disturbed landscapes) cannot be decoupled from one another. However, given the conservative flower and fruit morphology within the Magnoliaceae family and the extensive deforestation history of the studied populations, the landscape context is expected to be the main driver.
Unexpectedly, the two subspecies of M. cubensis express low genetic differentiation combined with a high geographic distance, while we find high structuring overall for the other Magnolias. Here, the hypothesis of relatively recent long-distance dispersal is put forward as the most likely explanation to be tested in further research. Similarly, MAR and GUA, the “populations” of M. dodecapetala, were expected to have a higher degree of genetic differentiation compared to the other infraspecific genetic differentiation regardless of the Wahlund effect, given that the populations are separated by ocean and that a “population” on Dominica lies in between that of Guadeloupe and Martinique.
Population statistics: infraspecific level
The high amount of allelic association found in three populations (ROD, LAC, YAJ) is most likely due to a major reduction in population size: a recent bottleneck. This is concluded given that (a) there is genome-wide allelic association for all three populations, in contradicting strengths when compared across populations pairs per species; and (b) the visited locations had a high degree of disturbance. The samples studied of the ROD and LAC populations indicate that they have not been able to recombine their genetic material since the bottleneck. For the YAJ population it cannot be excluded that a high degree of kinship between the samples produced the results. The 20 samples of this population could only be collected at the border of, what is expected to be, a much larger population and include two adults and 18 juveniles. It is recommended to either exclude the population from species-focused analyses, or to recollect a better representation of the population.
We cannot easily label the observed genetic diversity (Table 2) to be healthy, high or low, as there is no related, non-threatened Magnolia species studied for comparison (Spielman et al. 2004). However, comparisons of the population statistics between the studied threatened species can be made. Firstly, when comparing the statistics of the taxon-datasets, the two populations of M. hamorii from the Dominican Republic show a high mean number of alleles (A), in the same extent as the three populations suspected to experience the Wahlund effect. They also have the highest reported values of Ho and He compared with the other Magnolias of this dataset. In the Splendentes-normalized-dataset (dataset 3), the statistics of M. hamorii do not stand out anymore. However, they remain in the higher range of values, now similar to the statistics found for M. cubensis, M. portoricensis and M. splendens. The latter three species also show A- and H-values in the higher range of values in the calculations of their full taxon-datasets.
Secondly, GRA, MAN and ROD report the three lowest A values in their taxon-datasets, and MAN and ROD show lower A and H values than the GRA and BAR populations, respectively. The lower statistics of the GRA and MAN populations confirm that conservation management of Magnolias in the last remaining forests of Haiti is urgent. Interestingly, even though MAN appeared deforested in an equal, or even higher extent than the ROD population, its alleles tested to be independently associated. LD decreases after recombination events at a rate that depends on the recombination frequency and generally takes more than one generation of random mating to restore, even for (physically) unlinked loci (Slatkin 2008). Hence, the combination of highly disturbed forest and independently associated alleles indicates successful pollination events and surviving new recruits for the MAN population.
Thirdly, the population inbreeding coefficients (FIS) of the 14 populations not suspected to be under the Wahlund effect, do not significantly differ from zero. Taking the reproduction biology of Magnolias into consideration, both arguments in favour and against this result can be listed. No (apparent) inbreeding seems likely given that (1) Magnolia flowers are reported to be protogynous (Gibbs et al. 1977; Gottsberger 1977; Thien 1974); (2) trees have characteristics that promote outcrossing (Petit and Hampe 2006); and (3) high outcrossing rates have been found in other Magnolia species (Tamaki et al. 2009). However, (some degree of) inbreeding was expected given that (1) geitonogamy is theoretically possible (Gibbs et al. 1977; Ishida et al. 2003) provided that they express asynchronous flowering and no self-incompatibility mechanisms; (2) the species are classified as threatened due to small population sizes, high disturbance, and small estimations of extent of occurrence (Rivers et al. 2016); and (3) significant inbreeding has been reported for other Magnolias (Kikuchi and Isagi 2002; Sun et al. 2011). It is possible that recent inbreeding remains undetected due to a time-lag (Kramer et al. 2008; Lugo et al. 1981).
In conclusion, the data showed structuring on three different levels. Firstly, the supraspecific structuring confirms high species integrity with no extensive gene flow between species. Secondly, species sets within islands express lower genetic structuring but no signs of current gene flow, which is interpreted as a more recent shared ancestry. Thirdly, the populations within species also show moderate to strong differentiation, uncorrelated with the distance between the population pairs. The generalisation of extensive gene flow in trees does not withhold in the studied species. Our data support the hypothesis that the generalized concept of extensive gene flow in trees mainly applies to wind pollinated trees or trees that have larger animal vectors such as mammals (Dick et al. 2008). In contrast to the strong structuring, there is no sign of inbreeding, indicating ample gene flow within populations and mechanisms favouring outcrossing. Hence, the reproductive biology of the Neotropical Magnolias appears resilient yet limited in their animal mediated dispersal. A fragmented landscape is expected to strengthen this limitation. Hence, in terms of forest conservation, maintenance of – or preferably: an increase of – connectivity between forest patches would be the most effective strategy to ensure the survival of the species. To practically outline and further investigate the forest connectivity for Magnoliaceae, Magnolia SSR research would benefit from studying (1) the reproductive biology of the Magnolia trees (pollinators, seed dispersers and phenology) and its limits, shaping the high genetic differentiation between, and high gene flow within populations; (2) the genetic diversity of closely related non-threatened Magnolia species, either in fragmented or continuous landscapes, placing past and future SSR Magnolia studies on threatened populations in perspective; and (3) splitting Magnolia conservation genetic studies according to age, to exclude this potential time-lag and detect whether or not the younger generation of Magnolia trees are genetically depauperate (e.g. Graignic et al. 2016; Watanabe et al. 2017).
Data archiving
Data available from Dryad: https://doi.org/10.5061/dryad.0m625h4.
Genbank accession numbers for the 63 original sequences on which the primers were developed range from MH923371 to MH923433.
References
Aitken SN, Yeaman S, Holliday JA, Wang T, Curtis-McLane S (2008) Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol Appl 1(1):95–111
Aparicio A, Hampe A, Fernández-Carrillo L, Albaladejo RG (2012) Fragmentation and comparative genetic structure of four mediterranean woody species: complex interactions between life history traits and the landscape context. Divers Distrib 18(3):226–235
Austerlitz F, Mariette S, Machon N, Gouyon PH, Godelle B (2000) Effects of colonization processes on genetic diversity: differences between annual plants and tree species. Genetics 154(3):1309–1321
Bousquet J, Strauss SH, Doerksen AH, Price RA (1992) Extensive variation in evolutionary rate of rbcL gene sequences among seed plants. Proc Natl Acad Sci USA 89(16):7844–7848
Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20(6):1004–1006. 1008–1010
Calonje M, Martín-Bravo S, Dobeš C, Gong W, Jordon-Thaden I, Kiefer C et al. (2009) Non-coding nuclear DNA markers in phylogenetic reconstruction. Plant Syst Evol 282(3):257–280
Cires E, De Smet Y, Cuesta C, Goetghebeur P, Sharrock S, Gibbs D et al. (2013) Gap analyses to support ex situ conservation of genetic diversity in Magnolia, a flagship group. Biodivers Conserv 22(3):567–590
de Queiroz K (1998) The general lineage concept of species, species criteria, and the process of speciation. A conceptual unification and terminological recommendations. In: Howard DJ, Berlocher SH (eds) Endless forms. Species and speciation. Oxford University Press, New York, NY, p 57–75
Dick CW, Hardy OJ, Jones FA, Petit RJ (2008) Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Trop Plant Biol 1(1):20–33
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Ellstrand NC, Whitkus R, Rieseberg LH (1996) Distribution of spontaneous plant hybrids. Proc Natl Acad Sci USA 93(10):5090–5093
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Figlar RB, Nooteboom HP (2004) Notes on Magnoliaceae IV. Blumea 49(1):87–100
Frankham R, Ballou JD, Briscoe DA (2010) Introduction to Conservation Genetics, 2nd edn. Cambridge University Press, Cambridge
García-Morales LJ, Iamonico D, García Jiménez J (2017) Nomenclatural remarks on Magnolia sect. Macrophylla (Magnoliaceae), with description of a new species from North America (Tamaulipas, Mexico). Phytotaxa 309(3):238–244
Gibbs PE, Semir J, da Cruz ND (1977) Floral biology of Talauma ovata St. Hil. (Magnoliaceae). Cienc Cult 29:1436–1441
González Torres LR, Palmarola A, González Oliva L, Bécquer ER, Testé E, Barrios D (2016) Lista roja de la flora de Cuba. Bissea 10(número especial1):1–352
Gottsberger G (1977) Some aspects of beetle pollination in the evolution of flowering plants. Plant Syst Evol Suppl 1:211–226
Graignic N, Tremblay F, Bergeron Y (2016) Genetic consequences of selection cutting on sugar maple (Acer saccharum Marshall). Evol Appl 9(6):777–790
Guichoux E, Lagache L, Wagner S, Chaumeil P, Léger P, Lepais O et al. (2011) Current trends in microsatellite genotyping. Mol Ecol Resour 11(4):591–611
Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48(2):361–372
Hamrick JL (2004) Response of forest trees to global environmental changes. Ecol Manag 197(1–3):323–335
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New (Dordr) 6(1–4):95–124
Hartl DL, Clark AG (1997) Principles of Population Genetics, 3rd edn. Sinauer Associates, Inc, Sunderland, MA
Hoban S, Gaggiotti O, ConGRESS Consortium, Bertorelle G (2013) Sample Planning Optimization Tool for conservation and population Genetics (SPOTG): a software for choosing the appropriate number of markers and samples. Methods Ecol Evol 4(3):299–303
Hodel RG, Segovia-Salcedo MC, Landis JB, Crowl AA, Sun M, Liu X et al. (2016) The report of my death was an exaggeration: A review for researchers using microsatellites in the 21st century. Appl Plant Sci 4(6):1600025
Holleley CE, Geerts PG (2009) Multiplex Manager 1.0: a cross-platform computer program that plans and optimizes multiplex PCR. Biotechniques 46(7):511–517
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand Stat Theory Appl 6(2):65–70
Howard RA (1948) The morphology and systematics of the West Indian Magnoliaceae. Bull Torre Bot Club 75(4):335–357
Ishida K, Yoshimaru H, Itô H (2003) Effects of geitonogamy on the seed set of Magnolia obovata Thunb. (Magnoliaceae). Int J Plant Sci 164(5):729–735
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177(3):309–334
Kalinowski ST, Taper ML (2006) Maximum likelihood estimation of the frequency of null alleles at microsatellite loci. Conserv Genet 7(6):991–995
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4(8):782–788
Kikuchi S, Isagi Y (2002) Microsatellite genetic variation in small and isolated populations of Magnolia sieboldii ssp. japonica. Hered (Edinb) 88(4):313–321
Kramer AT, Ison JL, Ashley MV, Howe HF (2008) The paradox of forest fragmentation genetics. Conserv Biol 22(4):878–885
Kremer A, Le Corre V (2012) Decoupling of differentiation between traits and their underlying genes in response to divergent selection. Hered (Edinb) 108(4):375–385
Larridon I, Walter HE, Guerrero PC, Duarte M, Cisternas MA, Peña Hernández C et al. (2015) An integrative approach to understanding the evolution and diversity of Copiapoa (Cactaceae), a threatened endemic Chilean genus from the Atacama Desert. Am J Bot 102(9):1506–1520
Lewontin RC, Kojima K (1960) The evolutionary dynamics of complex polymorphisms. Evolution 14(4):458–472
Louis EJ, Dempster ER (1987) An exact test for Hardy-Weinberg and multiple alleles. Biometrics 43(4):805–811
Lugo AE, Schmidt R, Brown S (1981) Tropical forests in the Caribbean. Ambio 10(6):318–324
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12(2):111–122
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. II. Gene Freq data J Mol Evol 19(2):153–170
Palmarola A, Romanov MS, Bobrov AVFCh, González-Torres LR (2016) Las magnolias de Cuba: Talauma – taxonomía y nomenclatura. Rev Del Jardín Botánico Nac 37:1–10
Pautasso M (2009) Geographical genetics and the conservation of forest trees. Perspect Plant Ecol Syst 11(3):157–189
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28(19):2537–2539
Pérez ÁJ, Arroyo F, Neill DA, Vázquez-García JA (2016) Magnolia chiguila and M. mashpi (Magnoliaceae): two new species and a new subsection (Chocotalauma, sect. Talauma) from the Chocó biogeographic region of Colombia and Ecuador Phytotaxa 286:267–276
Petit RJ, Hampe A (2006) Some evolutionary consequences of being a tree. Annu Rev Ecol Evol Syst 37:187–214
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
R Core Team (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R–project.org/ URL
Rivers M, Beech E, Murphy L, Oldfield S (2016) The Red List of Magnoliaceae - revised and extended. BGCI, Richmond, UK
Roberge J, Angelstam P (2004) Usefulness of the umbrella species concept as a conservation tool. Conserv Biol 18(1):76–85
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4(1):137–138
Rousset F (2008) GENEPOP'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8(1):103–106
Schlötterer C (2004) The evolution of molecular markers — just a matter of fashion? Nat Rev Genet 5(1):63–69
Selkoe KA, Toonen RJ (2006) Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett 9(5):615–629
Setsuko S, Ishida K, Ueno S, Tsumura Y, Tomaru N (2007) Population differentiation and gene flow within a metapopulation of a threatened tree, Magnolia stellata (Magnoliaceae). Am J Bot 94(1):128–136
Slatkin M (2008) Linkage disequilibrium — understanding the evolutionary past and mapping the medical future. Nat Rev Genet 9(6):477–485
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci USA 101(42):15261–15264
Sun Y, Wen X, Huang H (2011) Genetic diversity and differentiation of Michelia maudiae (Magnoliaceae) revealed by nuclear and chloroplast microsatellite markers. Genetica 139(11-12):1439–1447
Tamaki I, Setsuko S, Tomaru N (2009) Estimation of outcrossing rates at hierarchical levels of fruits, individuals, populations and species in Magnolia stellata. Hered (Edinb) 102(4):381–388
Thien LB (1974) Floral Biology of Magnolia. Am J Bot 61(10):1037–1045
Thiers, B. [continuously updated]. Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/science/ih/.
Tozaki T, Mashima S, Hirota K, Miura N, Choi-Miura N, Tomita M (2001) Characterization of equine microsatellites and microsatellite-linked repetitive elements (eMLREs) by efficient cloning and genotyping methods. DNA Res 8(1):33–45
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4(3):535–538
Vartia S, Collins PC, Cross TF, Fitzgerald RD, Gauthier DT, McGinnity P et al. (2014) Multiplexing with three-primer PCR for rapid and economical microsatellite validation. Hereditas 151(2–3):43–54
Vavrek MJ (2011) fossil: Palaeoecological and palaeogeographic analysis tools. Palaeontol Electron 14(1):1–16
Vázquez-García JA, Véliz-Pérez ME, Tribouillier-Navas E, Muñiz-Castro MA (2013a) Magnolia quetzal and Magnolia mayae, a new species and a new record, respectively, for the flora of Guatemala. Phytotaxa 76(1):1–6
Vázquez-García JA, Pérez-Farrera MÁ, Martínez-Camilo R, Muñiz-Castro MÁ, Martínez-Meléndez N (2013b) Magnolia lacandonica (subsection Talauma, Magnoliaceae), a new rainforest species from Chiapas, Mexico Phytotaxa 79:30–36
Waples RS (2015) Testing for Hardy-Weinberg proportions: have we lost the plot? J Hered 106(1):1–19
Ward SM, Jasieniuk M (2009) Sampling weedy and invasive plant populations for genetic diversity analysis. Weed Sci 57(6):593–602
Watanabe S, Kaneko Y, Maesako Y, Noma N (2017) Detecting the early genetic effects of habitat degradation in small size remnant populations of Machilus thunbergii Sieb. et Zucc. (Lauraceae). Int J For Res 2017:1–7
Weber JL (1990) Informativeness of human (dC-dA)n · (dG-dT)n polymorphisms. Genomics 7(4):524–530
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370
Wheeler GL, Dorman HE, Buchanan A, Challagundla L, Wallace LE (2014) A review of the prevalence, utility, and caveats of using chloroplast simple sequence repeats for studies of plant biology. Appl Plant Sci 2(12):1400059
Whittle CA, Johnston MO (2003) Broad-scale analysis contradicts the theory that generation time affects molecular evolutionary rates in plants. J Mol Evol 56(2):223–233
Xin Z, Chen J (2012). A high throughput DNA extraction method with high yield and quality. Plant Methods 8(26).
Zhao X, Ma Y, Sun W, Wen X, Milne R (2012) High genetic diversity and low differentiation of Michelia coriacea (Magnoliaceae), a Critically Endangered endemic in Southeast Yunnan, China. Int J Mol Sci 13(4):4396–4411
Acknowledgements
We are grateful to the Cuban, Dominican, Haitian, Mexican, Puerto Rican, and US authorities for issuing the permits CICA-CITMA 15/16, DQCSPAP/SIC/365, VAPB-01488, SGPA/DGGFS/712/1643/13, 2015-EPE-007 and 2016-EPE-011, and CNF-2122, respectively. This work would not have been accomplished without the support of Fondation Franklinia, Arboretum Wespelaar, King Leopold III Fund for Nature Exploration and Conservation, Research Foundation Flanders (V445315N; K210116N; V431816N), the Instituto de Ecología, A.C., the Magnolia Society International, and Ghent University (BOF.DOC.2014.0062.01). Lastly, we are most grateful for all the helping hands during (organising) field work and lab work: James Ackerman, Fabiola Areces Berazaín, Franklin Axelrod, Félix Bastaraud, Jean-Francois Orilién Beauduy, William Cinéa, Michel Delblond, Emmanuel Giraud, Francisco Hernández Najarro, Jean Lubin, Hervé Magnin, Victor Martin-Velez, Esteban Martínez, Ruud Merkx, Rubén Padrón Vélez, Guy-André Pitoula, Rosa Rodríguez Peña, Ranza Rancé, Gerardo Rodríguez Guzmán, Alain Rousteau, Stéphanie Saint-Aimé, Eugenio Santiago-Valentin, Joel Timyan, Christian Torres Santana, Roland Trézil and Guy Van-Laere.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Veltjen, E., Asselman, P., Hernández Rodríguez, M. et al. Genetic patterns in Neotropical Magnolias (Magnoliaceae) using de novo developed microsatellite markers. Heredity 122, 485–500 (2019). https://doi.org/10.1038/s41437-018-0151-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-018-0151-5
This article is cited by
-
Genome-wide assessment of population genetic and demographic history in Magnolia odoratissima based on SLAF-seq
Conservation Genetics (2023)
-
High genetic diversity but spatially restricted genetic connectivity in a tropical montane cloud forest tree (Magnolia schiedeana)
Tree Genetics & Genomes (2023)
-
An integrative study of species distribution modelling and conservation genetics: Magnolia in Hispaniola
Biodiversity and Conservation (2023)
-
Conservation genetics of relict tropical species of Magnolia (section Macrophylla)
Conservation Genetics (2021)
-
Effect of the landscape on functional and spatial connectivity in Magnolia cubensis (Magnoliaceae) in two mountain massifs of Cuba
Conservation Genetics (2021)