Abstract
The silkworm cocoon colour has attracted researchers involved in genetics, physiology and ecology for a long time. ‘Ryokuken’ cocoons are yellowish green in colour due to unusual flavonoids, prolinylflavonols, while ‘Sasamayu’ cocoons are light green and contain only simple flavonol glucosides. We found a novel gene associated with the cocoon colour change resulting from a change in flavonoid composition and named it Lg (light green cocoon). In the middle silk glands of the +Lg/+Lg larvae, 1-pyrroline-5-carboxylic acid (P5C) was found to accumulate due to a decrease in the activity of pyrroline-5-carboxylate reductase (P5CR), an enzyme reducing P5C to proline. Sequence analysis of BmP5CR1, the candidate gene for Lg, revealed a 1.9 kb insertion and a 4 bp deletion within the 1st intron, a 97 bp deletion within the 4th intron, and a > 300 bp insertion within the 3′-UTR, in addition to two amino acid changes on exons 3 and 4 in +Lg/+Lg compared to Lg/Lg. Decreased expression of BmP5CR1 was observed in all of the investigated tissues, including the middle silk glands in +Lg/+Lg, which was probably caused by structural changes in the intronic regions of BmP5CR1. Furthermore, a BmP5CR1 knockout strain exhibited a yellowish green cocoon with the formation of prolinylflavonols. These results indicate that the yellowish green cocoon is produced by a BmP5CR1 deficiency. To our knowledge, this is the first report showing that the defect of an enzyme associated with intermediate metabolism promotes the conjugation of phytochemicals derived from foods with endogenously accumulating metabolites in animal tissues.
Similar content being viewed by others
Introduction
There are many cocoon colour mutants among the silkworm strain Bombyx mori (Fujii et al. 1998; Banno et al. 2005). It has been shown that green cocoon shells contain flavonoids (Fujimoto and Hayashiya 1972; Tamura et al. 2002; Kurioka and Yamazaki 2002; Hirayama et al. 2006), and yellow and pinkish cocoon shells contain carotenoids (Harizuka 1953; Tazima 1964), both of which are derived from the leaves of the mulberry tree (Morus alba), the host plant of the silkworm. These phytochemicals are absorbed in the midgut, transported to silk glands through the haemolymph, and then secreted into the sericin layer of the cocoon filament (Harizuka et al. 1960). It has been shown that the phenotypes of the yellow and cream yellow (flesh-coloured) cocoons, the constituents of which are lutein and β-carotene, respectively, are regulated by the Y, C, and F genes, following a classical Mendelian pattern (Tazima 1964; Sakudoh et al. 2007). The molecular mechanisms of these genes have already been clarified (Sakudoh et al. 2007, 2010, 2013). The inheritance pattern for the flavonoid cocoon is much more complicated than that for the carotenoid cocoon, mainly because the metabolic process of flavonoids ingested from diet is much more complex than that of carotenoids. In the case of cocoon carotenoids, the main constituents are lutein and β-carotene, both of which originally existed in mulberry leaves (Harizuka et al. 1960; Tazima 1964). Therefore, carotenoids are metabolised very little inside the insect. However, in the case of flavonoids, host plant constituents are rarely present in the cocoons, showing the active metabolism of flavonoids in the insect tissues. In mulberry leaves, flavonol glucosides with a sugar group at the 3-O-position in the C-ring, such as quercetin-3-O-glucoside, quercetin-3-O-malonylglucoside, and quercetin-3-O-rutinoside, are naturally occurring (Naito 1968; Onogi et al. 1993; Doi et al. 2001; Katsube et al. 2006). However, these glucosides are hydrolysed and modified in the absorption process. The aglycon form of quercetin was preferably glucosylated at the 5-O-position in the midgut, which is the first step in the biosynthesis of cocoon flavonoids of the Sasamayu strains (light green cocoons) (Hirayama et al. 2008). Recently we found that this region-specific glucosylation of quercetin is regulated by the Gb locus and that Bm-UGT10286, a UDP-glucosyltransferase gene with preferred 5-O-regioselectivity for quercetin, is responsible for Gb (Daimon et al. 2010). Quercetin 5-O-glucoside is transported into the silk glands and further glucosylated mainly at the 4′-O position (Hirayama et al. 2008). It has been considered that a more complicated flavonoid metabolism must occur in Ryokuken strains (yellowish green cocoons), such as Daizo, since the cocoons of these strains contain >30 kinds of flavonoids, while the cocoons of the Sasamayu strains have a small repertoire of flavonoids, which are all simple flavonol glucosides (Hirayama et al. 2009). Many flavonoids in the Ryokuken cocoon are considered to be abnormal, with the proline moiety in their molecules, which is related to C-prolinylquercetins, prolinalin A and B (Hirayama et al. 2006). We named these unique flavonoids prolinylflavonols (Hirayama et al. 2009).
In the process of making chromosomal substitution lines (CSLs) or ‘consomics’ of the silkworm using two strains, Daizo (Ryokuken cocoon) and J01 (white cocoon), we realised that the cocoon colour changed from yellowish green to light green due to a significant reduction in the prolinylflavonols when the Daizo chromosome 6 was replaced by that of J01. By backcross analysis using progeny generated by the mating of Daizo with DH6, a strain that has a chromosome 6 pair derived from J01 with the genetic background of Daizo, we confirmed that chromosome 6 carries a gene involved in the regulation of prolinylflavonol synthesis and named this gene Lg, for light green cocoon. In this study, we report the characterisation and identification of the Lg responsible for producing a light green cocoon. We demonstrate that Lg encodes BmP5CR1, a pyrroline-5-carboxyrate reductase 1, which catalyses the reduction of 1-pyrroline-5-carboxylic acid (P5C) to L-proline, and that prolinylflavonols, the substances responsible for the yellowish green cocoons, are produced by a defect in BmP5CR1. Our results provide new insight on the interaction between intermediary metabolites, such as P5C, and exogenous substances, such as flavonoids derived from foods in animals.
Materials and Methods
Insects
Daizo (Matsumura), J01 and DH6 are strains maintained at the National Agriculture and Food Research Organization, NARO. Daizo is a strain with a yellowish green cocoon, while J01 is a strain with a white cocoon (Mase et al. 2011). DH6 is a strain of the DH series, which is chromosomal substitution line (CSL), or consomic, with the Daizo genetic background (Fig. S1). Each strain of the DH series carries each pair of chromosomes substituted by successive backcrossing with Daizo and its sib-mating. The substituted chromosomes are marked with three DNA markers. Therefore, DH6 has a chromosome 6 pair derived from J01. These silkworms were reared on mulberry leaves or a semi-synthetic diet (Table S1) at 25 °C.
Analysis of flavonoids
Flavonoids were extracted from tissues and cocoons and analysed using an LC/MS system as described previously (Hirayama et al. 2008). Briefly, an aliquot (10 µl) of a sample was injected into an HP 1100 series HPLC equipped with an 1100 MSD mass spectrometer (Agilent Technologies, CA, USA) and separated by a C18 reverse phase column, 100 × 2.0 mm i.d. (Sunfire C18, Waters, MA, USA), at a flow rate of 0.3 ml/min. The column temperature was maintained at 40 °C. The mobile phase consisted of solvents A (0.2% aq. formic acid) and B (0.2% formic acid in acetonitrile). Flavonoids were separated using a linear gradient from 7% B to 40% B over 40 min and then to 100% B for 5 min. We followed the other analysis conditions described previously (Hirayama et al. 2013).
In the present study, we also used a simple method to roughly estimate the amount and composition of the cocoon flavonoids. The amount of flavonoids was estimated from the absorbance at 365 nm of the cocoon extracts using quercetin as a standard. In addition, we determined the rough chemical composition of cocoons based on the absorbance ratio A420/A365 because its value was correlated with the ratio of prolinylflavonols to total flavonoids in the cocoons (Hirayama and Okada 2014).
Measurement of P5C
Daizo and DH6 larvae were reared on a semi-synthetic diet (Table S1). Five-day-old fifth instar larvae were dissected to collect posterior and middle silk glands. These tissues were rinsed well with an ice-cold 0.85% KCl solution, then rapidly frozen and stored at −80 °C until analysis.
The tissues were homogenised in four volumes of 70% (V/V) methanol and centrifuged for 5 min at 20,000 × g at 4 °C. The supernatant was mixed with four volumes of o-aminobenzaldehyde solution (1.25 mg/ml ethanol) and incubated at 37 °C for 10 min. Absorbance at 440 nm was recorded, and P5C contents in the samples were calculated using a commercially available DL-P5C as a standard.
Enzyme assays
Day 5 fifth instar larvae were dissected to collect tissues and organs. Results were expressed as µmol product/min per mg protein. Protein in the tissue extracts was determined using a commercial assay kit (Coomassie Plus, Pierce, Rockford, IL, USA) with BSA as a standard.
For the P5CR assay, the collected tissues were homogenised in nine volumes of 50 mM potassium phosphate and 0.25 M sucrose (pH 7.0). The homogenates were centrifuged at 20,000 × g at 4 °C for 15 min, and the supernatant was used for the enzyme assays. The tissue extracts were incubated in 50 mM potassium phosphate (pH 6.8), 0.5 mM NADH, and 1.0 mM P5C at 37 °C to monitor the decrease in the absorbance of NADH at 340 nm.
P5CDH, pyrroline 5-carboxylate dehydrogenase, was assayed according to the previous paper with some modifications (Herzfeld et al. 1977). The enzyme extracts were the same as those used in the P5CR assay. The reaction mixture consisted of 50 mM potassium phosphate (pH 7.5), 2.0 mM NAD and 0.5 mM P5C. NADH formation at 37 °C was monitored at 340 nm.
OAT, ornithine aminotransferase, activity was also determined according to the previous method (Rosenthal and Dahlman 1990). The collected tissues were homogenised in four volumes of 0.1 M phosphate buffer (pH 7.5). The supernatant obtained after centrifugation at 15,000 × g for 10 min was used for the enzyme assay. An assay mixture of 0.5 ml containing 25 mM ornithine, 20 mM α-ketoglutarate, 1 mM pyridoxal phosphate, and enzyme extract was used. After incubation at 37 °C for 60 min, the reaction was stopped with the addition of 0.1 ml of 50% TCA solution. Then, the absorbance at 440 nm was recorded after 10 min of incubation at 37 °C with 0.4 ml of o-aminobenzaldehyde solution.
Oral administration of gabaculine, an inhibitor of OAT
To investigate the effect of P5C deficiency on the formation of prolinylflavonols in +Lg/+Lg individuals, gabaculine, a potent inhibitor of OAT, was orally supplied to Daizo (+Lg/+Lg) larvae. Day 0 fifth instar Daizo larvae were reared on two diets. One was a semi-synthetic diet supplemented with rutin, the main flavonoid in mulberry leaves (Table S1). The other was the same as the rutin-supplemented diet but contained 20 mg of gabaculine/100 g of diet. After 5 days of feeding with the test diets, the larvae were dissected, and the middle silk glands were collected to analyse the flavonoids in the tissues using the LC/MS system as described above.
Linkage analysis between Lg and BmP5CR1
To evaluate the relationship between the Lg phenotype and the BmP5CR1 genotype, a linkage analysis of Daizo (+Lg/+Lg) and DH6 (Lg/Lg) was performed. The male moth of the F1 progeny between Daizo and DH6 was crossed with a Daizo female. The phenotypes of the cocoons obtained from the backcrossed individuals (BC1) were confirmed using a simple method for the determination of flavonoid composition as described above (Hirayama and Okada 2014). Genomic DNA was isolated using DNAzol (Molecular Research Centre, Cincinnati, USA) from an individual pupa after distinguishing the flavonoid compositions. The individual BmP5CR1 genotype was analysed via genomic PCR using a specific primer set, 9658 F as a forward primer and 9063R as a reverse primer (Table S2). The PCR reaction was carried out with 2.5U Ex Taq polymerase (TaKaRa, Japan) under 35 cycles of denaturation at 95 °C, annealing at 60 °C, and extension at 72 °C for 30, 60 and 60 s, respectively, after hot starting at 95 °C for 3 min using a Thermal Cycler 9800 Fast (Applied Biosystems, Foster City, CA, USA). To discriminate between the genotypes, the specific bands were detected using 1.2% agarose gel electrophoresis in TBE-buffer (50 mM Tris base, 48 mM Boric acid, 2 mM EDTA, pH 8.0) at 50 V for 1 h and stained with 1 µg/ml ethidium bromide.
Cloning and sequencing of the 5′- and 3′-UTRs of BmP5CR1
Total RNA was extracted from the middle silk gland of Daizo and DH6 in 3-day-old fifth instar larvae using ISOGEN (Nippon Gene, Japan). The poly(A)+RNA was prepared from 200 µg of total RNA using an Oligotex-dT30<Super>mRNA purification kit (TaKaRa, Japan). The first and second-strand complementary DNA (cDNA) was generated from this poly(A)+RNA using a Marathon cDNA Amplification Kit (Clontech, CA, USA), and 5′- and 3′-RACE were also performed according to this manuscript. The 5′-upstream and 3′-downstream regions were amplified by touchdown PCR (denature: 94 °C, 5 s; anneal/elongate: 74 °C, 2 s, 5 cycles; A/E: 72 °C, 2 s, 5 cycles; and A/E: 70 °C, 2 s, 20 cycles) using adapter primer 1 and 0269R on exon 3 or 9827 F on exon 4 and then increased by nested PCR with the same conditions using adapter primer 2 and 1317R on exon 2 or 9765F on exon 4.
A clear single band of the amplified PCR fragment was removed with a laser blade from a 1% GTG agarose gel (Lonza, Switzerland) after electrophoresis at 50 V for 1 h and chopped into small pieces. These pieces were crushed down into the lower microtube through the small pore at the bottom of the upper microtube by centrifugation at 9500 × g for 5 min. The amplified DNA fragment was extracted with an equal volume of phenol (pH 7.5) from the crushed gel via the process of freezing and melting, purified by phenol-chloroform, and precipitated with 0.3 M sodium acetate/2.5×volume of ethanol at 21,500 × g for 20 min. The purified DNA fragment was cloned into a pGEM-T easy vector (Promega, Madison, WI, USA). Cloned DNAs were sequenced with the ABI PRISM Big Dye Terminator Cycle Sequencing Kit and the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Genomic PCR polymorphism and sequencing of BmP5CR1 intron
Genomic DNA was extracted from the posterior silk glands of Daizo and DH6 strains in 2-day-old fifth instar larvae according to the previously described phenol-chloroform methods (Hara 1996). The specific primers, 1F, 1317F and 9063R, were designated based on the full-length cDNA sequence, including the 5′-upstream region of BmP5CR1. The BmP5CR1-specific region was amplified from 10 ng of genomic DNA using 2.5 U Ex Taq or LA Taq HS polymerase (TaKaRa, Japan). The PCR parameters were 35 cycles of D: 95 °C, A: 60 °C and E: 72 °C for 30, 60 and 60 s after hot start at 95 °C for 3 min. The PCR products were detected on 1% agarose gels by electrophoresis and ethidium bromide staining. The clear single band of the PCR fragment was cloned into a pGEM-T easy vector and sequenced, as described above. The purified DNA fragments were also sequenced directly with a specific primer when it was difficult to clone them into the vector.
RT-PCR
Total RNA from each tissue, midgut, fat body, muscle, Malpighian tubule, testis, ovary, and three parts of the silk gland in 3-day-old fifth instar larvae was also prepared using ISOGEN (Nippon Gene, Japan). The first-strand cDNA was generated from 4 µg of total RNA of each using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare, Buckinghamshire, UK) with an oligo dT primer. The region from transcription initiation to terminator (1F-9063R) and to the 1st intronic region on the Daizo genome (1F-5924R) of BmP5CR1 was amplified by 2.5 U Ex Taq polymerase or LA Taq polymerase (TaKaRa, Japan). The PCR parameters were 35 or 28 cycles at 95 °C for 30 s, 60 °C for 1 min and 72 °C for 1 min after hot starting at 95 °C for 3 min, and the products were detected using 1% agarose gel electrophoresis. The cDNA region of BmP5CR2 (413F-252R) was also amplified using 28 cycles with the same condition described above.
Northern hybridisation
In total 2 and 20 µg of total RNA from the middle silk glands in 3-day-old fifth instar larvae of Daizo and DH6 were also used for Northern blot analysis using a DIG Northern Starter Kit (Roche, Switzerland). The total RNA was denatured at 65 °C for 10 min in loading buffer (MOPS, 50% formamide, 6.17% formaldehyde, 10% glycerol and 0.05% BPB) and electrophoresed on a 1.2% agarose gel containing 2% formaldehyde. After blotting onto positively charged nylon membranes (Roche, Switzerland) with 20XSSC, the RNA was hybridised with DIG-labelled BmP5CR1 riboprobe at 68 °C overnight. The membrane was reacted with an anti-DIG-AP conjugate in 1% blocking solution after adequately washing it with 2XSSC at RT and 0.1XSSC at 68 °C for 15 min. The hybridisation signal was chemically detected by CDP-Star on X-ray film, Super RX (Fuji film, Japan).
Generation of the BmP5CR1 knockout silkworm
Construction of TALENs and generation of a BmP5CR1 knockout silkworm strain was performed as reported previously (Takasu et al. 2013; 2014). A pair of TALENs was designed to bind the 5′-GATGTTGTTAACAACTGT-3′ sequence of the sense strand and 5′-GGCATAAAGCCAAAT-3′ sequence of the antisense strand in the 2nd exon of BmP5CR1, as the region was predicted to be within the NADH binding domain. The repeat variable diresidue (RVD)-encoding sequence was constructed using a Golden Gate TALEN and TAL Effector Kit (Addgene, Cambridge, MA, USA) and inserted into the backbone vector pBlue-TAL (Takasu et al. 2013). The constructed plasmids were purified using a HiSpeed Plasmid Midi Kit (Qiagen), linearised by Xba I, treated with proteinase K, extracted with phenol/chloroform/isoamyl alcohol (25:24:1) and chloroform, precipitated with ethanol, and washed with 70% ethanol three times. The TALEN mRNA was transcribed using an mMESSAGE mMACHINE Kit (Applied Biosystems, Foster City, CA, USA), followed by lithium chloride precipitation, and was then washed three times with 70% ethanol. The resulting TALEN mRNA was dissolved in an injection buffer (0.5 mM phosphate buffer (pH 7.0) and 5 mM KCl) to achieve a final concentration of 0.5 mg/ml for each TALEN mRNA and was microinjected into non-diapause DH6 eggs 3–5 h after laying.
The G0 female moth, the only individual that we obtained from 192 injected eggs, was crossed to a DH6 male moth. The G1 adult moths that produced yellowish green cocoons were crossed with each other to obtain G2 eggs, and then, the genotype of each G1 moth was determined. The genomic DNA was extracted from G1 moths by phenol-chloroform methods, the targeted region was amplified by PCR with the specific primers 1313 F and 1130 R (Table S2), and the PCR product was directly sequenced. The yellowish green cocoons in the G2 population were selected again, and G3 eggs were produced. The mutation caused by TALENs was confirmed by the sequence of the BmP5CR1 transcript extracted from G3 larvae.
Phylogenic analysis
Five kinds of animals out of four orders were selected as vertebrates, and four orders of insects and a nematode were selected as invertebrates for the phylogenic analysis of P5CR. The deduced amino acid sequences of the P5CR homologues were aligned with BmP5CR1 and BmP5CR2 using the Muscle program implemented in MEGA6 (Tamura et al. 2013). The P5CR sequences of rice and Chlamydomonas were also added for this analysis. The molecular phylogenic analysis was conducted via the neighbour-joining method using MEGA6. The evolutionary distances were computed using a JTT matrix-based method. The confidence of the phylogenic lineages was assessed by bootstrap analyses of 1000 resamplings.
Results
Effect of the Lg gene on the flavonoid composition of the cocoon and tissues
As shown in Fig. 1a, cocoons of the DH6 line (Lg/Lg) are light green (Sasamayu), and Daizo cocoons (+Lg/+Lg) are yellowish green (Ryokuken). As the DH6 line is a Daizo CSL in which a chromosome 6 pair was replaced with that of J01 by successive backcrossing and DNA marker assisted selection, this finding apparently indicates that Lg is located on chromosome 6 (Fig. S1). This cocoon colour difference is attributable to the difference in flavonoid composition of the cocoons. Almost all flavonoids in the cocoons of DH6 (Lg/Lg) are simple flavonol glucosides, such as quercetin glucosides, while the cocoons of Daizo (+Lg/+Lg) contain unique prolinylflavonols as the major constituents (Fig. S2), convincing us that the Lg gene on chromosome 6 suppresses the synthesis of prolinylflavonols in insect tissues. To investigate the effect of the Lg gene on the profile of tissue flavonoids, we analysed flavonoids in the midgut, haemolymph, middle and posterior silk glands of Daizo (+Lg/+Lg) and DH6 (Lg/Lg). No prolinylflavonols were found in the midgut, haemolymph, or posterior silk glands of the +Lg/+Lg or Lg/Lg larvae. However, these unusual compounds were present specifically in the middle silk glands of the +Lg/+Lg larvae (Fig. 1b) suggesting that these flavonoids were synthesised specifically in the middle silk glands of +Lg/+Lg larvae.
(a) Yellowish green cocoon (Ryokuken) of Daizo, and light green cocoon (Sasamayu) of DH6. The cocoon colour depends on the flavonoid composition in the cocoon (see Fig. S2). (b) Tissue distribution of flavonoids in the 5-day-old fifth instar larvae. For determination of the tissue flavonoids, HPLC–ESI–MS analysis was carried out (see text and Fig. S2). The bars indicate the means ± SD (n = 5). **P < 0.001
Metabolism of P5C in the middle silk glands of Daizo (+Lg/+Lg) and DH6 (Lg/Lg) larvae
Since extracts from the middle silk glands of Daizo (+Lg/+Lg), but not DH6 (Lg/Lg), contain prolinylflavonols (Fig. 1b), we speculated that 1-pyrroline 5-carboxylate (P5C) could also be accumulated in the middle silk glands of +Lg/+Lg larvae, enhancing the formation of prolinylflavonols. To evaluate the hypothesis that prolinylflavonols could be formed by the accumulation of P5C, the P5C content was investigated in the middle and posterior silk glands of the Lg/Lg and +Lg/+Lg larvae reared on a semi-synthetic diet that did not contain any flavonoids (Table S1), and P5C was found to be specifically accumulated in the middle silk glands of the +Lg/+Lg larvae (Fig. S3).
As shown in Fig. 2a, P5C is an intermediary metabolite in the proline synthesis process. The activities of several enzymes involved in this metabolism were assayed to investigate the cause of P5C accumulation in the middle silk glands of the +Lg/+Lg larvae. Among the enzymes investigated in the present study, P5CR activity in the middle silk glands of the +Lg/+Lg larvae was significantly lower than that of the Lg/Lg larvae, although the ornithine aminotransferase (OAT) and pyrroline 5-carboxylate dehydrogenase (P5CDH) activities did not show any differences between the two genotypes (Fig. 2b).Since P5CR is the enzyme catalysing the P5C reduction to proline, a defect in this enzyme could cause an accumulation of P5C in the middle silk glands. In addition, the activities of OAT, P5CDH, and P5CR in the posterior silk glands were also investigated, and we found that only P5CR activity was significantly decreased in the +Lg/+Lg larvae (data not shown). This result suggests that the ornithine supply is very low in the posterior silk glands, as P5C was not accumulated in the posterior silk glands even in the +Lg/+Lg larvae.
(a) A schematic diagram for the metabolism of 1-pyrroline 5-carboxylate in the middle silk glands of Bombyx mori. GSA; glutamate semialdehyde, P5C; 1-pyrroline 5-carboxylate. Dotted line indicates the deficiency of P5CR in Daizo (+Lg/+Lg). Frame indicates the accumulation of the corresponding compounds in Daizo (+Lg/+Lg). (b) Activity of the enzymes in the middle silk glands of Daizo (+Lg/+Lg) and DH6 (Lg/Lg). OAT ornithine aminotransferase, P5CDH pyrroline 5-carboxylate dehydrogenase, P5CR pyrroline 5-carboxylate reductase. The bars indicate the means ± SD (n = 5). **P < 0.001
Furthermore, to confirm that an excess accumulation of P5C was responsible for the formation of prolinylflavonols in the Daizo (+Lg/+Lg) larvae, we attempted to inhibit P5C accumulation by oral administration of gabaculine, an inhibitor of OAT. OAT is the enzyme that supplies P5C via the transamination of ornithine to glutamate semi-aldehyde (GSA), which can be easily converted to P5C by a non-enzymatic reaction (Fig. 2a). As a result of the feeding experiment using gabaculine, the proline flavonol content in the middle silk glands of the Daizo (+Lg/+Lg) larvae significantly decreased (Fig. S4), strongly suggesting that the formation of prolinylflavonols was controlled by the supply of and demand for P5C in the middle silk glands.
Linkage analysis between the Lg phenotype and BmP5CR1 genotype
Based on the obtained biochemical data, we speculated that a substantial reduction of P5CR activity was responsible for the accumulation of P5C, causing the formation of unusual flavonoids with a proline residue. A search of the integrated genome database of the silkworm (KAIKObase, http://sgp.dna.affrc.go.jp/KAIKObase/; Shimomura et al. 2009) revealed the existence of two putative P5CR-like genes named BmP5CR1 and BmP5CR2. The sequence of BmP5CR1 is located on Bm_scaf 11, which is a scaffold belonging to chromosome 6, while that of BmP5CR2 is located on Bm_scaf 123, a scaffold belonging to chromosome 27, convincing us that BmP5CR1 is a candidate gene for Lg. Thus, we focused on BmP5CR1 in subsequent experiments.
First, we designed several specific primer sets based on the sequence data of the full-length BmP5CR1 cDNA [AK384198] and compared its amplified fragment size to the genomic PCR experiment between Daizo (+Lg/+Lg) and DH6 (Lg/Lg) (Table S2). The band sizes of both strains were distinguishable (Daizo: 600 bp; DH6: 700 bp) when the specific primer set (9658F and 9063R) on exons 4 and 5 of BmP5CR1 was used, indicating that this primer set is available as a genetic marker (Figs. 3b and 4b).
(a) Classification of the cocoons of BC1 individuals, Daizo x (Daizo×DH6), based on the absorbance ratio A420/A365 of the cocoon extracts. BC1 cocoons were clearly divided into two groups, showing that chromosome 6 of DH6 carries a single gene, Lg, which regulates the flavonoid composition in the cocoon. (b) Different size bands of the fragment amplified by the specific primer set (9658F and 9063R; see Fig. 1b and Table S2) of BmP5CR1 between the two parental strains, Daizo (+Lg/+Lg) and DH6 (Lg/Lg). M is the DNA ladder marker. (c) Coincident segregation between cocoon colour distinguished by A420/A365 and the banding pattern of the amplified BmP5CR1 in the BC1 population
(a) Genomic PCR analysis of the three parts of BmP5CR1 by Ex or LA Taq polymerase HS. M is the DNA ladder marker. (b) Schematic representation of the BmP5CR1 gene structure. Lines and rectangular boxes show introns and exons, respectively. White boxes indicate the untranslated regions (UTRs), and grey boxes are the coding regions. Arrows indicate differences between Daizo and DH6. Arrowheads are primer sites used in this study. Numbers show the length (bp) of each region
To evaluate the genetic relationship between the Lg phenotype and the BmP5CR1 genotype, we performed linkage analysis using the band-size difference described above. We obtained F1 progenies between Daizo (+Lg/+Lg) and DH6 (Lg/Lg) and backcrossed the F1 male with the Daizo (+Lg/+Lg) female because genetic recombination only occurs in the testis of B. mori (Sturtevant 1915). The cocoon colour phenotypes were estimated by the absorbance ratio A420/A365 of methanolic cocoon extracts because it was sometimes difficult to distinguish the cocoon colours by mere visual observation (Fig. S5; Hirayama and Okada 2014). Then, the phenotypes of 124 individuals in the BC1 progenies were confirmed based on the absorbance ratio A420/A365, resulting in yellowish green cocoon (+Lg/+Lg; A420/A365 > 0.2) and almost light green cocoon (Lg/+Lg; A420/A365 < 0.15) phenotypes segregated at a ratio of 70:54, significantly fitting a 1:1 ratio (Fig. 3a and c; χ2 = 2.065, P = 0.1508) . Next, we isolated the genomic DNA from pieces of individual pupae of the 124 BC1 progenies and detected the PCR fragment size polymorphism by the specific primer set (9658F and 9063R) of BmP5CR1. The yellowish green cocoons showed all the Daizo band patterns (600 bp), whereas the band patterns of most light green phenotypes were the F1 type (600 + 700 bp), indicating no genetic recombination between the cocoon colour and band pattern (Fig. 3c). This result suggested that Lg is a single gene regulating the flavonoid composition in the cocoon and is probably BmP5CR1.
Comparison of the genomic structure of BmP5CR1 between +Lg/+Lg and Lg/Lg
The difference between both genotypes was investigated by genomic PCR analysis using the specific primer set, 1F (transcription initiation site) on exon 1 and 1317R on exon 2 or 1F and 9063R, which includes the stop codon of BmP5CR1. The longer 1.9 kb band compared to that of DH6 was detected in Daizo, suggesting a 1.9 kb insert sequence in the 1st intron of Daizo BmP5CR1 (Fig. 4a).
Based on the full-length cDNA sequence of BmP5CR1in the genome database, we determined the exons and introns and both sides of the untranslated regions (UTRs) of the +Lg/+Lg and Lg/Lg genotypes. Comparing these sequences between both genotypes, we found four base changes in the coding region, including two amino acid changes, threonine to proline and alanine to valine (Fig. 4b and S6). An approximately 300 bp insertion, a small insertion, a small deletion, and several base changes within the 3’-UTR were also recognised in the Daizo (+Lg/+Lg) genome. Furthermore, the 99 bp deletion in the 4th intron, which is the band-size difference used for the linkage analysis, a 1903 bp insertion and a 4-base (TAAC) deletion in the 1st intron were recognised in the Daizo (+Lg/+Lg) genome (Fig. 4b and S6). Similar sequences with this large insertion in the 1st intron were found to be present on chromosomes 4, 9 and 18 using BLASTn. The insertion also has sequences in common with some non-LTR retrotransposons, suggesting that the sequence may be a movable genetic element. Notably, the 4-base (TAAC) deletion in the 1st intron was observed only at the eukaryote branch site (YNCURAC) of the region upstream of the 3′ splice site in the intron.
Expression analysis of BmP5CR1 in the +Lg/+Lg and Lg/Lg larvae
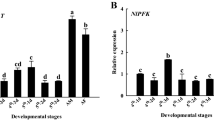
We then compared the mRNA expression of BmP5CR1 in several Daizo (+Lg/+Lg) and DH6 (Lg/Lg) tissues by RT-PCR analysis using the specific primer set (1F and 9063R). BmP5CR1 was strongly expressed in the muscle, Malpighian tubules, testis, ovary, and each part of the silk glands of DH6 (Lg/Lg) larvae (Fig. 5a). Weaker expression was also detected in the midgut and fat body of DH6 (Lg/Lg) larvae. On the other hand, no expression was shown in the tissues of Daizo (+Lg/+Lg) larvae except for a slight band in the testis. This result supports the theory that BmP5CR1 is responsible for Lg.
(a) RT-PCR patterns obtained by using the specific primer sets of BmP5CR1 (1 F and 9063 R) and BmP5CR2 (413 F and 252 R) on each different tissue; MG midgut, FB fat body, ML muscle, MT Malpighian tubules, ASG anterior silk glands, MSG middle silk glands, PSG posterior silk glands, TES testis, OV ovary. Each tissue was collected from 3-day-old fifth instar larvae of the two strains, Daizo (+Lg/+Lg) and DH6 (Lg/Lg). The PCR conditions for BmP5CR1 and BmP5CR2 are Denature: 94 °C-30 s, Anneal: 60 °C-60 s, and Elongate: 72 °C-60 s, 35 cycles, and D: 94 °C-30 s, A: 60 °C-60 s and E: 72 °C-60 s, 27 cycles, respectively. M is the DNA ladder marker. (b) Northern blot analysis using DIG-labelled BmP5CR1 cDNA riboprobe. (c) The transcript (1300 bp) that failed in splicing was detected by the specific primer set (1F and 5924R; see Fig. 4b and Table S2) on exon 1 and the 1st intron of Daizo (+Lg/+Lg) BmP5CR1
We also compared the expression patterns of BmP5CR2 between Daizo (+Lg/+Lg) and DH6 (Lg/Lg) in the same tissues. The results showed strong BmP5CR2 expression in the fat body, muscle, testis, and ovary and weak expression in the midgut at similar levels in both strains (Fig. 5a). Interestingly, expression was seen in the Malpighian tubule and weakly in the silk gland of Daizo (+Lg/+Lg) larvae, but not in those of DH6 (Lg/Lg). These results also suggest that the lower P5CR enzymatic activity in the middle silk glands of Daizo (+Lg/+Lg) larvae is attributed to the reduction of the BmP5CR1 transcripts and that BmP5CR2 is not involved in the formation of prolinylflavonols.
With respect to the BmP5CR1 expression, Northern blot analysis showed that a single band of 1.4 kb, corresponding to the predicted molecular size of its mRNA (1329 bp), was clearly detected in the middle silk glands of only DH6 (Lg/Lg) larvae (Fig. 5b), whereas a larger size band (~3 kb) was slightly detected in the middle silk glands of Daizo (+Lg/+Lg), in addition to a faint 1.4 kb band when the amount of total RNA was increased to 20 µg/lane. This larger transcript is reflected in the 1st intron, involving a 1.9 kb insertion into the BmP5CR1 of Daizo, because the fragment between exon 1 and this insert region (1F-5924R; 1300 bp) could be specifically amplified by RT-PCR in the middle silk glands of only the Daizo genome, whereas the 1F-1317R spliced fragment (260 bp) from exon 1 to exon 2 was detected in both strains (Fig. 5c). The base sequence of the 1F-5924R amplified fragment was coincident with that in the 1st intron (data not shown). These results suggest that the immature mRNA derived from the failure of normal splicing in the 1st intron remarkably reduces the transcription level of BmP5CR1.
Functional analysis of BmP5CR1 by knockout experiment
To obtain direct evidence showing that the formation of prolinylflavonols is caused by the disruption of BmP5CR1, a knockout experiment of BmP5CR1 was carried out using a genome editing method, called TALEN technology (Fig. 6a). Only one female moth was obtained from 192 DH6 eggs injected with TALEN mRNA and crossed to a DH6 male moth. In the G1 individuals, deletions of five to seven nucleotides were detected in the 2nd exon, which encodes the NAD(P)H binding domain of P5CR (Fig. 6b). The adult G1 moths carrying a 7 bp deletion allele were sib-mated with each other, and the cocoons produced by the G2 larvae were classified into three groups by colour: yellowish green, intermediate colour, and light green. We tested two broods of eggs and revealed a segregation ratio of 1:2:1 for both broods for the three colour groups, which indicated the correspondence of the genotype to the cocoon colour phenotype (Fig. 6c). To establish the BmP5CR1 knockout line, the moths from the yellowish green cocoons were sib-mated, as the homozygous disruption of the gene was supposed to cause the yellowish green cocoon phenotype. As expected, all the cocoons obtained in the G3 generation displayed a yellowish green colour (Fig. 6d). Sequence analysis of the BmP5CR1 transcripts from the G3 progenies revealed that they carried a homozygous mutant BmP5CR1 in which seven nucleotides (AATATTT) were deleted in exon 2 (Fig. S7), and additionally, the generation of prolinylflavonols in the G3 cocoons was confirmed by flavonoid composition analysis (Fig. 6e). Furthermore, enzyme activity assays revealed a complete extinction of P5CR activity in the silk gland of the G3 progenies (Table 1). Consequently, we concluded that the BmP5CR1 frame shift mutation disrupted the normal function of the gene, which resulted in the formation of prolinylflavonols in the silk gland.
(a)The genomic structure of the BmP5CR1 and TALEN banding sites on exon 2 (double underlines). Top lines and rectangular boxes represent introns and exons, respectively. Numbers show the length (bp) of each region. White boxes are untranslated regions (UTRs), and grey boxes are coding regions. (b) The deletion of five, six and seven nucleotides occurred in the G1 individuals. (c) Three cocoon colour degrees and segregations in the G2 population. (d) Cocoon colouration phenotype in the G3 generation of the BmP5CR1 knockout line. All cocoon shells were genetically fixed to the yellowish green colour. (e) Changing of the cocoon’s flavonoid composition by knockout experiment with BmP5CR1. The bars indicate the means ± SD (n = 6)
Discussion
In the present study, we first identified a gene associated with silkworm cocoon colour change resulting from the change in flavonoid composition. It has been shown that there are roughly two flavonoid cocoon phenotypes, Sasamayu (light green) and Ryokuken (yellowish green), according to the flavonoid composition (Hirayama et al. 2009). Ryokuken cocoons contain abnormal flavonoids named prolinylflavonols, while Sasamayu cocoons contain only simple flavonol glucosides, such as quercetin glucosides. However, it is very difficult to distinguish between the two cocoon phenotypes at a glance because the Ryokuken cocoons appear similar to Sasamayu cocoons, but Ryokuken cocoons have a low flavonoid content. This is the reason why the cocoon colour phenotype does not apparently follow Mendelian inheritance and is usually observed as a quantitative trait (i.e., it looks like a continuous variation from pale or light green to deep or yellowish green). Chromosomal substitution lines (CSLs), or consomics, are very useful for identifying the specific chromosome associated with quantitative traits or complicated segregation characteristics (Cowley et al. 2004; Ebitani et al. 2005; Gregorova et al. 2008; Takada et al. 2008), convincing us that the use of CSLs helps us understand the complicated genetic character of the silkworm, such as the flavonoid cocoons of Ryokuken and Sasamayu. In the present study, we used DH6, a strain of CSLs with a Daizo genetic background, which carries a chromosome 6 pair derived from J01. The cocoons of DH6 are light green, and those of the F1 (DH6×Daizo) are also almost light green because of the low or lack of prolinylflavonol content, while the Daizo cocoon is yellowish green and has a large amount of prolinylflavonols, allowing us to set up a novel gene located on chromosome 6, which is associated with the cocoon colour change resulting from the change in flavonoid composition (Fig. S5). Fujimoto et al. (1962) reported the presence of an incomplete dominant gene suppressing expression of the green colour of the cocoon in the Daizo-EKp strain, which has the set of Daizo chromosomes except for one chromosome 6 derived from the EKp mutant strain. They named the gene Ign-1 (Green inhibitor-1) and indicated that Ign-1 is located on chromosome 6 at 7.53 cM from the EKp gene. They also reported that the Ign-1 gene inhibits the permeability of the two kinds of fluorescent pigments (compounds such as flavonoids) from the haemolymph to the middle silk glands (Fig. 5b and c). The action of Ign-1 is apparently different from the action of the gene we reported in the present study; therefore, we named it Lg, a novel gene. However, it is possible that Lg and Ign-1 are the same gene on chromosome 6 because previous researchers may have misunderstood the function of Ign-1 due to the poor analytical flavonoid techniques at that time. It is necessary to investigate the relationship between Ign-1 and Lg in more detail.
Here, we presented data indicating that BmP5CR1 is the gene responsible for Lg. In +Lg/+Lg individuals, the expression level of BmP5CR1 was much lower than that in Lg/Lg, causing an accumulation of P5C, a substrate for the synthesis of prolinylflavonols in the middle silk glands. Among the structural differences between the two genotypes, a large insertion and/or TAAC deletion in the 1st intron probably caused the lower expression of BmP5CR1 because the transcripts containing this intron could be detected in Daizo (+Lg/+Lg) middle silk glands (Fig 5b and c). A lack of TAAC was found at the branching point (YNCURAC), which is a major splicing recognition site of U2 snRNP (Parker et al. 1987; Sharp 1994). Immature mRNA produced by imperfect splicing is usually degraded through the mRNA surveillance mechanism and nonsense-mediated mRNA decay (NMD) in the nuclei (Hentze and Kulozik 1999; Cartegni et al. 2002). In short, the incomplete spliced mRNA caused by the deletion of the branch site must be eliminated due to NMD. As a result, the BmP5CR1 transcript was hardly detected in the expression analysis. Moreover, the abnormal P5CR1 has two amino acid changes and may show insufficient activity, even if a small amount of this enzyme was produced (Figs. 4b and S6).
Decisively, the knockout of BmP5CR1 caused the complete disappearance of P5CR activity in the silk glands and promoted the synthesis of prolinylflavonols, changing the cocoon colour to yellowish green (Fig. 6e). Meanwhile, genome search clarified the presence of another gene encoding P5CR, BmP5CR2, which is only 34% homologous to BmP5CR1. Molecular phylogenetic analysis revealed that almost all insects investigated here have at least two P5CR genes, which are grouped into distinct phylogenetic trees (Fig. S8), showing that two P5CR genes in insects diverged, probably before the vertebrate and invertebrate separated. The present study showed that BmP5CR1 was expressed mainly in the muscle, Malpighian tubules, silk glands, and reproductive organs, while BmP5CR2 was expressed mainly in the muscle, fat body, and reproductive organs, suggesting the functional differentiation of these P5CRs (Fig. 5a). In the present study, the defect of BmP5CR1 apparently had no effect on growth and development, besides cocoon colour change, because the Daizo larvae, which do not have BmP5CR1 expression, can normally grow to adulthood and lay fertile eggs. We consider that the need for proline in the silk glands is not significant, although the silk glands require a large amount of amino acids for silk protein synthesis during the fifth instar (Horie et al. 1978). Indeed, the ratio of proline in the amino acids of fibroin and sericin, the two major silk proteins, is <0.6% (Sprague 1975; Takasu et al. 2002), suggesting the low demand for this amino acid. Sufficient proline is thought to be supplied from the diet or produced due to the action of BmP5CR2. On the other hand, BmP5CR2 is strongly expressed in the fat body, which is an important organ for amino acid metabolism in insects. It is interesting to investigate the effect of the knockout of BmP5CR2.
It was reported that pyrydoxal-5-phosphate (PLP; vitamin B6 coenzyme) could be deactivated by P5C, which accumulated endogenously in patients with hyperprolinaemia type II (Farrant et al. 2001), which is an inherited disorder due to the lack of P5C dehydrogenase (EC 1.2.1.88). The accumulating P5C reacts with PLP by Knoevenagel condensation, causing a deficiency in PLP, which provides an explanation for the observed seizures in hyperprolinaemia type II. Since it has been reported that P5C also condenses other aromatic and aliphatic aldehydes and ketones (Walker et al. 2003), we speculated that flavonoids that have aromatic ketones in their molecules, such as quercetin and kaempferol, could react with P5C in a similar manner. In fact, our preliminary experiments indicated that several complexes, which were considered to be prolinylflavonols, are formed between quercetin and P5C without catalysts at physiological pH and temperature (37 °C) in vitro (Hirayama et al, unpublished data). This finding may be important for clinical reasons. Flavonoids absorbed from foods would easily react with accumulating P5C in patients with inherited disorders, such as hyperprolinaemia type II, and could prevent the depletion of PLP, which is an essential cofactor for many enzymatic reactions, including those involved in neurotransmitter metabolism. Furthermore, intake of dietary flavonoids may be useful for patients with pyridoxine-dependent seizures who have mutations in the ALDH7A1 gene, causing an accumulation of 1-piperideine-6-carboxylayte (P6C), which condenses PLP and inactivates this essential cofactor in the same manner as described above (Mills et al. 2006; Plecko and Stockler 2009). Thus, our findings suggest that flavonoids could neutralise harmful intermediary metabolites accumulating in patients with some genetic disorders by producing complexes such as prolinylflavonols, leading to a reconsideration of the guidelines for the nutritional management of inherited diseases.
In conclusion, we demonstrated that the defect of BmP5CR1, one of the P5CR genes of B. mori, produced a yellowish green cocoon (Ryokuken) by causing the formation of prolinylflavonols, which are presumably non-enzymatic products between flavonols and P5C that accumulate specifically in the middle silk glands. The fact that a defect of BmP5CR1 apparently has no severe effect on silkworm growth may be attributed to proline in the diet or to BmP5CR2, which may be essential for the endogenous synthesis of proline, and the elimination of accumulating harmful P5C by dietary flavonoids. Here, we first suggest that the defect of an enzyme associated with intermediary metabolism could promote the chemical modification of phytochemicals, such as flavonoids, providing a unique model to study the interaction between endogenous metabolites and exogenous substances derived from foods in animals.
Data archiving
Data available from the DDBJ repository under accession number LC276933 (http://getentry.ddbj.nig.ac.jp/top-j.html).
References
Banno Y, Fujii H, Kawaguchi Y, Yamamoto K, Nishikawa K, Nishizaka A et al. (2005) A Guide to the Silkworm Mutants 2005—Gene Name and Gene. Silkworm Genetics Division, Institute of Genetic Resorces, Kyushu Univ, Fukuoka, Japan
Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev 3:285–298
Cowley A, Liang M, Roman R, Greene A, Jacob H (2004) Consomic rat model systems for physiological genomics. Acta Physiol Scand 181:585–592
Daimon T, Hirayama C, Kanai M, Ruike Y, Meng Y, Kosegawa E et al. (2010) The silkworm Green b locus encodes a quercetin 5-O-glucosyltransferase that produces green cocoons with UV-shielding properties. Proc Natl Acad Sci USA 107:11471–11476
Doi K, Kojima T, Makino M, Kimura Y, Fujimoto Y (2001) Studies on the constituents of the leaves of Morus alba L. Chem Pharm Bull 49:151–153
Ebitani E, Takeuchi Y, Nonoue Y, Yamamoto T, Takeuchi K, Yano M et al. (2005) Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed Sci 55:65–73
Farrant R, Walker V, Mills G, Mellor J, Langley G (2001) Pyridoxal phosphate de-activation by pyrroline-5-carboxylic acid - Increased risk of vitamin B-6 deficiency and seizures in hyperprolinemia type II. J Biol Chem 276:15107–1511
Fujii H, Banno Y, Doira H, Kihara H (1998) Genetical Stocks and Mutations of Bombyx mori: Important Genetic Resources. Institute of Genetic Resources, Kyushu Univ, Fukuoka, Japan
Fujimoto N, Yoshitake N, Yamashita O (1962) On the Green inhibitor-I in the silkworm, Bombyx mori. J Seric Sci Jpn 31:239–244
Fujimoto N, Hayashiya K (1972) Studies on the pigments of cocoon. (IX) The precursor of the pigments of green cocoon in the silkworm, Bombyx mori. J Sericult Sci Jpn 41:383–386
Gregorova S, Divina P, Storchoval R, Trachtulec Z, Fotopulosova lV, Svenson KL et al. (2008) Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res 18:509–515
Hara W (1996) Linkage analysis and identification of strains by restriction fragment length polymorphism (RFLP) in the silkworm Bombyx mori. Bull Natl Inst Seric Entomol Sci 16:21–30
Harizuka M (1953) Physiological genetics of the carotenoids in Bombyx mori, with special reference to the pink cocoon. Bull Seric Exp Stn Jpn 14:141–156
Harizuka M, Ueda S, Hirao T (1960) Physiogenetic studies on carotenoids in the silkworm, Bombyx mori L. (2) On the origin of pigments and conditions of developing the cocoon color in pink-, and flesh-coloured strains. Bull Seric Exp Stn Jpn 16:1–34
Hentze MW, Kulozik A (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307–310
Herzfeld A, Mezl VA, Knox WE (1977) Enzymes metabolizing Δ1-pyrroline-5-carboxylate in rat tissues. Biochem J 166:95–103
Hirayama C, Ono H, Tamura Y, Nakamura M (2006) C-prolinylquercetins from the yellow cocoon shell of the silkworm Bombyx mori. Phytochemistry 67:579–583
Hirayama C, Ono H, Tamura Y, Konno K, Nakamura M (2008) Regioselective formation of quercetin 5-O-glucoside from orally administered quercetin in the silkworm, Bombyx mori. Phytochemistry 69:1141–1149
Hirayama C, Kosegawa E, Tamura Y (2009) Analysis of flavonoids in the cocoon layer of the silkworm regional races by LC-MS. Sanshi-Konchu Biotec 78:57–63
Hirayama C, Ono H, Meng Y, Shimada T, Daimon T (2013) Flavonoids from the cocoon of Rondotia menciana. Phytochem 94:108–112
Hirayama C, Okada E (2014) Simple spectrophotometric analysis of flavonoids from the cocoon shell of the silkworm, Bombyx mori. J Silk Sci Technol Jpn 22:65–70
Horie Y, Inokuchi T, Watanabe K (1978) Quantitative study of food utilization by the silkworm, Bombyx mori, through its life cycle II. Economy of nitrogen and amino acids. Bull Seric Exp Stn Jpn 27:531–578
Katsube T, Imawaka N, Kawano Y, Yamazaki Y, Shiwaku K, Yamane Y et al. (2006) Antioxidant flavonol glycosides in mulberry (Morus alba L.) leaves isolated based on LDL antioxidant activity. Food Chem 97:25–31
Kurioka A, Yamazaki M (2002) Purification and identification of flavonoids from the yellow green cocoon shell (Sasamayu) of the silkworm Bombyx mori. Biosci Biotechnol Biochem 66:1396–1399
Mase K, Hirayama C, Okada E, Iizuka T, Kadono-Okuda K et al. (2011) Reinstatement of the 27th linkage group in the classical linkage map of Bombyx mori by molecular linkage analysis of faint-green cocoon gene. J Insect Biotechnol Sericol 80:71–77
Mills P, Struys E, Jakobs C, Plecko B, Baxter P, Baumgartner M et al. (2006) Mutations in antiquitin in individuals with pyridoxine-dependent seizures Nat Med 12:307–309.
Naito K (1968) Studies on the micro constituent in mulberry leaves Part VII. Isolation of rutin and quercetin from mulberry leaves. Nippon Nôgeikagaku Kaishi 42:423–425
Onogi A, Osawa K, Yasuda H, Sakai A, Morita H, Itokawa H (1993) Flavonol glycosides from the leaves of Morus alba L. Shoyakugaku Zasshi 47:423–425
Parker R, Siliciano PG, Guthrie C (1987) Recognition of the TACTAAC box during mRNA splicing in yeast involves base paring to the U2-like snRNA. Cell 49:229–239
Plecko B, Stockler S (2009) Vitamin B-6 dependent seizures. Canad J Neurol Sci 36:S73–S77
Rosenthal GA, Dahlman DL (1990) Interaction of L-canaline with ornithine aminotransferase of the tobacco hornworm, Manduca sexta (Sphingidae). J Biol Chem 265:868–873
Sakudoh T, Sezutsu H, Nakashima T, Kobayashi I, Fujimoto H, Uchino K et al. (2007) Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the Yellow blood gene. Proc Natl Acad Sci USA 104:8941–8946
Sakudoh T, Iizuka T, Narukawa J, Sezutsu H, Kobayashi I, Kuwazaki S et al. (2010) A CD36-related transmembrane protein is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration. J Biol Chem 285:7739–7751
Sakudoh T, Kuwazaki S, Iizuka T, Narukawa J, Yamamoto K, Uchino K et al. (2013) CD36 homolog divergence is responsible for the selectivity of carotenoid species migration to the silk gland of the silkworm Bombyx mori. J Lipid Res 54:482–495
Sharp P (1994) A split genes and RNA splicing. Cell 77:805–815
Shimomura M, Minami H, Suetsugu Y, Ohyanagi H, Satoh C, Antonio B et al. (2009) KAIKObase: an integrated silkworm genome database and data mining tool. BMC Genome 10:486
Sprague K (1975) Bombyx-mori Silk proteins–characterization of large polypeptides. Biochemistry 14:925–931
Sturtevant AH (1915) No crossing over in the female of the silkworm moth. Am Nat 49:42–44
Takada T, Mita A, Maeno A, Sakai T, Shitara H, Kikkawa Y et al. (2008) Mouse inter-subspecific consomic strains for genetic dissection of quantitative complex traits. Genome Res 18:500–508.
Takasu Y, Yamada H, Tsubouchi K (2002) Isolation of three main sericin components from the cocoon of the silkworm Bombyx mori. Biosci Biotechnol Biochem 66:2715–2718
Takasu Y, Sajwan S, Daimon T, Osanai-Futahashi M, Uchino K, Sezutsu H et al. (2013) Efficient TALEN Construction for Bombyx mori Gene Targeting. PLoS ONE 8(9):e73458
Takasu Y, Tamura T, Sajwan S, Kobayashi I, Zurovec M (2014) The use of TALENs for nonhomologous end joining mutagenesis in silkworm and fruitfly. Methods 69(1):46–57
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tamura Y, Nakajima K, Nagayasu K, Takabayashi C (2002) Flavonoid 5-glucosides from the cocoon shell of the silkworm Bombyx mori. Phytochemistry 59:275–278
Tazima Y (1964) The Genetics of the silkworm. Logos Press, London
Walker V, Mills G, Mellor J, Langley G, Farrant R (2003) A novel pyrroline-5-carboxylic acid and acetoacetic acid adduct in hyperprolinaemia type II. Clin Chim Acta 331:7–17
Acknowledgements
We are grateful to Drs H. Machii (NARO) and H. Sawada (Nihon University) for supporting this research. We also thank Dr. T. Togawa (Nihon University) for technical advice in DNA sequencing. This work was financially supported by competitive Grants-in Aid from the National Institute of Agrobiological Sciences (NIAS) and JSPS KAKENHI Grant Number JP25450490.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hirayama, C., Mase, K., Iizuka, T. et al. Deficiency of a pyrroline-5-carboxylate reductase produces the yellowish green cocoon ‘Ryokuken’ of the silkworm, Bombyx mori. Heredity 120, 422–436 (2018). https://doi.org/10.1038/s41437-018-0051-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-018-0051-8