Abstract
Previously, American black bears (Ursus americanus) were thought to follow the pattern of female philopatry and male-biased dispersal. However, recent studies have identified deviations from this pattern. Such flexibility in dispersal patterns can allow individuals greater ability to acclimate to changing environments. We explored dispersal and spatial genetic relatedness patterns across ten black bear populations—including long established (historic), with known reproduction >50 years ago, and newly established (recent) populations, with reproduction recorded <50 years ago—in the Interior Highlands and Southern Appalachian Mountains, United States. We used spatially explicit, individual-based genetic simulations to model gene flow under scenarios with varying levels of population density, genetic diversity, and female philopatry. Using measures of genetic distance and spatial autocorrelation, we compared metrics between sexes, between population types (historic and recent), and among simulated scenarios which varied in density, genetic diversity, and sex-biased philopatry. In empirical populations, females in recent populations exhibited stronger patterns of isolation-by-distance (IBD) than females and males in historic populations. In simulated populations, low-density populations had a stronger indication of IBD than medium- to high-density populations; however, this effect varied in empirical populations. Condition-dependent dispersal strategies may permit species to cope with novel conditions and rapidly expand populations. Pattern-process modeling can provide qualitative and quantitative means to explore variable dispersal patterns, and could be employed in other species, particularly to anticipate range shifts in response to changing climate and habitat conditions.
Similar content being viewed by others
Introduction
Dispersal mediates population and genetic connectivity (Bacles et al. 2006; Bohonak 1999; Cushman and Landguth 2012) and influences population dynamics (Hestbeck 1982; Morales et al. 2010), making it central to understanding the survival and persistence of organisms across their range. With increasing habitat fragmentation, shifts in climate, and expanding human populations, the ability of individual animals to move across landscapes and disperse to new habitats is critical for maintaining populations (Clark et al. 2001; Cushman 2006; Cushman et al. 2012; Fahrig 2003; Lindenmayer and Fischer 2007; Neubert and Caswell 2000). Thus, improving understanding of dispersal strategies in fragmented landscapes can facilitate modeling movements of organisms and inform conservation and management efforts (Travis et al. 2013).
Dispersing individuals encounter costs associated with energy expenditure from movement, competition, and acquisition of resources, yet there are also benefits (Clobert et al. 2012; Travis et al. 2013). Specifically, sex-biased dispersal can reduce mate and resource competition (Greenwood 1980), limit inbreeding (Pusey 1987), and reduce kin competition (Lawson Handley and Perrin 2007). Further, condition-dependent dispersal strategies are often superior to static strategies, allowing organisms to respond to changing environments (Bowler and Benton 2005).
Since the 1980s, American black bears (Ursus americanus) have recolonized previously extirpated portions of their range (Pelton and van Manen 1994; Scheick and McCown 2014), as have other carnivores including cougars and wolves (LaRue et al. 2012; Pletscher et al. 1997). This recolonization into historic portions of their range by black bears provides an example of successful expansion into a changing landscape (Scheick and McCown 2014) and presents an opportunity to compare population processes between recently established and long-standing populations. Additionally, though bears typically exhibit female philopatry and male-biased dispersal (Costello 2010; Rogers 1987; Taberlet et al. 1995; Zedrosser et al. 2007), several recent studies observed variation in these processes (Costello et al. 2008; Jerina and Adamic 2008; Moore et al. 2014; Roy et al. 2012; Schenk et al. 1998). For instance, in the Lower Peninsula of Michigan, 21% of female and 76% of male black bears dispersed, suggesting variation in dispersal strategy within each sex (Moore et al. 2014). Female philopatry was not detected in a population of black bears in northern Ontario (Schenk et al. 1998), nor in a high-density population in Québec (Roy et al. 2012). Further, female brown (U. arctos) bears exhibited long distance dispersal into unoccupied habitat (Jerina and Adamic 2008; Swenson et al. 1998), indicating the potential for flexibility in dispersal, particularly into novel environments.
Variation in bear dispersal strategy has been attributed to density differences (Roy et al. 2012). However, a number of studies suggest that density alone does not explain variation in bear dispersal strategies (Costello et al. 2008; Moore et al. 2014). Indeed, the probability and distance of natal dispersal increased as density decreased for some populations of black and brown bears (Moore et al. 2014; Roy et al. 2012; Støen et al. 2006). Predation, in the form of harvest, may also play a role. When exposed to high levels of harvest, male black bears in a low-density population dispersed shorter distances than bears in a high-density population, likely increasing competition with related individuals of both sexes for resources (Costello et al. 2008). Therefore, additional factors may mediate the effects of density on philopatry and dispersal.
To characterize black bear dispersal, we focused on populations from two distinct regions in the United States, the Interior Highlands and the Southern Appalachian Mountains (Fig. 1). While these regions are geographically close, they are separated by unsuitable bear habitat and Puckett et al. (2015) estimated that these populations of bears diverged 11 kya. Black bears in the Interior Highlands and in the Kentucky portion of the Southern Appalachians were considered extirpated by the early twentieth century (Smith and Clark 1994; Unger et al. 2013), but reintroductions and natural recolonization have contributed to population growth in both regions (Bales et al. 2005; Smith and Clark 1994; Unger et al. 2013) (Fig. 1). Importantly, in each of these regions, both recently established (hereafter “recent”) and historic populations exist, allowing the unique opportunity to compare multiple recent and historic populations within two independently evolving geographic regions. Historic populations have existed longer and often served as a source of the individuals that contributed to forming the recent populations (Puckett et al. 2014; Unger et al. 2013). Recent populations were established by founding individuals from historic populations, by recovery from severe bottlenecks, or by reintroductions after 1995 (Puckett et al. 2014; Unger et al. 2013); substantial unoccupied habitat surrounds these populations, allowing for potential continued range expansion. To complement our empirical data, we used individual-based, spatially explicit genetic simulations with the goal of qualitatively comparing empirical and simulated data sets by simulating processes inferred from the observed patterns (i.e., pattern-process modeling; Cushman 2014, Landguth et al. 2014; Shirk et al. 2012).
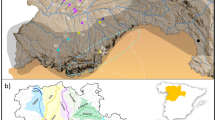
Location of black bear (Ursus americanus) samples collected in the Interior Highlands and Southern Appalachians (2005–2012). Samples from the Interior Highlands were: Arkansas Ouachita Mountains (OU; n = 77), Arkansas Ozark Mountains (OZ; n = 96), Oklahoma (OK; n = 20), and Missouri (MO; n = 110). Samples from the Southern Appalachians: were Big South Fork KY (BSF; n = 19), Pine Mountain KY (PM; n = 84), Tennessee (TN; n = 22), Virginia (VA; n = 8), and West Virginia (WV; n = 29). The range of American black bears (Scheick and McCown 2014) is displayed in the top left. In the main map, historic populations shown as blue circles; recent populations shown as green squares where the Missouri inset map identifies MO–MO and MO–OZ samples in light and dark green, respectively. Note that the arrows show approximate direction from historic to recent populations disregarding that sampling areas are a fraction of total population area. Base layer (ESRI 2009)
Our objective was to characterize variation in patterns of dispersal across historic and recent populations from two distinct regions. Specifically, we compared data from historic and recent populations by observing spatial genetic structure among dyads (i.e., female, male, all) to assess whether observed patterns were congruent with variable dispersal strategies. To further evaluate potential patterns in spatial genetic structure, we simulated the interacting effects of density, genetic diversity, and sex-biased philopatry on female and male dispersal patterns and subsequent spatial genetic structure. Pattern-process models that incorporate dispersal with demographic information can help us consider possible distribution patterns in response to changing environmental conditions.
Methods
Sample acquisition and genotyping
Interior Highlands—Samples from the Arkansas Ozark Mountains (OZ), Arkansas Ouachita Mountains (OU), and Missouri (MO) were collected from hair snare studies conducted in 2008–2011 (OU, OZ) (Kristensen 2013), 2011–2012 (MO) (Wilton et al. 2014), and Oklahoma (OK) from 2004–2006 (Gardner-Santana 2007), respectively.
Southern Appalachian Mountains—Samples from the Big South Fork (BSF) population in Kentucky were collected using hair snares in 2009; remaining Kentucky samples were collected from the Pine Mountain (PM) region through vehicle kill, nuisance bears, poaching cases, and live-trapping (Hast 2010). West Virginia (WV) hair samples were collected as part of routine population monitoring during 2009 (Hast 2010). Virginia (VA) samples were from harvested animals, vehicle kill, hair snares, and live-captures during 2009. Tennessee (TN) samples were a subset of samples from hair snare population monitoring in the Great Smoky Mountains National Park in 2004 (Hast 2010).
We classified populations as “historic” or “recent,” with those known to have formed breeding populations after 1970 considered recently established (Bales et al. 2005; Smith and Clark 1994; Unger et al. 2013). Both population types have dispersing individuals; however, recent populations have not fully recolonized the landscape and hence space may be less limited. Population genetic structure has been investigated in both regions and forms the basis for our population divisions (Hast 2010; Puckett et al. 2014). Because two distinct evolutionary clusters (MO–MO and MO–OZ) were detected in Missouri (Puckett et al. 2014), we separated those samples into populations to analyze separately. These individuals are sympatric and genotypes are expected to homogenize over time; however, as of 2014, a single F1 offspring had been genotyped (Puckett et al. 2014). Consequently, we split these groups due to (1) not recently shared ancestry for the ecological/evolutionary process of natal dispersal we are studying here and (2) we would overestimate unrelated dyads in the population thus skewing the summary statistics analyzed. In addition, population genetic structure analyses revealed that the MO–MO population has low genetic diversity and may represent a remnant population in the region (Puckett et al. 2014). However, because reproduction in MO–MO was not detected until recently (MDC 2008), population size is low (Wilton et al. 2014), and there is available unoccupied habitat in this region, we grouped this with recent populations for analysis. Sample sizes, sexes, and classifications are reported in Table 1.
Microsatellite genotyping
In the Interior Highlands, we genotyped samples at 15 microsatellite loci (Puckett et al. 2014): G1A, G10B, G10C, G1D, G10J, G10L, G10M, G10O, G10P, G10U, UarMU05, UarMU10, UarMU23, UarMU59, and P2H03 (Kristensen et al. 2011; Paetkau et al. 1998a; Sanderlin et al. 2009; Taberlet et al. 1997). We used the marker AmelFrag to determine sex (Kristensen et al. 2011). Samples from the Southern Appalachian Mountains were genotyped by Wildlife Genetics International (Nelson, British Columbia, CA) at 20 microsatellite loci: G10B, G10H, G10J, G10P, G10M, G10L, G1D, G1A, G10X, G10U, G10C, UarMU59, UarMU23, UarMU50, UarMU51, Cxx20, Cxx110, 145P07, 144A06, and CPH9 (Fredholm and Winterø 1995; Paetkau et al. 1998a; Taberlet et al. 1997). Sex was determined using sex-specific differences in the amelogenin gene (Ennis and Gallagher 1994). We used Microsatellite Toolkit in Excel (Park 2001) to calculate the number of alleles and standard deviation for population, and HP-Rare (Kalinowski 2005) to determine the number of private alleles (Table S1).
Empirical data analysis
We separated our genetic data analyses into dyad type within each population (all, female–female (FF), and male–male (MM)), as well as population type (historic and recent). We then used MLRelate (Kalinowski et al. 2006) to determine the likely relationship (parent–offspring, full sibling, half sibling, and unrelated) for each dyad within each population and to obtain the maximum likelihood estimate coefficient of relatedness. We tested for the degree of spatial autocorrelation across dyad and population type using Multiple Pop Subsets in GENALEX v6.501 (Peakall and Smouse 2012). Differences in spatial autocorrelation allow for detection of sex-biased dispersal (Banks and Peakall 2012). We tested significance at each distance class (1, 3, 6, 9, 15, 30, and 45 km) using 1000 bootstrap replicates to calculate the average and 95% confidence intervals of relatedness (Banks and Peakall, 2012). Distance classes indicate how far individuals are from one another. The distance classes were based on prior studies of spatial structuring in bears (Costello et al. 2008; Roy et al. 2012). The following classes each had biological significance, indicating points where effects of philopatry might drop off: 1 km distance class isolated potential mother–cub pairings, 3 km denoted the approximate dispersal distance of a female offspring under conditions of philopatry, 6 km was a typical mating distance, 45 km represented an estimated dispersal distance of male offspring, and the remaining distances provide intermediate values, allowing us to assess the pattern of spatial autocorrelation across the landscape (Costello et al. 2008; Roy et al. 2012). Because female relatedness rapidly declined by 6 km, we also ran an ANOVA to compare relative frequency of closely related individuals (parent–offspring, full sibling, and half siblings from MLRelate) of males vs. females in the 6 km distance class.
We performed isolation-by-distance (IBD) tests (i.e., Mantel test; (Mantel 1967) using matrices of genetic and Euclidean distance in the R library ‘ecodist’ v1.2.2 (Goslee and Urban 2007). At the individual level, we first calculated the proportion of shared alleles (Ps), with the distance measure calculated as 1-Ps (Dps; Bowcock et al. 1994) to obtain our genetic distance matrices for each dyad type within each population. To generate Euclidean distance matrices for each population, we determined a location for each bear. For bears from OK and the Southern Appalachians (BSF, PM, TN, VA, and WV), we used the coordinates where samples were collected. For bears from the Interior Highlands detected as part of a mark-recapture study using hair snares, we calculated activity centers using the ‘fxi’ function in the ‘secr’ package (Efford 2013) in program R (R Core Team 2016). If a bear from the Interior Highlands was only detected once, we used the location at which it was detected as the home range center. A subset of samples from MO were from GPS collared animals (Wilton et al. 2014); therefore, home range centers were calculated using fixed kernel density estimates in Geospatial Modeling Environment v0.7.2.0 with plug-in smoothing parameter (Beyer 2012). We calculated distances between locations of detection or home range centers for individuals within each population using the point distance analysis with a 10 km radius in ArcGIS v10.1 (ESRI, Redlands, CA). We plotted genetic distance vs. Euclidean distance for each combination, which also confirmed linearity before each Mantel test (Zeller et al. 2016) and determined that untransformed matrices produced the best fit. Virginia was not included in these calculations due to low sample size (n = 8). We used a Bonferroni correction to account for multiple comparisons.
To assess evidence of sex-biased philopatry through landscape-level spatial genetic structuring (Banks and Peakall 2012), we generated a linear regression of genetic distance (Dps) on Euclidean distance for each dyad combination within a population, then recorded the slope of the linear model, where a high slope indicated greater population structuring (Paquette et al. 2010). We compared slopes between sexes using a Mann–Whitney U test in program R (wilcox.test) within each population. Significant (P ≤ 0.05) differences were identified based upon Z-scores (Paquette et al. 2010).
Simulations
We used CDPOP v1.2.30 (Landguth and Cushman 2010), an individual-based simulator of population genetic processes. It simulates mating and dispersal in a finite population assigned to fixed locations, recording allele usage by all individuals per generation. In each generation, adult individuals mate according to a user-specified mating system and probability function based on proximity in Euclidean (or effective landscape) distance. Once mated, females give birth to a number of offspring determined by a user-specified probability function which can also control the sex ratio at birth. After birth, adult mortality occurs probabilistically based on user-specified demographic parameters. Finally, vacant locations where adults died are filled by dispersing offspring. Dispersal probabilities follow a user-specified function based on Euclidean or effective distances to the vacant locations. If all locations are occupied, any remaining offspring not yet assigned to a location are eliminated (Balloux 2001).
The goal of our simulation experiment was to identify explanatory variables that produced patterns in spatial genetic structure qualitatively similar to those observed in the empirical data by controlling for factors in the simulation design that cannot be extracted from empirical data (Cushman 2014). We created 12 models using a factorial design: (1) philopatry with two levels for female philopatry and no philopatry, (2) genetic diversity with levels low and high, and (3) density with three levels 2, 12, and 25 bears per 100 km2 in a 100 × 100 km neutral landscape model. Dispersal and mating probabilities were a function of the inverse square of the pairwise Euclidean distances between individuals, with the following given maximum distance thresholds. To simulate female philopatry, we constrained dispersal movements to a maximum threshold of 20 km for females and 60 km for males in the models with female philopatry, whereas for all other models, both sexes had a 60 km dispersal threshold (Costello 2010; Moore et al. 2014). For the genetic diversity factor, allele frequencies at generation 0 were equivalent to frequencies for 15 microsatellite loci from the OZ (high diversity) and MO–MO (low diversity) populations observed in Puckett et al. (2014); the mutation rate was set to 0.0005 per year (near the lower range of mammalian microsatellite mutation rates (Ellegren 2000). We set the maximum distance that bears were permitted to move to find a mate at 20 km (Costello et al. 2009). Age distribution and mortality were based on previous work (Clark 1991; Clark and Smith 1994). Each mated female produced offspring of an equal sex ratio drawn from a Poisson process of mean 2, which ensured a constant population each time step (year). Females and males were allowed to mate with replacement, allowing for multiple matings within a season (Kovach and Powell 2003). All simulations ran for 95 years (approximately 15 bear generations), with 10 replicate simulations of each factorial combination (hereafter scenario) to evaluate variation in this genetic process.
Simulated data analysis
To understand how philopatry, genetic diversity, and density interacted, we calculated slopes from individual linear regressions of genetic distance (Dps) on geographic distance, similar to the observed data (Paquette et al. 2010). The density factor produced uneven sample sizes between scenarios; therefore, we randomly subsampled 30 slopes from each repetition of each scenario for both female (FF) and male (MM) dyads for a final sample size of 7200. We used a Kolmogorov–Smirnov test in R (“ks.test’”) to confirm that each random sample was drawn from the same distribution as the full dataset. We ran a four-factor ANOVA (philopatry, genetic diversity, density, and sex) in R v3.2.4 (R Core Team 2016) on the slopes to understand how factors interacted with each other and which factors contributed to differences among the 12 scenarios. We also ran the tests of IBD as conducted with the observed data, but averaged these across 10 replicate simulations for each scenario and sex combination.
To understand how sampling variation may influence results, we conducted a simulation with philopatry, high genetic diversity, and 12 bears per 100 km2 on a 200 × 200 km landscape. To compare how landscape size from which samples were taken affected results, we sampled all individuals within the full landscape, then randomly subsampled at both 100 × 100 km and 50 × 50 km areas centered at the origin, thus measuring core habitat. We ran individual linear regressions then randomly subsampled 30 slopes to calculate means and variance. To investigate how the proportion of individuals sampled within a landscape affected slope estimates, we randomly subsampled the 50 × 50 km area at 50, 25, and 10% of total individuals; then ran linear regressions, and subsampled 30 slopes to calculate the means and variance. We compared scenarios using a two-factor ANOVA between the variable of interest (landscape area or proportion individuals sampled) and sex with an interaction term included.
Results
Empirical data
We observed differences in spatial genetic structure based on both sex (FF and MM dyads) and our classification of populations as historic or recent (Fig. 2 and Fig. S1). Specifically, there was a difference in spatial autocorrelation between sexes (Ω = 15.56, P = 0.005) when sex was compared across all populations (Fig. S1A). This difference was due to higher relatedness of females at 1 and 3 km (Ω = 62.08, P = 0.001) than in males (Ω = 15.88, P = 0.323); however, relatedness rapidly declined in females by 6 km (Fig. S1A). When both sex and population type were considered, at 1 and 3 km distance classes, females in recent populations had higher relatedness than females in historic populations and males in both population types (Figs. 2a, e).
Spatial autocorrelation for observed a, e and simulated b–d, f–h data between female (pink) and male (blue) dyads calculated in GenAlEx. Observed estimates were compared between recent a and historic e populations. Within the simulated data sets, model factors included low (2 bears km−2; b, f), medium (12 bears km−2; c, g), and high (25 bears km−2; d, h) density; female philopatry b–d and no philopatry f–h; and low (circles and light colors) and high (triangles and dark colors) genetic diversity
Spatial autocorrelation was significant for females in historic (Ω = 56.34, P = 0.001) and recent populations (Ω = 44.11, P = 0.001), and males (Ω = 34.23, P = 0.003) in historic populations. Spatial autocorrelation persisted only through 3 km for recent populations and 6 km for historic populations (Figs. 2a, e and Fig. S2). There was a greater relative frequency of related FF than MM dyads within close proximity (<6 km; F = 10.1, P = 0.007); relative frequency for females with close relatives in this distance class ranged from 24 to 100%, whereas for males it ranged from 0 to 63% (Tables S2 and S3). Thus while females were more likely to have relatives within close proximity, this is not exclusively the case, and also occurs in males.
Spatial genetic structure differed across populations for each sex (Table 2; Fig. 3). In three historic populations (OU, OZ, and WV) and three recent populations (MO–MO, PM, and OK) we detected a significant difference between FF and MM dyads (Table 1 and Figs. 4b, d). Thus, differences in sex-biased dispersal occurred between population types. Notably, MO–MO had higher DPS relatedness in both sexes than other populations (Fig. 3c) and a significantly higher percentage of related dyads than other populations (Tables S1, S2 and Figure S3). For all MO–MO dyads, even male dyads, we detected a significant correlation between genetic and geographic distance (Table 2). Due to the departure of this population from all other populations, we reran the spatial autocorrelation analysis without MO–MO (Figure S1B) and observed the same patterns of significance when comparing sex and population type (Figs. 2a, e).
Genetic distance (Dps) vs. Euclidean distance for dyad type (female (pink) and male (blue) dyads) and population type (recent (top row) or historic (bottom row)). The recent populations included Kentucky Big South Fork (BSF; a), and Kentucky Pine Mountain (PM; b), Missouri Ozark Mountains with MO genotypes (MO–MO; c), Missouri Ozark Mountains with OZ genotypes (MO–OZ; d), and Oklahoma Ouachita Mountains (OK; e). The historic populations included: Tennessee Appalachian Mountains (TN; f), West Virginia Appalachian Mountains (WV; g), Arkansas Ouachita Mountains (OU; h), Arkansas Ozark Mountains (OZ; i). Notes: There was variability in sample size and sampling scheme across populations, so caution should be used in interpreting results. Because patterns shift little after 60 km, data are displayed only through that distance though some populations had pairs farther apart; these are displayed in Figure S6
Mean and standard deviations of slope from linear regressions of genetic (Dps) and geographic distance in the simulated a, c and observed b, d data for female a, b and male c, d dyads. The x-axis in all panels is density, where densities for the simulated data included low, medium, and high densities equal to 2, 12, and 25 bears per 100 km2; population-specific densities for the observed data as reported in Table 1. For the simulated data, lighter colors indicated the philopatry scenarios and darker colors the no philopatry scenarios; circles and triangles represent low and high genetic diversity scenarios, respectively. Populations from the observed dataset included the following historic populations (squares; TN (yellow), WV (brown; U—unknown density), OZ (moss), and OU (blue)) and recent populations (circles; BSF (red), PM (orange), MO–MO (light green), MO–OZ (forest green), and OK (purple))
Simulated data
Spatial autocorrelation varied across simulated scenarios (Fig. 2b–d, f–h). The four-factor (philopatry, genetic diversity, density, and sex) ANOVA of linear regression slopes of genetic distance (Dps) on geographic distance, for each simulated scenario was significant (P < 0.0001; Fig. 4a, c, Fig. S4 and Table S4). There was a significant three-way interaction of philopatry–density–sex, with corresponding two-way interactions between philopatry–sex, and density–sex (Table S4). The philopatry by sex interaction was characterized by similar levels of spatial genetic structure within scenarios without philopatry, whereas, in philopatric scenarios, results differed between sexes. Mantel tests of IBD indicated that philopatric scenarios had greater spatial genetic structure than non-philopatric scenarios for all dyads, with FF dyads having greater spatial genetic structure than MM dyads (Table 3). The significant density by sex interaction identified that spatial genetic structure increased for males more rapidly than for females as population density increased. Genetic diversity, density, and sex were also significant main effects (Table S4). Scenarios with low density had greater spatial genetic structure than those with medium to high density in Mantel tests and FF dyads also had greater structure than MM dyads across all scenarios (Table 3).
We ran an independent simulation to investigate how sampling variation may influence estimates of sex-specific spatial genetic structure. We found an inverse relationship between landscape size and estimated slope (P < 0.0001; Figure S5A). However, there was not a significant difference (P = 0.883) between the proportion of individuals sampled within a fixed sized landscape (Figure S5B) using 10, 25, 50, and 100% of randomly sampled individuals.
Discussion
In our study, we observed variation in sex-biased dispersal of black bears and greater evidence for spatial genetic structure in recent than in historic populations (Figs. 2, 4 and Table 2). Simulations mirrored classical population genetics in that low density resulted in greater spatial genetic structure and corroborated our empirical findings by demonstrating that both density and female philopatry influenced spatial genetic structuring. Specifically, our simulations suggested that higher densities decreased the spatial genetic structure, while females under philopatric conditions counteracted this result by increasing spatial genetic structure. Greater spatial genetic structure in females has traditionally been viewed as a result of female philopatry and male-biased dispersal (Costello 2010; Rogers 1987). Our empirical results support this finding, as well as demonstrate variation in effect for each of these factors (Figs. 2 and 4 and Table S1).
Effects of density on dispersal
Although less common than positive density-dependent dispersal, some species exhibit negative density-dependent dispersal in response to improved access to resources and reduced competition. This occurs when settling near relatives confers fitness benefits (Bowler and Benton 2005; Matthysen 2005), and has been observed in black and brown bears (Moore et al. 2014; Roy et al. 2012; Støen et al. 2006). Results from our simulations showed spatial genetic structure across all densities, but with greater structure in low-density populations (Table 3). In the observed high-density populations (OK, OZ, TN), females were not spatially structured (Table 2). These results, together with previous findings of negative density-dependent dispersal in female black bears (Roy et al. 2012), lack of detectible female philopatry (Schenk et al. 1998), and reduced sex-biased dispersal in a low-density population in New Mexico (Costello et al. 2008), indicate that factors other than density (e.g., landscape matrix and local environmental conditions; Pflüger and Balkenhol 2014) may contribute to distribution. Local factors including habitat suitability and connectivity may be more important or interact with density to influence spatial genetic structure (Baguette et al. 2013; Clobert et al. 2009; Rodrigues and Johnstone 2014; White et al. 2012).
In our simulations, low diversity, which has often been observed in isolated populations (Keyghobadi 2007; Paetkau et al. 1998b), did not by itself result in higher levels of spatial genetic structure (Table 3). However, increased costs of movement due to distance or landscape complexity may further affect spatial relatedness (Bowler and Benton 2005; Ricketts 2001). Two populations in this study (MO–MO and BSF) were geographically isolated (Hast 2010; Puckett et al. 2014). MO–MO, in particular, displayed higher overall relatedness and males exhibited spatial genetic structure. The high relatedness of male dyads could have resulted from genetic drift following a bottleneck or losses given long-term low population size (Puckett et al. 2014). Harvest of male black bears in low-density populations also could reduce the number of unrelated mating pairs, increasing genetic relatedness of male black bears (Costello et al. 2008; Moore et al. 2014, 2015). However, neither of the isolated populations in our study were exposed to harvest. Lower competition as a result of low density may have reduced pressure on males to disperse (Costello et al. 2008), or habitat fragmentation (Clark and Smith 1994) may have restricted movement in these populations.
Female philopatry
In high-density populations, dispersal reduces competition with conspecifics (Bowler and Benton 2005; Levin and Kerster 1969; Matthysen 2005). We did not detect evidence of philopatry in the empirical high-density populations (OK, OZ, and TN), suggestive of a fitness benefit for dispersers. There is likely a limit to the number of female offspring that can establish home ranges close to the maternal home range; thus, the level of realized female fitness may determine whether female offspring obtain fitness benefits by dispersing from their natal range.
We found stronger evidence of spatial genetic structure in recent than in historic populations (Fig. 2). Remaining near the maternal home range may provide an advantage in food acquisition, particularly in novel environments where females can acquire knowledge about local food availability from their mothers (Rogers 1987). However, our empirical data suggest that this benefit exists only up to a distance of 3–6 km, similar to Costello et al. (2008), who found that female spatial genetic structure was limited to approximately 6 km in black bears in New Mexico.
Overall, our results support a growing body of evidence demonstrating variability in female philopatry in black bears. There were exceptions to the trends mentioned above, specifically: the high-density population OU displayed female spatial genetic structure (Table 2 and Fig. 3i); and two recent populations lacked detectable female spatial genetic structure (BSF and OK; Table 2 and Figs. 3a, e). Further, distance between female parent–offspring pairs ranged from 0 to 27 km, suggesting large variation in dispersal distances. In the Lower Peninsula of Michigan, 21% of female black bears dispersed an average of 48.9 km and up to 187 km (Moore et al. 2014) and female brown bears have dispersed long distances in recent populations (Jerina and Adamic 2008); therefore, significant variation in dispersal distance for female bears exists. Thus, philopatry is not the sole dispersal strategy for female black and brown bears (Costello et al. 2008; Moore et al. 2014). Having flexibility in dispersal patterns may confer fitness benefits, particularly in a generalist species that exists in a varied and complex landscape matrix.
Caveats, limitations, and future research
Different sampling schemes and methodologies for home range estimation can influence landscape genetic inferences (Landguth et al. 2012; Oyler-McCance et al. 2012; Wagner and Fortin 2005) and may have influenced our empirical findings. However, we think that bias was unlikely to strongly influence our results because the geographic location of samples was not likely to be systematically biased in a particular direction within populations. For the analysis of spatial genetic structure, we note that populations with estimates similar to the simulated data (PM, MO–MO, MO–OZ) were sampled across the full extent of the landscape, whereas other populations were limited to a subset of the continuous bear habitat (Fig. 1). Perhaps not surprisingly, simulations were less accurate when the sampling area was smaller compared to the full landscape (Figure S5A); thus, it is possible that spatial genetic structure may be overestimated for particularly small areas (Fig. 4b, d and Fig. S4). This has implications for our empirical results, as sampling at several sites was spatially limited, thus we may be overestimating the spatial structure, where full spatial sampling may show results more similar to the simulated values. Variation in sample size and bias in the sampled sex ratio could also contribute to observed differences between and within populations, particularly where sample sizes were low and individual variation is high (i.e. patterns are noisy) (Fig. 3). However, simulations demonstrated that estimates were robust to decreasing proportions of individuals sampled (Figure S5B), as reported previously (Landguth et al. 2012). As most of our data were analyzed separately for males and females, variation in the sampled sex ratios between populations would not be expected to bias comparisons between populations or sexes within a population.
Simulations helped to improve our understanding of empirically observed patterns and our ability to predict future distribution and sex ratio of populations. While we qualitatively assessed the effects of density, philopatry, and inbreeding, simulations that include a greater range of complexity to better reflect real systems should be considered. The goal would be to shift from qualitatively representing patterns from observed data to quantitatively representing data on the same scale (Fig. 4). For example, considering density-dependent responses to resource availability and space use could improve our understanding of how competition influences dispersal (Pflüger and Balkenhol 2014). In addition, our simulation models included movement based on IBD. While this is a first step in helping to qualitatively understand life-history mechanisms and the effects of density, philopatry, and inbreeding, future simulations (and empirical analyses) could include a wider range of landscape genetics analyses, including movement linked to more complex landscape resistance scenarios, gene flow from adjacent populations, and putatively adaptive loci associated with traits for migration and behavior responses. Cushman et al. (2012) argued that high landscape resistance, particularly due to a patchy distribution of high quality habitat, could increase spatial genetic structure if dispersal among patches is limited, as compared to a landscape with even resistance, as in our simulation scenarios. Understanding the potential tradeoffs between population dynamics, density effects, and landscape influences is an exciting and promising future research area in landscape genetics.
Conclusions
We demonstrated evidence supporting sex- and population-specific variation in dispersal by black bears and suggest that variation in this behavioral response is influenced by population density and additional, unsampled extrinsic factors. Overall, black bear forays into novel areas are likely to be male-biased due to the overall greater propensity and distances of male dispersal (Moore et al. 2014; Støen et al. 2006), with spatial genetic structure more likely in females in these low-density areas. However, females can exhibit long distance dispersal and variable dispersal patterns (Moore et al. 2014; Schenk et al. 1998), making rapid expansion and establishment of new populations possible. With many black bear populations increasing and recolonizing portions of their historic range, variation in dispersal will contribute to colonization patterns.
Variation in dispersal strategies among invasive species has similarly facilitated colonization in novel landscapes (Burton et al. 2010; Phillips et al. 2006). Our study offers support for multiple dispersal strategies of a native large carnivore species and its potential for recolonization through population expansion or reintroduction. Condition-dependent dispersal strategies permit organisms to respond to environmental change and facilitate mobility across increasingly complex landscapes (Bowler and Benton 2005). As habitat conditions are altered at increasing rates, the importance of understanding species range shifts has similarly increased (Princé and Zuckerberg 2015; Struebig et al. 2015). The pattern-process modeling used here, in which observed data are compared with simulations, provides a framework with which to extend our findings to a wide range of species.
Data archiving
Genotypes from the Interior Highlands may be accessed at http://datadryad.org/resource/doi:10.5061/dryad.q405j. Genotypes from the Southern Appalachians, script for regression of slopes, and CDPOP input may be accessed at http://dx.doi.org/10.5061/dryad.pc053.
References
Bacles CF, Lowe AJ, Ennos RA (2006) Effective seed dispersal across a fragmented landscape. Science 311(5761):628
Baguette M, Blanchet S, Legrand D, Stevens VM, Turlure C (2013) Individual dispersal, landscape connectivity and ecological networks. Biol Rev Camb Philos Soc 88(2):310–326
Bales SL, Hellgren EC, Leslie DMJ, Hemphill JJ (2005) Dynamics of a recolonizing population of black bears in the Ouachita Mountains of Oklahoma. Wildl Soc Bull 33(4):1342–1351
Banks SC, Peakall ROD (2012) Genetic spatial autocorrelation can readily detect sex-biased dispersal. Molecular Ecology 21(9): 2092-2105
Balloux F (2001) EASYPOP (version 1.7): a computer program for population genetics simulations. J Hered 92(3):301–302
Beyer HL (2012). Vol. 0.7.2. http://www.spatialecology.com/gme/.
Bohonak AJ (1999) Dispersal, gene flow, and population structure. Q Rev Biol 74(1):21–45
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368(6470): 455-457
Bowler DE, Benton TG (2005) Causes and consequences of animal dispersal strategies: relating individual behaviour to spatial dynamics. Biol Rev 80(2):205–225
Burton OJ, Phillips BL, Travis JM (2010) Trade‐offs and the evolution of life‐histories during range expansion. Ecol Lett 13(10):1210–1220
Castillo JA, Epps CW, Davis AR, Cushman SA (2014) Landscape effects on gene flow for a climate-sensitive montane species, the American pika. Mol Ecol 23(4):843–856
Clark JD (1991) Ecology of two black bear (Ursus americanus) populations in the Interior Highlands of Arkansas. Doctor of philosophy thesis, University of Arkansas, Fayetteville, AR, USA
Clark JD, Smith KG (1994) A demographic comparison of two black bear populations in the Interior Highlands of Arkansas. Wildl Soc Bull 22:593–603
Clark JS, Lewis M, Horvath L (2001) Invasion by extremes: population spread with variation in dispersal and reproduction. Am Nat 157:537–554
Clobert J, Baguette M, Benton TG, Bullock JM (2012) Dispersal ecology and evolution. Oxford University Press, Oxford
Clobert J, Le Galliard JF, Cote J, Meylan S, Massot M (2009) Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecol Lett 12(3):197–209
Costello CM (2010) Estimates of dispersal and home-range fidelity in American black bears. J Mammal 91(1):116–121
Costello CM, Creel SR, Kalinowski ST, Vu NV, Quigley HB (2008) Sex-biased natal dispersal and inbreeding avoidance in American black bears as revealed by spatial genetic analyses. Mol Ecol 17(21):4713–4723
Costello CM, Creel SR, Kalinowski ST, Vu NV, Quigley HB (2009) Determinants of male reproductive success in American black bears. Behav Ecol Sociobiol 64(1):125–134
Cushman SA (2006) Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biol Conserv 128:231–240
Cushman SA (2014) Grand challenges in evolutionary and population genetics: the importance of integrating epigenetics, genomics, modeling, and experimentation. Front Genet 5:197
Cushman SA, Landguth EL (2012) Multi-taxa population connectivity in the Northern Rocky Mountains. Ecol Modell 231:101–112
Cushman SA, Shirk A, Landguth EL (2012) Separating the effects of habitat area, fragmentation and matrix resistance on genetic differentiation in complex landscapes. Landscape Ecol 27(3):369–380
Efford MG (2013) Spatially explicit capture-recapture. http://cran.r-project.org/web/packages/secr/index.html.
Ellegren H (2000) Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet 16(12):551–558
Ennis S, Gallagher TF (1994) PCR-based sex determination assay in cattle based on the bovine amelogenin locus. Anim Genet 25:425–427
ESRI (2009) World shaded relief. https://www.arcgis.com/home/item.html?id=9c5370d0b54f4de1b48a3792d7377ff2
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Syst 34:487–515
Fredholm M, Winterø AK (1995) Variation of short tandem repeats within and between species belonging to the Canidae family. Mamm Genome 6(1):11–18
Gardner-Santana LC (2007) Patterns of genetic diversity in black bears (Ursus americanus) during a range expansion into Oklahoma. Master of Science thesis, Oklahoma State University, Stillwater, OK, USA
Goslee SC, Urban DL (2007) The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22(7):1–19
Greenwood PJ (1980) Mating systems, philopatry and dispersal in birds and mammals. Anim Behav 28:1140–1162
Hast J (2010) Genetic diversity, structure, and recolonization patterns of Kentucky black bears. Master of Science thesis, University of Kentucky, Lexington, Kentucky, USA
Hestbeck JB (1982) Population regulation of cyclic mammals: the social fence. Oikos 39:157–163
Jerina K, Adamic M (2008) Fifty years of brown bear population expansion: effects of sex-biased dispersal on rate of expansion and population structure. J Mammal 89(6):1491–1501
Kalinowski S, Wagner A, Taper M (2006) ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes 6:576–579
Kalinowski ST (2005) HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5:187–189
Keyghobadi N (2007) The genetic implications of habitat fragmentation for animals. This review is one of a series dealing with some aspects of the impact of habitat fragmentation on animals and plants. This series is one of several virtual symposia focussing on ecological topics that will be published in the journal from time to time. Can J Zool 85(10):1049–1064
Kovach AI, Powell RA (2003) Effects of body size on male mating tactics and paternity in black bears Ursus americanus. Canad J Zool 81(7):1257–1268
Kristensen TV (2013) Ecology and structure of black bear (Ursus americanus) populations in the Interior Highlands of Arkansas. Ph.D. thesis, University of Arkansas, Fayetteville.
Kristensen TV, Faries KM, White D, Eggert LS (2011) Optimized methods for high-throughput analysis of hair samples for American black bears (Ursus americanus). Wildlife Biol Pract 7(1):123–128
Landguth E, Cushman SA, Balkenhol N (2014) Simulation modeling in landscape genetics. In: Balkenhol N, Cushman SA, Storfer AT, Waits LP (eds) Landscape genetics: concepts, methods, applications. John Wiley & Sons, Ltd, Chichester, UK, p 99–113
Landguth EL, Cushman SA (2010) cdpop: a spatially explicit cost distance population genetics program. Mol Ecol Resour 10(1):156–161
Landguth EL, Fedy BC, Oyler-McCance SJ, Garey AL, Emel SL, Mumma M et al. (2012) Effects of sample size, number of markers, and allelic richness on the detection of spatial genetic pattern. Mol Ecol Resour 12(2):276–284
LaRue MA, Nielsen CK, Dowling M, Miller K, Wilson B, Shaw H et al. (2012) Cougars are recolonizing the Midwest: analysis of courager confirmations during 1990-2008. J Wildl Manage 76(7):1364–1369
Lawson Handley LJ, Perrin N (2007) Advances in our understanding of mammalian sex-biased dispersal. Mol Ecol 16(8):1559–1578
Levin DA, Kerster H (1969) Density-dependent gene dispersal in Liatris. Am Nat 103(929):61–74
Lindenmayer DB, Fischer J (2007) Tackling the habitat fragmentation pancheston. Trends Ecol Evol 22:111–166
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27(2 Part 1):209–220
Matthysen E (2005) Density-dependent dispersal in birds and mammals. Ecography 28(3):403–416
McLean PK, Pelton MR (1994) Estimates of population density and growth of black bears in the Smoky Mountains. Bears Biol Manage 9(1):253–261
MDC (2008) Management plan for the black bear in Missouri. Department of Conservation, Jefferson City, MO, USA
Moore JA, Draheim HM, Etter D, Winterstein S, Scribner KT (2014) Application of large-scale parentage analysis for investigating natal dispersal in highly vagile vertebrates: a case study of American black bears (Ursus americanus). PLoS ONE 9(3):e91168
Moore JA, Xu R, Frank K, Draheim H, Scribner KT (2015) Social network analysis of mating patterns in American black bears (Ursus americanus). Mol Ecol 24(15):4010–4022
Morales JM, Moorcroft PR, Matthiopoulos J, Frair JL, Kie JG, Powell RA et al. (2010) Building the bridge between animal movement and population dynamics. Philos Trans R Soc Lond B Biol Sci 365:2289–2301
Murphy WJ, Hearn AJ, Ross J, Bernard H, Bakar SA, Hunter LTB et al. (2016) The first estimates of marbled cat Pardofelis marmorata population density from bornean primary and selectively logged forest. PLoS ONE 11(3):e0151046
Neubert MG, Caswell H (2000) Demography and dispersal: calculation and sensitivity analysis of invasion speed for structured populations. Ecology 81:1613–1628
Oyler-McCance SJ, Fedy BC, Landguth EL (2012) Sample design effects in landscape genetics. Conserv Genet 14(2):275–285
Paetkau D, Shields GF, Strobeck C (1998a) Gene flow between insular, coastal and interior populations of brown bears in Alaska. Mol Ecol 7:1283–1292
Paetkau D, Waits LP, Clarkson PL, Craighead L, Vyse E, Ward R et al. (1998b) Variation in genetic diversity across the range of North American brown bears. Conserv Biol 12(2):418–429
Paquette SR, Louis Jr EE, Lapointe FJ (2010) Microsatellite analyses provide evidence of male-biased dispersal in the radiated tortoise Astrochelys radiata (Chelonia: Testudinidae). J Hered 101(4):403–412
Park SDE (2001) Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. thesis, University of Dublin, Dublin, Ireland.
Pelton MR, van Manen FT (1994) Distribution of black bears in North America. Eastern Black Bear Workshop for Research and Management 12:133–138
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28 (19): 2537-2539
Pflüger FJ, Balkenhol N (2014) A plea for simultaneously considering matrix quality and local environmental conditions when analysing landscape impacts on effective dispersal. Mol Ecol 23:2146–2156
Phillips BL, Brown GP, Webb JK, Shine R (2006) Invasion and the evolution of speed in toads. Nature 439(7078):803–803
Pletscher DH, Ream RR, Boyd DK, Fairchild MW, Kunkel KE (1997) Population dynamics of a recolonizing wolf population. J Wildl Manage 61(2):459–465
Princé K, Zuckerberg B (2015) Climate change in our backyards: the reshuffling of North America’s winter bird communities. Global Change Biol 21(2):572–585
Puckett EE, Etter PD, Johnson EA, Eggert LS (2015) Phylogeographic analyses of American black bears (Ursus americanus) suggest four glacial refugia and complex patterns of postglacial admixture. Mol Biol Evol 32(9):2338–2350
Puckett EE, Kristensen TV, Wilton CM, Lyda SB, Noyce KV, Holahan PM et al. (2014) Influence of drift and admixture on population structure of American black bears (Ursus americanus) in the Central Interior Highlands, USA, 50 years after translocation. Mol Ecol 23(10):2414–2427
Pusey AE (1987) Sex-biased dispersal and inbreeding avoidance in birds and mammals. Trends Ecol Evol 2(10):295–299
R Core Team (2016) R Foundation for Statistical Computing, Vienna, Austria.
Ricketts TH (2001) The matrix matters: effective isolation in fragmented landscapes. Am Nat 158(1):87–99
Rodrigues AMM, Johnstone RA (2014) Evolution of positive and negative density-dependent dispersal. Proc R Soc Lond B Biol Sci 281(1791):20141226
Rogers LL (1987) Effects of food supply and kinship on social behavior, movements, and population growth of black bears in Northeastern Minnesota. Wildl Monogr 97:1–72
Roy J, Yannic G, Côté SD, Bernatchez L (2012) Negative density-dependent dispersal in the American black bear (Ursus americanus) revealed by noninvasive sampling and genotyping. Ecol Evol 2(3):525–537
Sanderlin JS, Faircloth BC, Shamblin B, Conroy MJ (2009) Tetranucleotide microsatellite loci from the black bear (Ursus americanus). Mol Ecol Resour 9(1):288–291
Scheick BK, McCown W (2014) Geographic distribution of American black bears in North America. Ursus 25(1):24–33
Schenk A, Obbard ME, Kovacs KM (1998) Genetic relatedness and home-range overlap among female black bears (Ursus americanus) in northern Ontario, Canada. Can J Zool 76:1511–1519
Shirk AJ, Cushman SA, Landguth EL (2012) Simulating pattern-process relationships to validate landscape genetic models. Int J Ecol 2012:1–8
Smith KG, Clark JD (1994) Black bears in Arkansas: characteristics of a successful translocation. J Mammal 75(2):309–320
Støen O-G, Zedrosser A, Sæbø S, Swenson JE (2006) Inversely density-dependent natal dispersal in brown bears Ursus arctos. Oecologia 148(2):356–364
Struebig MJ, Wilting A, Gaveau DLA, Meijaard E, Smith RJ, Borneo Mammal Distribution Consortium et al. (2015). Targeted conservation to safeguard a biodiversity hotspot from climate and land-cover change. Curr Biol 25(3): 372–378
Swenson JE, Sandegren F, Söderberg A (1998) Geographic expansion of an increasing brown bear population: evidence for presaturation dispersal. J Anim Ecol 67:819–826
Taberlet P, Camarra JJ, Griffin S, Uhrés E, Hanotte O, Waits LP et al. (1997) Noninvasive genetic tracking of the endangered Pyrenean brown bear population. Mol Ecol 6:869–876
Taberlet P, Swenson JE, Sandegren F, Bjarvall A (1995) Location of contact zone between two highly divergent mitochondrial DNA lineages of the brown bear Ursus arctos in Scandinavia. Conserv Biol 9(5):1255–1261
Travis JMJ, Delgado M, Bocedi G, Baguette M, Bartoń KA, Bonte D (2013) Dispersal and species’ responses to climate change. Oikos 122:1532–1540
Tredick CA, Vaughan MR (2009) DNA-based population demographics of black bears in coastal North Carolina and Virginia. J Wildl Manage 73(7):1031–1039
Unger D, Cox JJ, Harris HB, Larkin JL, Augistine B, Dobey S et al. (2013) History and current status of the black bear in Kentucky. Northeast Nat 20(2):289–308
Wagner HH, Fortin M-J (2005) Spatial analysis of landscapes: concepts and statistics. Ecology 86(8):1975–1987
White TA, Lundy MG, Montgomery WI, Montgomery S, Perkins SE, Lawton C et al. (2012) Range expansion in an invasive small mammal: influence of life-history and habitat quality. Biol Invasions 14(10):2203–2215
Wilton CM, Puckett EE, Beringer J, Gardner B, Eggert LS, Belant JL (2014) Trap array configuration influences estimates and precision of black bear density and abundance. PLoS ONE 9(10):2414–2427
Zedrosser A, Støen O-G, Sæbø S, Swenson JE (2007) Should I stay or should I go? Natal dispersal in the brown bear. Anim Behav 74(3):369–376
Zeller KA, Creech TG, Millette KL, Crowhurst RS, Long RA, Wagner HH et al. (2016) Using simulations to evaluate Mantel-based methods for assessing landscape resistance to gene flow. Ecol Evol 6(12):4115–28
Acknowledgements
We thank Frank van Manen, Jennapher Teunissen van Manen, and Ronald A. Van Den Bussche for sample contributions; Sébastien R. Paquette for R code for linear regressions; and Edward Gbur for statistical advice. We thank Jill S. Miller, Jason Munshi-South, three anonymous reviewers, and the editor for their comments that improved the manuscript. Funding sources for sample collection and analysis included Arkansas Game and Fish Commission, Federal Aid in Wildlife Restoration, Safari Club International Foundation, the University of Tennessee, the United States Geological Survey, the University of Arkansas, Oklahoma State University, West Virginia Department of Natural Resources, Virginia Department of Game and Inland Fisheries, the University of Kentucky, Kentucky Department of Fish and Wildlife, Missouri Department of Conservation, the University of Missouri, and Mississippi State University. T.V.K. was supported by the Distinguished Doctoral Fellowship at the University of Arkansas and EEP was supported by a University of Missouri Life Sciences Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Kristensen, T.V., Puckett, E.E., Landguth, E.L. et al. Spatial genetic structure in American black bears (Ursus americanus): female philopatry is variable and related to population history. Heredity 120, 329–341 (2018). https://doi.org/10.1038/s41437-017-0019-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-017-0019-0
This article is cited by
-
Connecting mountains and desert valleys for black bears in northern Mexico
Landscape Ecology (2021)