Abstract
Purpose
Copy-number variant (CNV) assessment is recommended for patients undergoing prenatal diagnostic testing. Noninvasive screening tests have not been extensively validated for CNV detection. The objective of this study was to compare the ability of genome-wide noninvasive prenatal screening (NIPS) to chromosomal microarray to detect clinically significant findings.
Methods
We prospectively enrolled 198 subjects at the time of consent for diagnostic prenatal testing. Genome-wide NIPS results were compared with diagnostic testing results to assess NIPS test performance (n = 160, 38 subjects without microarray results excluded). Cohen’s kappa statistic was used to assess test agreement.
Results
Genome-wide NIPS did not detect clinically significant chromosomal abnormalities at the same rate as diagnostic testing, κ = 0.75 (95% confidence interval [CI], 0.62–0.87). When excluding CNVs <7 Mb and findings outside the limits of genome-wide NIPS, test agreement improved, κ = 0.88 (0.79–0.97) driven by agreement for common aneuploidies (κ = 1.0). However, among patients with an abnormal fetal survey, agreement was only fair, κ = 0.38 (0.08–0.67).
Conclusion
While NIPS is an excellent screening test for common aneuploidies, genome-wide NIPS misses clinically significant findings detected on routine diagnostic testing. False positive and false negative cases highlight the importance of pretest counseling regarding NIPS limitations, especially in the setting of fetal anomalies.
Similar content being viewed by others
INTRODUCTION
Prenatal genetic testing has witnessed unprecedented advances over the last decade, including the introduction of noninvasive prenatal screening (NIPS) and chromosomal microarray. Given its excellent clinical performance, NIPS rapidly replaced traditional serum screening methods for detection of trisomy 13, 18, 211 and is now endorsed by the American College of Obstetricians and Gynecologists (ACOG) and the Society for Maternal Fetal Medicine (SMFM) for aneuploidy screening in both high- and low-risk pregnancies.2
Although NIPS can accurately screen for common aneuploidies, pregnancies in average-risk women are more commonly affected by pathogenic copy-number variants (CNVs) than whole-chromosome abnormalities.3 Pathogenic CNVs affect 1–3% of all structurally normal pregnancies and are not detected by standard genetic screening in early pregnancy.4,5 Therefore, ACOG recommends that chromosomal microarray be routinely offered to patients undergoing prenatal diagnostic testing due to its increased diagnostic yield,6 particularly in pregnancies with fetal structural abnormalities.4
Given the importance of CNV detection, genome-wide NIPS was introduced in 2015 as a new screening test designed to detect CNVs >7 Mb and CNVs within seven microdeletion regions associated with common microdeletion syndromes.7 Initial validation studies of expanded NIPS have demonstrated mixed results, with some suggesting poor positive predictive value (PPV) for microdeletion syndromes and others suggesting higher PPVs.8,9,10,11 Test performance is influenced by a number of factors, including prevalence of each disorder, maternal CNVs, placental mosaicism, fetal fraction, sequencing depth (i.e., the number of unique reads that include a given nucleotide in the reconstructed sequence), and CNV size, among other factors. Although there has been considerable excitement and implementation of these new expanded noninvasive screening tests, prospective clinical validation data are lacking.12
Coinciding with the launch of genome-wide NIPS, one validation study was published reporting a sensitivity of 97.7% and specificity of 99.9% for detection of whole-chromosome and subchromosomal abnormalities other than trisomies 13, 18, 21, and the sex chromosome aneuploidies, by genome-wide NIPS (n = 42 cases, specific chromosomal anomalies not all specified).13 Although promising, this study has limited clinical application, as it lacked confirmation of screening results by diagnostic chromosomal microarray and follow-up of presumed negative cases. Studies on other genome-wide NIPS platforms have raised concern about balancing the marginal detection rate of fetal chromosomal aberrations against the rate of false positive results.14
In this study, we sought to conduct an independent clinical validation study of genome-wide NIPS and estimate the proportion of patients in which expanded NIPS would yield results concordant with clinical chromosomal microarray. For patients with imperfect agreement between NIPS and diagnostic testing, we aimed to identify cases where the discrepancy had clinically meaningful consequences.
MATERIALS AND METHODS
Study design and subjects
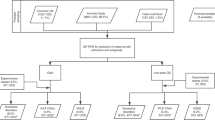
We conducted an institutional review board (IRB)–approved prospective cohort study of patients with singleton pregnancies undergoing diagnostic genetic testing (amniocentesis, chorionic villus sampling, percutaneous umbilical cord sampling [PUBS], or cord blood testing) with a chromosomal microarray in the Center for Fetal Medicine and Reproductive Genetics at Brigham and Women’s Hospital (Boston, MA) and in the antenatal diagnostic centers at Massachusetts General Hospital (Boston, MA) and Newton-Wellesley Hospital (Newton, MA) over 24 months (2017–19). We excluded patients with prior diagnostic testing, as well as those with diagnostic testing whose cytogenetic results could not later be confirmed and those with normal karyotypes who declined chromosomal microarray.
Following clinical consent for diagnostic testing, study participants were approached and consented by study investigators (genetic counselors and obstetricians). Prior to diagnostic testing, maternal blood was drawn and sent for evaluation with the genome-wide cell-free fetal DNA test, MaterniT Genome® (Sequenom), on a research basis. Sequenom entered a collaborative agreement with the principal investigators in which Sequenom agreed to run and interpret their genome-wide test per their standard protocol at no cost. Staff at Sequenom did not have access to the results of the clinical chromosomal microarray. Patients received prenatal clinical diagnostic testing results similar to any patient not enrolled in the study and did not receive results from the NIPS test.
Diagnostic testing (karyotype or chromosomal microarray) was performed by independent CLIA-certified molecular cytogenetic laboratories per standard protocol. All laboratories in this study used the Affymetrix Cytoscan HD assay, a high density, whole-genome microarray with 2,696,550 probes. The array consists of 1,953,246 nonpolymorphic regions and 743,304 single-nucleotide polymorphisms, with an average genome-wide spacing of 1.1 kb. Laboratory staff were unaware of patients’ involvement in this clinical study. At the time of study enrollment, subjects were also consented for genetic testing on postdelivery placental samples if noninvasive and clinical diagnostic testing results were discordant and otherwise unexplained. These results were for research purposes only and were not returned to enrolled subjects.
Assessment of agreement between tests
Results of clinical diagnostic testing were compared to those obtained by genome-wide NIPS by study investigators, including MFM geneticists and genetic counselors familiar with interpretation of microarray analyses. The primary outcome was overall agreement between the two tests as measured by a kappa statistic (κ) with 95% confidence intervals (CI).
The κ statistic is an index of agreement, with 1.0 indicating perfect agreement and 0 indicating agreement equivalent to chance. Interpretations of kappa statistic ranges include 0.01–0.20 as none to slight, 0.21–0.40 as fair, 0.41–0.60 as moderate, 0.61–0.80 as substantial, and 0.81–1.0 as near perfect. We defined “agreement” as identification of any deletion or duplication overlapping with that discovered on diagnostic testing. For example, a positive call of a 16.8-Mb duplication by genome-wide NIPS that overlapped with a 17.5-Mb duplication identified on diagnostic testing would be considered in agreement.
We calculated the minimum sample size necessary to detect 90% agreement between genome-wide NIPS and chromosomal microarray testing, represented by κ = 0.95. Given that our institutional rate of clinically significant findings on diagnostic testing is 15% and assuming a two-sided alpha of 0.05 and power of 80%, at least 55 study participants were needed to evaluate agreement between the two tests.
Simulated estimates of risk modification
To understand NIPS clinical utility, we then calculated the baseline pretest risk, as well as the post-test residual risk, of having a clinically significant finding on chromosomal microarray in patients with positive and negative NIPS results in three clinically relevant cohorts: all patients referred for diagnostic testing, patients referred for testing who were not found to have a common aneuploidy or sex chromosome aneuploidy, and patients referred for diagnostic testing with an abnormal ultrasound. The cohort of patients in whom we excluded common aneuploidies and sex chromosome aneuploidies was designed to simulate the added benefit of genome-wide screening over that of NIPS for 13, 18, 21, X, and Y only.
Data collection
Participant data, including demographic information, genetic screening, and pregnancy information including sonographic findings and indication for diagnostic testing, were collected by chart abstraction.
RESULTS
Study population
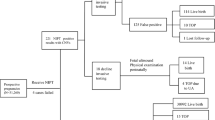
In this prospective study, 198 participants had maternal blood collected for genome-wide NIPS at the time of consent for either a diagnostic procedure or cord blood collection with subsequent karyotype and/or chromosomal microarray testing. Of the 198 enrolled participants, 14 were excluded due to lack of confirmatory diagnostic testing, which was planned at delivery and not performed due to circumstances including patient preference, clinical scenario, or change in indication. An additional 24 subjects were excluded who had a normal karyotype and declined chromosomal microarray. The demographic characteristics of the remaining 160 participants are shown in Table 1. The average age of study participants was 34.5 (+/-5.2) years and participants were recruited at Brigham and Women’s (78.8%), Massachusetts General (13.8%), and Newton-Wellesley (7.5%) Hospitals. Pregnancy and prenatal testing characteristics of the cohort are depicted in Table 2; notably 54.8% of participants had an abnormal fetal survey with at least one structural anomaly (excluding soft markers). At the time of diagnostic testing, consenting providers documented one or more indications for testing. Fetal anomaly suspected on ultrasound was the most common documented indication for diagnostic testing in our cohort (43.1% of participants). Other common indications for diagnostic testing were increased nuchal translucency (18.1%) and abnormal aneuploidy screen (14.4%). Enrollment occurred at the time of diagnostic testing after consent for amniocentesis (46.3%), chorionic villus sampling (42.5%), cord blood collection (10.6%), or PUBS (0.6%).
Assessment of agreement between tests
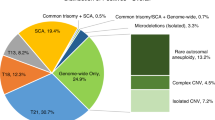
Performance of genome-wide NIPS was assessed by comparing NIPS results with the results of clinical diagnostic testing. Detailed findings are highlighted in Table 3 with discordant results detailed in Supplemental Table 1. Common aneuploidies were detected by genome-wide NIPS in 21 cases, with accurate identification of 100% of cases of trisomies 21, 18, and 13 in our cohort. There were no false positives or false negatives among these common aneuploidies.
For the sex chromosome abnormalities (SCA), 3 of 5 (60%) identified by genome-wide NIPS were consistent with diagnostic testing results, including two cases of Klinefelter syndrome (XXY) and one case of monosomy X. For the other two abnormal NIPS sex chromosome results, there was one false positive case of monosomy X (interpreted as possible mosaic monosomy X) and one case where sex chromosome analysis by genome-wide NIPS was inconclusive (but was normal on microarray). False negatives included one case with normal genome-wide NIPS but monosomy X on diagnostic testing.
In terms of genome-wide abnormalities beyond the detection capabilities of the genome-wide NIPS platform utilized, two cases of triploidy and one case with multiple areas of homozygosity were identified by diagnostic testing and were not ascertained by genome-wide NIPS.
For CNVs, ten cases with CNVs were detected by genome-wide NIPS. One microdeletion syndrome, 5p minus (cri-du-chat) syndrome, with targeted coverage on genome-wide NIPS was correctly identified. No other targeted microdeletion syndromes were detected in this cohort. The remainder of the copy-number results were split into those >7 Mb and those <7 Mb, as this is the targeted threshold of detection by the genome-wide NIPS testing platform (Sequenom®). Among the large CNVs, two false positive calls (10 Mb and 24 Mb in size) were reported by genome-wide NIPS and were not confirmed on diagnostic testing. The 10-Mb deletion on genome-wide NIPS was on 10q in a pregnancy undergoing diagnostic testing due to a fetal right-sided aortic arch. Microarray on amniotic fluid in this pregnancy demonstrated an isolated 110-kb deletion on 2q, classified as a variant of uncertain significance (VUS). As part of the study, a microarray was performed on the placenta following delivery given the discordant results and there was no evidence of the 10-Mb loss on 10q reported by genome-wide NIPS. In the second false positive case of a CNV >7 Mb, a 24-Mb gain on 18q was identified by genome-wide NIPS in a fetus with tetralogy of Fallot and was not confirmed by diagnostic microarray performed on a cord blood sample after birth. In this case, diagnostic microarray on cord blood revealed a 22.5-Mb pathogenic deletion on 18q and no other significant findings. Follow-up placental microarray revealed the same 18q 22.2-Mb deletion in addition to a 15.9-Mb duplication on 18q (overlapping with the 24-Mb gain detected on genome-wide NIPS). Karyotype was not performed due to sample limitations and follow-up parental testing was clinically recommended, but remains unavailable. A false negative genome-wide NIPS result occurred in one case where a large (7.7 Mb) pathogenic CNV was detected on diagnostic testing, but was not reported on genome-wide NIPS.
Although the targeted threshold for CNV detection by genome-wide NIPS is >7 Mb, there were three cases in which NIPS reported smaller CNVs (ranging in size from 2 to 5 Mb) that were not confirmed on diagnostic testing. In each of these cases, a microarray was performed as part of this study on placental tissue following delivery and did not show evidence of the deletions or duplications reported by genome-wide NIPS. In contrast, by diagnostic microarray, two smaller clinically significant CNVs (104 kb and 813 kb) were identified, but were not detected by genome-wide NIPS and were below the resolution of the test.
Cohen’s kappa statistic was used to assess the agreement between genome-wide NIPS and diagnostic testing. This analysis demonstrated that expanded noninvasive genome-wide screening does not detect the same rate of clinically significant chromosomal abnormalities when compared with chromosomal microarray, with a kappa statistic of κ = 0.75 (95% CI, 0.62–0.87), as illustrated in Fig. 1. This measure of agreement reflects 29 concordant positive results, 108 concordant negative results, 7 false positives, and 7 false negatives. Excluding cases of triploidy, areas of homozygosity, and CNVs <7 Mb (all abnormalities not expected to be detected by this genome-wide NIPS platform), the agreement between noninvasive genome-wide screening and chromosomal microarray was improved with κ = 0.88 (95% CI, 0.79–0.97). In this subanalysis, there were only four false positives and two false negatives. This improvement was driven by the accurate detection of all 21 cases of common aneuploidies in our cohort, translating to a kappa statistic of κ = 1.0 for this subset of cases.
Forest plot of the kappa statistics with 95% confidence intervals (CI) calculated in our cohort. The first data point illustrates the ability of genome-wide NIPS to detect all clinically significant findings identified on chromosomal microarray, κ = 0.75 (95% CI, 0.62–0.87). The second data point illustrates the same comparison, but excludes copy-number variants (CNVs) <7 Mb and other findings not detectable by genome-wide NIPS. In this scenario, the agreement between genome-wide NIPS and diagnostic testing improves, κ = 0.88 (95% CI, 0.79–0.97). If variants of uncertain significance (VUS) are included as clinically significant findings, the agreement between genome-wide NIPS and diagnostic testing is moderate illustrated in the third data point, κ = 0.56 (95% CI, 0.42–0.71). Finally, the red data point illustrates only fair agreement between genome-wide NIPS and the clinically significant findings identified through diagnostic testing among patients with an abnormal fetal ultrasound, κ = 0.38 (95% CI, 0.08–0.67).
To address the agreement between tests beyond that of the common aneuploidies, we also calculated a kappa statistic excluding the common aneuploidies and sex chromosome aneuploidies, which would be evaluated on common NIPS platforms. The agreement between genome-wide NIPS and chromosomal microarray beyond trisomies 13, 18, 21, and the sex chromosome aneuploidies was more limited with κ = 0.43 (95% CI, 0.15–0.71) indicating moderate agreement.
An additional 13 CNVs <7 Mb classified as VUS were detected by chromosomal microarray (see Supplemental Table 2). When including these 13 CNVs in our calculation, the agreement between noninvasive genome-wide screening and chromosomal microarray inclusive of all CNVs was also more limited with κ = 0.56 (95% CI, 0.42–0.71) indicating moderate agreement. This was driven by 27 discordant cases including 7 false positives and 20 false negatives.
Among cases in which diagnostic testing was pursued in the setting of an abnormal fetal survey, noninvasive genome-wide screening demonstrated the most limitations compared with diagnostic testing in detecting clinically significant chromosomal abnormalities with κ = 0.38 (95% CI, 0.08–0.67) indicating only fair agreement. This agreement reflects 5 concordant positives, 4 false positives, 7 false negatives, and 48 concordant negative results.
Simulated estimates of risk modification
To translate the Cohen’s kappa statistics into more tangible counseling advice, we simulated the effect of a positive or negative genome-wide NIPS result on the risk of detection of a chromosomal abnormality on diagnostic testing. In our cohort, the baseline post hoc risk of a clinically significant chromosomal abnormality in all participants presenting for diagnostic testing and enrolled in the study was 23.8%. Among those participants with a positive genome-wide NIPS result, the risk of a fetal chromosomal abnormality was 80.6%. Therefore, a positive genome-wide NIPS result increased the risk of a clinically significant finding on diagnostic testing by more than threefold, from 23.8% to 80.6%, as shown in Fig. 2a. In contrast, a negative genome-wide NIPS result reduced the risk of a significant chromosomal abnormality from the baseline risk of 23.8% to 6.1%.
Flow diagram, moving left to right, depicts the change in baseline risk for a patient referred to our institution. (a) Entire cohort, in which 23.8% of patients enrolled had a clinically significant chromosomal abnormality. A hypothetical patient referred to us with a positive genome-wide NIPS would have their residual risk increased from 23.8% to 80.6%, which may serve to support confirmatory diagnostic testing for patients of a given risk preference profile. In contrast, a negative genome-wide NIPS serves to reduce residual risk from 23.8% to 6.1%, which for patients of a given risk profile may serve as a measure of reassurance. (b) Subset of patients who did not have a common aneuploidy or sex chromosome aneuploidy diagnosed by NIPS. In this group, 50% of patients with a positive genome-wide NIPS result were found to have a clinically relevant finding on chromosomal microarray. Those with a negative genome-wide NIPS result had their risk of a clinically significant finding on microarray reduced from 8.7% to 5.2%. (c) Subset of subjects with an abnormal fetal ultrasound. Note that the baseline risk of 18.8% is lower than in the first simulation likely because aneuploidy is screened out in the first trimester. A hypothetical patient referred with an abnormal fetal ultrasound who was found to have a positive genome-wide NIPS test would have their risk of a clinically significant chromosomal abnormality increased from 18.8% to 55.6%, which may serve to support confirmatory diagnostic testing for patients of a given risk preference profile. In contrast, a negative genome-wide NIPS in this population serves to reduce residual risk only marginally from 18.8% to 12.7%.
Next, we examined the group of patients who did not have a common aneuploidy or sex chromosome aneuploidy diagnosed by NIPS in Fig. 2b. This subset of our cohort had an 8.7% baseline risk of having a clinically significant finding on chromosomal microarray. In those with a positive genome-wide NIPS result, this risk increased to 50%. Those with a negative genome-wide NIPS result had a residual risk of 5.2%.
Finally, we simulated the effect of genome-wide NIPS results in the population of pregnant subjects whose documented indication for diagnostic testing was an abnormal anatomic fetal survey. In this subpopulation, the baseline risk of a clinically significant chromosomal abnormality was 18.8% and a positive genome-wide NIPS finding increased that risk to 55.6%, as shown in Fig. 2c. Those with an abnormal ultrasound and a negative genome-wide NIPS had their risk of a clinically significant finding reduced minimally from 18.8% to 12.7%.
Evaluation of nonreportable results
Among the 160 subjects in our study, 9 (5.6%) had genome-wide NIPS results that were not evaluable, as detailed in Supplemental Table 3. Six of those were reported as quantity not sufficient (QNS) in the setting of a fetal fraction <4%. Two of these six cases were found to have triploidy on diagnostic testing at 15 and 18 weeks gestational age. The other four cases that were reported as QNS ranged in gestational age from 12 to 21 weeks and had normal microarrays following diagnostic testing. The remaining three nonreportable genome-wide NIPS tests failed quality control in the setting of excessive noise and were drawn at the gestational ages of 12, 16, and 30 weeks; two had small VUS diagnosed on microarray and one had a normal microarray.
DISCUSSION
Chromosomal microarray is recognized as the standard of care for patients undergoing diagnostic testing during pregnancy.5,6 Genome-wide testing with a chromosomal microarray has the ability to detect clinically significant CNVs that occur independent of maternal age and may be present in the setting of a normal fetal anatomic survey. Currently, many patients who present for genetic counseling looking for genome-wide information due to fetal structural anomalies or other indications opt for noninvasive testing using cell-free fetal DNA, despite recommendations to proceed directly to a diagnostic test (e.g., chorionic villus sampling or amniocentesis).15 Patients may decline diagnostic testing due to their perception of risk associated with diagnostic testing and the availability and attractiveness of the genome-wide noninvasive cell-free DNA test. Concerningly, some patients with negative noninvasive testing results may incorrectly conclude that a normal result means that there is not a genetic disorder affecting the fetus.16 Despite its frequent use in clinical care, to date, no clinical validation of genome-wide NIPS has been performed. Our project was designed to begin to address this gap.
As expected, we found that genome-wide NIPS does not detect the same rate of clinically significant findings as diagnostic testing using chromosomal microarray. This finding has important implications for patient care, with the utility of these results highlighted in hypothetical decision trees in Fig. 2a–c. In Fig. 2a, a patient in our cohort having diagnostic testing with a positive genome-wide NIPS finding would have their risk of a clinically significant finding in the fetus increased from 23.8% to 80.6%. Although this might help some patients make a decision about diagnostic testing, it is certainly not a replacement for diagnostic testing and might delay the definitive diagnosis, potentially limiting reproductive options. On the other hand, a patient with a negative genome-wide NIPS would have their risk of a clinically significant finding reduced from 23.8% to 6.1%, which provides incomplete reassurance and is above many patient and provider thresholds for proceeding with diagnostic testing. Notably, the risk modification represented in these decision trees is driven by high specificity of NIPS for common aneuploidies.
To evaluate for other chromosomal abnormalities beyond those detected in the most commonly used NIPS, we next excluded the common aneuploidies and sex chromosome aneuploidies from our decision tree analysis (Fig. 2b). After excluding these conditions, a positive genome-wide NIPS would increase the risk of a clinically significant finding in our cohort from 8.7% to 50% and a negative result would decrease the risk of a clinically significant finding from 8.7% to 5.2%. In other words, a positive genome-wide NIPS result has a 50% chance of being a true positive and a negative result does not significantly change the risk stratification. Given this, patients desiring definitive information about the risk of a chromosomal abnormality in pregnancy should be strongly encouraged to pursue diagnostic testing.
We next generated a decision tree in the population of study subjects having diagnostic testing due to an abnormal fetal anatomic survey, as this represents a large proportion of patients who have genome-wide NIPS sent clinically.6 As depicted in Fig. 2c, a patient with an abnormal ultrasound and a positive genome-wide NIPS finding would have their risk of a clinically significant finding increased from 18.8% to 55.6%. Those with an abnormal ultrasound and a negative genome-wide NIPS would have their risk of a clinically significant finding reduced from 18.8% to 12.7%. Although not all patients may agree, each of these risk adjustments is unlikely to meaningfully impact clinical care, either in the pursuit of diagnostic testing, decision making, or delivery planning. These data strongly support societal recommendations for diagnostic testing with evaluation by chromosomal microarray in the setting of fetal anomalies.17
It is worth noting that these hypothetical models include results on diagnostic testing that are not designed to be captured by genome-wide NIPS, including triploidy, areas of homozygosity, and CNVs <7 Mb. These were included in one of the calculations of agreement as our primary aim was to compare genome-wide NIPS to standard diagnostic testing, which now includes chromosomal microarray.5,18,19 Furthermore, prenatal counseling resources are limited in many settings and, thus, patients may be unaware of the differences in the detection capabilities of genome-wide noninvasive testing compared with prenatal diagnostic testing. To aid in patient counseling, the question of overall concordance of these testing modalities is, therefore, intended to answer the question of agreement between noninvasive and diagnostic testing.
Not surprisingly, after limiting the evaluation of agreement to findings specifically designed to be detected by genome-wide NIPS, the kappa statistic increased to 0.88, which in some interpretations is defined as “almost perfect.” In other words, genome-wide NIPS shows “almost perfect” agreement with findings that would be detected by karyotype, not chromosomal microarray. As common trisomies and sex chromosome aneuploidies were 82.8% of the cytogenetic abnormalities correctly identified in these cases, expanded genome-wide NIPS performed well. However, when considering only the added value of including select microdeletions and CNVs >7 Mb present in genome-wide NIPS but not standard NIPS platforms, there were three false positive or negative calls and only five true positive results, which is problematic. This result suggests that, with enough pretest counseling on the limitations of genome-wide NIPS including conditions that this noninvasive test will not detect, there may be a role for this screening in patients who are interested in limited additional information, but are not willing to assume the risks (albeit small) associated with diagnostic testing. However, counseling should always include discussion of the benefits of diagnostic testing with chromosomal microarray compared with currently available screening technologies.
Other results not included in our evaluations of agreement include the 13 VUS detected on chromosomal microarray. Although some percentage of these may later be classified as pathogenic and, therefore, be additional clinically significant results that were “missed,” many will also later be classified as benign and are therefore unlikely to alter clinical management of the pregnancy or neonatal care. Such VUS might also contribute to parental anxiety during pregnancy and, thus, the exclusion of these from the results of noninvasive testing is one potential benefit for patients.
Finally, similar to other commercial companies offering noninvasive screening, 9 of 160 (5.6%) subjects in our study had a nonreportable result. Similar to other reports, this subset was found to have a significant rate of aneuploidy, with 2 of 9 (22.2%) cases diagnosed with triploidy on diagnostic testing.20 With this confirmed increased risk of aneuploidy among nonreportable cases, patients with these results should be encouraged to pursue diagnostic testing if information about aneuploidy during pregnancy is desired.
In interpreting this study, we must keep in mind a number of limitations. We have limited ability to determine concordance for specific CNVs given the relatively small sample size and the rarity of each individual finding. Our cohort also includes only those patients who consented to diagnostic testing and, therefore, represents a population at increased risk for genetic abnormalities. This cohort also may not be completely generalizable to a population of patients who decline diagnostic testing, toward whom genome-wide NIPS is primarily marketed. Despite these limitations, this study has several strengths. This is the first study of genome-wide NIPS with independent clinical validation by chromosomal microarray, the gold standard test for CNVs detection. Additionally, this study involved multicenter recruitment and a prospective design with patients pursuing diagnostic testing for a range of indications. Unlike previously published studies, the negative cases in this cohort were validated with a microarray.
In summary, the results of this prospective cohort study highlight the absolute importance of prenatal genetic counseling. In particular, for patients with a fetal anatomic abnormality, diagnostic testing with chromosomal microarray and availability of fetal DNA for targeted genetic testing or exome sequencing should continue to be strongly encouraged if the patient desires a prenatal genetic diagnosis.14 Patients’ motivations are of utmost importance when proceeding with prenatal screening or testing and can help guide testing choice, whether it be noninvasive or diagnostic. Understanding these motivations will help inform the most high-yield testing for each specific patient depending on the clinical scenario. Additionally, the results of this study emphasize the need for ongoing technology and test development to address discordance between NIPS and clinical diagnostic testing and improve noninvasive genetic testing modalities.
Data availability
Study data are available from the corresponding author on request.
References
Norton, M. E. & Rink, B. D. Changing indications for invasive testing in an era of improved screening. Semin. Perinatol. 40, 56–66, https://doi.org/10.1053/j.semperi.2015.11.008 (2016).
Rose, N. C. et al. Screening for fetal chromosomal abnormalities: ACOG practice bulletin, number 226. Obstet. Gynecol. 136, e48–e69, https://doi.org/10.1097/AOG.0000000000004084 (2020).
Srebniak, M. I. et al. Frequency of submicroscopic chromosomal aberrations in pregnancies without increased risk for structural chromosomal aberrations: systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 51, 445–452, https://doi.org/10.1002/uog.17533 (2018).
Wapner, R. J. et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 367, 2175–2184, https://doi.org/10.1056/NEJMoa1203382 (2012).
Hay, S. B. et al. ACOG and SMFM guidelines for prenatal diagnosis: Is karyotyping really sufficient? Prenat. Diagn. 38, 184–189, https://doi.org/10.1002/pd.5212 (2018).
Practice bulletin no. 162: prenatal diagnostic testing for genetic disorders. Obstet. Gynecol. 127, e108–e122, https://doi.org/10.1097/AOG.0000000000001405 (2016).
Srebniak, M. I. et al. Social and medical need for whole genome high resolution NIPT. Mol. Genet. Genomic Med. 8, e1062, https://doi.org/10.1002/mgg3.1062 (2020).
Helgeson, J. et al. Clinical outcome of subchromosomal events detected by whole-genome noninvasive prenatal testing. Prenat. Diagn. 35, 999–1004, https://doi.org/10.1002/pd.4640 (2015).
Snyder, M. W. et al. Copy-number variation and false positive prenatal aneuploidy screening results. N. Engl. J. Med. 372, 1639–1645, https://doi.org/10.1056/NEJMoa1408408 (2015).
Schwartz, S. et al. Clinical experience of laboratory follow-up with noninvasive prenatal testing using cell-free DNA and positive microdeletion results in 349 cases. Prenat. Diagn. 38, 210–218, https://doi.org/10.1002/pd.5217 (2018).
Hu, H. et al. Noninvasive prenatal testing for chromosome aneuploidies and subchromosomal microdeletions/microduplications in a cohort of 8141 single pregnancies. Hum. Genomics. 13, 14, https://doi.org/10.1186/s40246-019-0198-2 (2019).
Ehrich, M. et al. Genome-wide cfDNA screening: clinical laboratory experience with the first 10,000 cases. Genet. Med. 19, 1332–1337, https://doi.org/10.1038/gim.2017.56 (2017).
Lefkowitz, R. B. et al. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. Am. J. Obstet. Gynecol. 215, 227.e1–227.e16, https://doi.org/10.1016/j.ajog.2016.02.030 (2016).
van der Meij, K. R. M. et al. TRIDENT-2: national implementation of genome-wide noninvasive prenatal testing as a first-tier screening test in the Netherlands. Am. J. Hum. Genet. 105, 1091–1101, https://doi.org/10.1016/j.ajhg.2019.10.005 (2019).
Beulen, L., Faas, B. H. W., Feenstra, I., van Vugt, J. M. G. & Bekker, M. N. Clinical utility of noninvasive prenatal testing in pregnancies with ultrasound anomalies. Ultrasound Obstet. Gynecol. 49, 721–728, https://doi.org/10.1002/uog.17228 (2017).
Wittman, A. T., Hashmi, S. S., Mendez-Figueroa, H., Nassef, S., Stevens, B. & Singletary, C. N. Patient perception of negative noninvasive prenatal testing results. AJP Rep. 6, e391–e406, https://doi.org/10.1055/s-0036-1594243 (2016).
Society for Maternal-Fetal Medicine (SMFM), Dugoff, L., Norton, M. E. & Kuller, J. A. The use of chromosomal microarray for prenatal diagnosis. Am. J. Obstet. Gynecol. 215, B2-9, https://doi.org/10.1016/j.ajog.2016.07.016 (2016).
Fiorentino, F. et al. Chromosomal microarray analysis as a first-line test in pregnancies with a priori low risk for the detection of submicroscopic chromosomal abnormalities. Eur. J. Hum. Genet. 21, 725–730, https://doi.org/10.1038/ejhg.2012.253 (2013).
Wu, X. et al. Chromosomal microarray analysis for pregnancies with or without ultrasound abnormalities in women of advanced maternal age. J. Clin. Lab. Anal. 34, e23117, https://doi.org/10.1002/jcla.23117 (2020).
Yaron, Y. The implications of noninvasive prenatal testing failures: a review of an under-discussed phenomenon. Prenat. Diagn. 36, 391–396, https://doi.org/10.1002/pd.4804 (2016).
Acknowledgements
We acknowledge our collaborators at Sequenom who ran MaterniT Genome® tests for this study at no cost on a research basis. Sequenom collaborators had the right to review the manuscript prior to publication; however, the authors retained the right to final manuscript preparation and submission. This study was funded by an Expanding the Boundaries grant from the Department of Obstetrics & Gynecology, Brigham and Women’s Hospital.
Author information
Authors and Affiliations
Contributions
Conceptualization: S. Guseh, L.D., K.G. Data curation: S. Guseh, K.G. Formal analysis: S. Guseh, K.G. Funding acquisition: S. Guseh, K.G. Investigation: S. Guseh, K.G. Project administration: S. Guseh Resources: S. Guseh, S.A., S.C., M.D., L.D., M.B., J.F., S. Gbur, H.G., N.H., C.M., M.P., P.R., A.S., K.S., C.S. Supervision: S. Guseh, L.W.-H., A.K., L.D.-A., K.G. Writing—original draft: S. Guseh, K.G. Writing—reviewing & editing: S. Guseh, L.W.-H., A.K., L.D., K.G., S.A., S.C., M.D., L.D.-A., M.B., S. Gbur, H.G., N.H., C.M., M.P., K.S., C.S.
Corresponding author
Ethics declarations
Ethics declaration
The Mass General Brigham Human Research Committee approved this study. Informed consent was obtained from all participants as required by the IRB.
Competing interests
L.W.-H. is an editor for the Prenatal Genetics section of UpToDate. S.A. received funding from BillionToOne outside the scope of the submitted work. K.G. receives funding from the NIH and has consulted for Aetion, Illumina, and BillionToOne outside the scope of the submitted work. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Guseh, S., Wilkins-Haug, L., Kaimal, A. et al. Utility of noninvasive genome-wide screening: a prospective cohort of obstetric patients undergoing diagnostic testing. Genet Med 23, 1341–1348 (2021). https://doi.org/10.1038/s41436-021-01147-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-021-01147-4