Abstract
Purpose
Biallelic variants in LARS1, coding for the cytosolic leucyl-tRNA synthetase, cause infantile liver failure syndrome 1 (ILFS1). Since its description in 2012, there has been no systematic analysis of the clinical spectrum and genetic findings.
Methods
Individuals with biallelic variants in LARS1 were included through an international, multicenter collaboration including novel and previously published patients. Clinical variables were analyzed and functional studies were performed in patient-derived fibroblasts.
Results
Twenty-five individuals from 15 families were ascertained including 12 novel patients with eight previously unreported variants. The most prominent clinical findings are recurrent elevation of liver transaminases up to liver failure and encephalopathic episodes, both triggered by febrile illness. Magnetic resonance image (MRI) changes during an encephalopathic episode can be consistent with metabolic stroke. Furthermore, growth retardation, microcytic anemia, neurodevelopmental delay, muscular hypotonia, and infection-related seizures are prevalent. Aminoacylation activity is significantly decreased in all patient cells studied upon temperature elevation in vitro.
Conclusion
ILFS1 is characterized by recurrent elevation of liver transaminases up to liver failure in conjunction with abnormalities of growth, blood, nervous system, and musculature. Encephalopathic episodes with seizures can occur independently from liver crises and may present with metabolic stroke.
Similar content being viewed by others
INTRODUCTION
Acute liver failure (ALF) in infancy is a rare but life-threatening event.1 The etiology is heterogeneous: in Europe, pediatric ALF is predominantly caused by infections and inherited metabolic diseases, whereas it remains unresolved in approximately 50% of all cases.1,2 Specific diagnosis can aid decision-making on appropriate treatment strategies and counseling of affected families.
In recent years, exome sequencing (ES) has unraveled “new” genetic causes of pediatric liver disease such as disorders of Golgi homeostasis3 and congenital disorders of intracellular trafficking.4,5,6,7,8 Another emerging group involves defects of cytosolic aminoacyl-tRNA synthetases (ARS).9,10,11,12,13 Although they are exclusively encoded by nuclear genes, ARS play an important role in protein translation, either in the cytoplasm or in the mitochondria, where they catalyze the aminoacylation of transfer RNA (tRNA) by their cognate amino acid.14 Some ARS have acquired additional domains throughout evolution that have been linked to noncanonical functions, including translation control, transcription regulation, signal transduction, and cell migration.15,16 While monoallelic variants in ARS are associated with autosomal dominant Charcot–Marie–Tooth disease,16,17 autosomal recessive ARS deficiencies due to biallelic variants can cause multisystemic disorders mostly affecting growth and the nervous system, and, in a variable manner, other organs such as the lung, liver, gut, bone marrow, kidney, and musculature,9 regardless of cytosolic or mitochondrial localization of the affected ARS. Due to their multiorgan presentation, ARS deficiencies mimic features of mitochondrial diseases and clinical differentiation can be difficult.18 To facilitate recognition of these disorders, accurate phenotyping is of pivotal importance but has remained challenging due to the low number of individuals identified.
Variants in LARS1 were first described in 2012 in six individuals belonging to the Irish Traveller community with a predominant hepatic phenotype (infantile liver failure syndrome 1 [ILFS1], MIM 615438).19 Since then, further cases were reported,9,10,20,21,22,23,24 but no systematic analysis of the phenotypic spectrum nor allelic series analyses were performed; little is known about disease mechanisms.
We systematically analyzed the clinical phenotype of subjects with pathogenic LARS1 biallelic variants, including 13 previously published cases and 12 novel patients. For the previously published individuals from the Irish Traveller community,10 follow-up data are provided. This study explores the clinical spectrum of the disease and provides biological insights into the disease mechanism via protein and enzymatic in vitro studies.
MATERIALS AND METHODS
Study design, patient ascertainment, and data acquisition
Individuals were included within an international, multicenter study. Inclusion criteria were rare biallelic variants in LARS1 classified as pathogenic or likely pathogenic according to the American College of Medical Genetics and Genomics guidelines for the interpretation of sequence variants. Inclusion was via one of the following options: (1) individuals followed by one of the coauthors, or (2) previously published patients. For option 1, clinical data were retrieved via case report forms and stored in a disease-specific database located at the University Hospital Heidelberg. For option 2, data were retrieved from the published reports identified by a comprehensive bibliographic search via PubMed and Google Scholar. Queries were based on the terms “LARS,” “LARS1,” “infantile liver failure syndrome type 1,” and “ILFS1.” For phenotyping, the following variables were analyzed within this study: individual’s country of origin, sex, age at last assessment, and vital status. Additionally, clinical features of the main organ symptoms involved were scrutinized according to Human Phenotype Ontology terminology (Supplementary information, including definitions). The cutoff date for data acquisition was 10 December 2019.
Ethics statement
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2013. Informed consent to participate in the study was obtained from all patients or their parents in case of minor patients. The study was approved by the ethical committee of the University Hospital Heidelberg.
Exome sequencing and variant filtering
LARS1 variants of hitherto unreported individuals were identified using ES; parental carrier status was confirmed via family-based genomics, trio-based ES, or Sanger sequencing (details available upon request).
In silico effect prediction of LARS1 missense variants
To assess the deleterious effect of missense variants, six commonly used prediction scores (CADD v1.4, M-CAP v.3.5a, PolyPhen-2 v.2.2.2, PROVEAN v.3.5a, REVEL v.3.5.a, and SIFT v.5.2.2) were calculated for all possible LARS1 missense variants and subsequently mapped onto a two-dimensional representation of the LARS1 protein, as methodically shown by Hebebrand et al.25 We generated all possible base exchanges referring to the LARS1 coding sequence (transcript NM_020117.9) and matched the resulting variant call format file to the GRCh37/hg19 reference genome using the Mutalyzer Position Converter. Prediction scores of resulting genomic sequence variants were calculated using the Ensembl variant effect prediction tool.26 We used the ggplot2 package within the RStudio framework (Version 1.2.1335, RStudio Team (2018) to build a generalized additive model by a quadratically penalized likelihood type approach (formula = y ~ s[x, bs = “cs”]) of the geom_smooth function to enable improved pattern detection by plotting a smoothened line and the confidence interval (CI) around the calculated scores for each alternate amino acid position.
In vitro studies in patient fibroblasts and controls
Fibroblasts were collected from affected individuals after informed consent was obtained. A fibroblast control cell line (human foreskin, not age matched) was purchased from Merck (SCC058, Darmstadt, Germany). All cells were tested for mycoplasma contamination by polymerase chain reaction (PCR). Fibroblasts were cultivated in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin at 37 °C and 5% CO2. Before harvesting the cells for aminoacylation measurement and immunoblotting, they were cultured in duplicates for 48 hours at 37 °C and 40 °C to mimic febrile conditions.
LARS1 enzyme assay
Aminoacylation was assessed by measuring LARS1 activity in cultured fibroblasts. Fibroblast lysates (cytosolic fraction) were incubated at 37 °C for 10 minutes in a reaction buffer containing 50 mmol/L Tris buffer pH 7.5, 12 mmol/L MgCl2, 25 mmol/L KCl, 1 mg/mL bovine serum albumin, 0.5 mmol/L spermine, 1 mmol/L ATP, 0.2 mmol/L yeast total tRNA, 1 mmol/L dithiothreitol, 0.3 mmol/L [13C2]-leucine, [13C6,15N]-isoleucine, [D4]-lysine, and [15N2]-arginine. The reaction was terminated using trichloroacetic acid. After sample washing with trichloroacetic acid, ammonia was added to release the labeled amino acids from the tRNAs. [D3]-leucine and [13C6]-arginine were added as internal standards, and the labeled amino acids were quantified by liquid chromatography–tandem mass spectrometry (LC-MS/MS). Intra-assay and interassay variation were <15%. Isoleucyl-tRNA synthetase (IARS1), lysyl-tRNA synthetase (KARS1), and arginyl-tRNA synthetase (RARS1) activities were simultaneously detected as control enzymes. Statistical significance was assessed using Student's t-test, CI 95% (SPSS 26.0.0.1).
Western blot
For immunoblotting, cells were washed in phosphate buffered saline with tween (0.1%), and resolved in radioimmunoprecipitation assay buffer. Then, 20 µg of protein of every sample was separated on a 10% polyacrylamide gel. Primary antibodies against LARS1 (Abcam, Berlin, Germany) and β-actin (Santa Cruz Biotechnology, Heidelberg, Germany) were incubated overnight. Secondary horseradish peroxidase–coupled antibodies (goat antirabbit or rabbit antimouse) were obtained from Dianova (Hamburg, Germany). Enhanced chemiluminescence of proteins was detected using Vilberscan Fusion FX7.
RESULTS
Study population/genetics
Literature search identified five peer-reviewed publications on individuals with biallelic pathogenic LARS1 variants, published since April 2012, reporting 13 patients from six families with a total of nine different variants. Twelve further patients from nine families harboring eight novel variants were identified by ES and Sanger sequencing by one of the coauthors (Tables 1, S1, and S2). Of those, one individual (LARS1-10) had been presented at a conference previously.23 Two further patients had been reported in abstract books of conferences,20,22 but as they were not published in peer-reviewed journals and had not been identified by one of the coauthors, they were not included in this study.
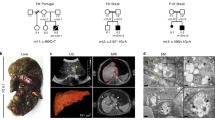
In total, 25 individuals from 15 families with biallelic pathogenic LARS1 variants were identified. There were a total of 17 different variants: 12 missense variants, 1 frameshift variant, 2 in-frame deletions, 1 nonsense variant, and 1 splice site variant. Variants are distributed throughout the gene with an increased density in the catalytic and editing domain with variant c.(1292T>A); p.(Val431Asp) being the most frequent one, present in six families (Fig. 1b). All patients carry at least one missense variant affecting the catalytic or editing domain, except two individuals from family V (LARS1-06; LARS1-07) carrying a homozygous nonsense variant affecting the UNE-L domain in the C-terminal part of the protein (Fig. 1a, Table S1). Interestingly, variants localizing to the highly conserved N-terminal editing and catalytic domain tend to result in higher REVEL scores compared with variants in the C-terminal region of the protein (Figs. 1c and S1).
(a) All known pathogenic variants including affected region of LARS1 protein. Novel variants are displayed in red, previously published variants present in our patients in bold. (b) Density plot of known pathogenic variants with respect to affected protein domains. (c) In silico effect prediction of LARS1 missense variants using REVEL score. (d) Most prevalent clinical signs within our cohort present in at least ten patients and 50% of analyzed individuals. Recurrent elevation of transaminases includes episodes of acute liver failure; neurodevelopmental delay includes motor and language delay as well as learning disabilities. IUGR intrauterine growth restriction, SGA small for gestational age.
Phenotypic spectrum
The cohort comprises 15 male and 10 female individuals originating from Europe, North America, and Asia. Twenty patients were alive at the time of this report while five patients had died, all due to severe multiorgan failure triggered by febrile illness (median age of death: 4.0 years, range: 1.7 months–8 years). Age at presentation was at birth for 23 patients (presenting small for gestational age). For siblings LARS1-06 and LARS1-07, anthropometric data at birth are unknown and age at presentation was at one month of age. Median age at diagnosis was 3 years (range: first month–23 years) and age at last visit ranged from 2 months to 36 years (median: 7.8 years).
Hereafter, clinical features of the cohort are listed, organized by organ system involvement. For an overview of affected organ systems and most prevalent symptoms, see Tables 1, S1, and Fig. 1d.
Abnormality of growth (HP:0001507)
Intrauterine growth retardation (IUGR) was noted in all patients during pregnancy where data were available (n = 11). In line with this, all newborns were reported small for gestational age (SGA) when birth data were available (n = 23; mean birth weight -2.13 SDS). Fifteen individuals were born preterm (15/25; range: 28–36 weeks). In concordance with the high prevalence of SGA, 24/25 patients presented with failure to thrive during infancy associated with poor feeding in 18 cases. Short stature was present in nine individuals at last follow-up visit.
Abnormality of the nervous system (HP:0000707)
Presence of neurological symptoms was not addressed in LARS1-23, whereas neurological abnormalities were noted in all other patients (n = 24). Twenty-two of these 24 individuals showed neurodevelopmental delay: gross motor delay in 18/24, and delayed speech and language development in 10/13. Cognitive impairment was present in 13/17 patients, ranging from mild to severe, whereas most patients presented with learning disabilities.
Nineteen of 24 patients presented with seizures, commonly associated with infections. Thirteen patients developed infection-associated encephalopathic episodes ranging from one to seven. These encephalopathic episodes were accompanied by seizures up to status epilepticus, lasted up to five days and also occurred independently of hepatic decompensation (Table 2). As far as we can deduct from MRI reports and from inspection of follow-up MRI of LARS1-01 and MRIs in LARS1-03, findings encompass a broad spectrum ranging from normal findings, nonspecific atrophy, and signal changes of periventricular white matter to transient edema, lasting lesions of white and/or gray matter, and malformations. MRI changes in LARS1-03 with transient, extensive, and symmetric edema of the brainstem and cerebellum including dentate nuclei, basal ganglia, thalami, and hippocampi might be consistent with so-called metabolic stroke, namely the association of clinical signs of an acute neurological syndrome with stroke-like lesions (Fig. S4). However, cortical changes might be associated with seizures/postictal and should be carefully interpreted, whereas signal changes of periventricular white matter could be related to prematurity.
Despite severe neurologic involvement during decompensations, all patients experienced good recovery with exception of LARS1-04, who improved only gradually and incompletely; on discharge two months after the first encephalopathic episode, there still was a left-sided hemiparesis with slight hypertonia and spasticity of the extremities. Further follow-up was not possible as he died at 22 months during a second encephalopathic episode. LARS1-09, after being liver transplanted aged 4 months, experienced one encephalopathic episode without liver decompensation. He experienced developmental delay and attends special needs education. Additional neurological and neuroradiological findings included postural instability, microcephaly, sensineuronal deafness, agenesis of the corpus callosum, and cerebral palsy (Table S1). Interestingly, despite early neurodevelopmental delay, follow-up of the Irish Traveller cohort reveals positive neurodevelopment in most of the patients some years later. Only LARS1-21 attends a special needs school, while others attend mainstream education (median age at last follow-up 8.4 years, range 4–36 years).
Abnormality of the blood and blood forming system (HP:0001871)
Microcytic anemia was noted in 24/25 individuals and specified as hypochromic in 18, but also microangiopathic hemolytic anemia (2) and siderocytic anemia (1) were reported. Anemia could already be present during the neonatal period and was resistant to iron supplementation. Minimum measured hemoglobin value was 5.2 g/dL (52.0 g/L), necessitating repeated erythrocyte transfusions. Thrombocytopenia was reported in five patients.
Abnormality of the liver (HP:0001392)
Twenty-four individuals showed liver involvement with the most frequent finding being recurrent elevation of liver transaminases (n = 21). Episodes ranged from elevated liver transaminases up to full-blown liver failure (n = 17) and hepatic transaminases normalized within the interval. Even during liver failure episodes, hyperammonemia and hypoglycemia were present only in a few cases (Table 2). Age at onset of hepatic decompensations was within the first two years of life (range: 1–13 months; median: 8.0), and the number of decompensations ranged from one to eight episodes (median: 4.0). Median age of the most severe episode was 11 months, and the last episode of elevated liver transaminases occurred between 11 months and 16 years (median age: 4.9 years). All decompensations were triggered by febrile illnesses. For laboratory values and further clinical signs during acute disease crises see Table 2. ALF was often accompanied by encephalopathy and hepatic transaminases were mostly elevated during encephalopathic crises, but there were encephalopathic episodes without signs of liver affection. Liver biopsy revealed steatosis, fibrosis, or cirrhosis in 12/13 patients (Table S1).
Only the two siblings LARS1-06 and LARS1-07 did not experience any hepatic decompensation. Individual LARS1-09 was liver transplanted at the age of 4 months after a severe episode of ALF and did not experience another hepatic decompensation afterward. At time of this report he is 8 years old, given a post-transplant follow-up time of seven years.
Abnormality of the musculature (HP:0003011)
Muscular hypotonia was present in 17/21 patients. LARS1-04 presented with muscular hypertonia due to flexion spasms of the left arm and leg following his first encephalopathic episode. Exercise intolerance was noted in two patients. Follow-up of the Irish Traveller cohort showed that muscular hypotonia diagnosed during the first years of life can normalize over time (LARS1-15 to 18, age at last visit 8, 8, and 17 years).
Abnormality of the immune system (HP:0002715)
Frequent infections were reported in 17 individuals. However, quantification of infections was not performed and anamnestic hints of severe infections or atypical pathogens were not reported. Hypogammaglobulinemia was found in three patients. Interestingly, viral infections of the respiratory and intestinal tract were responsible for triggering severe decompensations including liver failure and encephalopathy. Among identified pathogens, influenza A or B were found to be the most frequent pathogens, evoking severe decompensations in five patients.
Additional affected organ systems, hypoalbuminemia and dietary preferences
Other organs affected to a lesser extent included the skeletal (n = 7) and renal systems (n = 6), nonspecific facial dysmorphology (abnormalities of head or neck) (n = 5), and the lung (n = 5). Postnatal hypoglycemia that required intravenous glucose infusion was reported in five patients, with short fasting tolerance and recurrent hypoglycemia over the first six months of age in two of them. Hypoalbuminemia was noted in 24/25, worsening during febrile illness and resistant to higher nutritional intake. Interestingly, parents of four individuals described a preference for meat, milk, eggs, and, in general, products with high protein content when asked specifically (Table S1).
In vitro analyses
For functional analyses, fibroblasts of eight patients were available (Table S1). All studies were performed as biological triplicates in comparison with control fibroblasts.
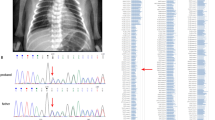
Aminoacylation assay showed reduced enzyme activity in all available patient fibroblasts to different extents (35–85% of controls), significantly reduced (p < 0.05) in four of eight cases. After temperature elevation to 40 °C, aminoacylation activity was reduced significantly in all samples (11–49% of controls, p < 0.05 Fig. 2a; Table S3). Reduced enzyme activity was specific for LARS1; activities of KARS1 and RARS1 did not differ from the control cells at both conditions. For IARS1 there was also no significant difference for all patients except patient LARS1-02; the reason remains unclear, but due to structural similarity there may be an interference of Leu and Ile in our assay (Fig. S2, Table S3).
(a) Aminoacylation measured in fibroblasts of patients and controls cultured at 37 °C or 40 °C for 48 hours. Error bars indicate standard deviations. Enzyme activity measured at 40 °C differed significantly compared with controls (p values Table S3) in all patient samples. Comparison of 40 °C aminoacylation with the activity at 37 °C in the same sample showed significant decrease upon temperature shift in three patient cells (see asterisks). (b) LARS1 protein expression by western blotting in fibroblasts of patients and controls cultured at 37 °C or 40 °C during 48 hours.
Western blot analysis showed reduced protein levels in most patient fibroblasts (Fig. 2b), decreasing upon temperature elevation to 40 °C. LARS1-03, LARS1-06, and LARS1-10 did not show reduced protein levels at 37 °C, but levels decreased on temperature shift. LARS1-06 showed a protein band reduced in size, possibly corresponding to the truncated product induced by the homozygous nonsense variant c.(3313C>T), p.(Arg1105*), that seems to escape nonsense-mediated messenger RNA (mRNA) decay (NMD) (Figs. 2b and S3).
DISCUSSION
We present an international, multicenter cross-sectional study on the clinical phenotype of individuals with ILFS1 in a cohort of 25 patients from 15 families, including 12 novel patients. Genotype–phenotype associations are explored; aminoacylation assays and protein expression analysis contribute to the understanding of the underlying pathomechanism.
The predominant affections were abnormalities of growth (25/25), nervous system (24/24), blood (24/25), liver (24/25), and musculature (18/21). Specific clinical symptoms prevalent in at least 75% of the patients are SGA/IUGR (23/23), microcytic anemia (24/25), failure to thrive (24/25), hypoalbuminemia (24/25), neurodevelopmental delay (22/24), recurrent elevation of liver transaminases (21/25), muscular hypotonia (17/21), and seizures (19/24, mostly associated with infections) (Fig. 1d). Our study primarily confirms10,21,24 and further clarifies the clinical phenotype of ILFS1 with this systematic approach applied to an increased number of patients. While growth retardation, motor delay, muscular hypotonia, seizures, and hypoalbuminemia are common in cytosolic ARS deficiencies,9 recurrent elevation of liver transaminases up to liver failure but also microcytic anemia are more specific and may guide physicians in the diagnostic work-up.
Recurrent hepatic crises with elevated transaminases up to liver failure can be life-threatening and often are the most prominently noticed clinical sign of acute illness. Liver biopsies show signs of chronic liver disease, and the recurrent crises may therefore reflect acute-on-chronic liver failure episodes. However, dynamics of tissue reorganization is unclear, as liver biopsies were only available in some patients and taken at different time points. Follow-up data of the Irish Traveller cohort are encouraging, with some of the individuals having reached adulthood with normal liver function tests, pointing at clinical variability.
Interestingly, four individuals did not show recurrent elevation of liver transaminases. For LARS1-11, this may be due to young age at last assessment (1 year). LARS1-09 underwent liver transplant and showed no further episodes afterward. The other two individuals are the siblings LARS1-06 and LARS1-07, sharing homozygosity for the nonsense variant c.(3313C>T), p.(Arg1105*). Western blot suggests that this variant leads to a truncated isoform, likely affecting the UNE-L domain in the C-terminal part of the protein, which is a unique sequence motif acquired later in evolution with no link to their aminoacylation activity.27 In contrast, all other individuals carry at least one missense variant affecting the catalytic domain. In silico analysis of independent pathogenicity-computation methods revealed a high impact of missense variants within the highly conserved N-terminal region, which include the catalytic and editing domains, underlining its functional relevance (Figs. 1c and S1).
Due to the acute phase response, demand for protein in the liver is increased during febrile illness. Incorporation of incorrect amino acids into nascent polypeptides can cause misfolding and production of defective proteins, eventually leading to endoplasmic reticulum (ER) stress, a mechanism known to be involved in infection-associated liver disease.4,28,29 While there is a relatively high aminoacylation activity in all patient samples after culturing at 37 °C, a temperature shift to 40 °C reduces the activity significantly in all patient samples compared with controls (Fig. 2a, Table S3). In line with this, immunoblotting shows reduced expression levels after temperature shift, implicating temperature-sensitive transcripts of mutant LARS1 proteins (Fig. 2b). Reduced enzymatic activity during conditions with elevated temperature could be an underlying mechanism of fever-related episodes of liver and cerebral dysfunction in ILFS1. However, we observed temperature dependent reduced aminoacylation activity also in LARS1-06, who did not show episodic dysfunction of liver or brain, while presenting a more chronic phenotype with severe global developmental delay and growth retardation. Due to the different clinical presentation of the siblings LARS1-06 and LARS1-07 and consanguinity in their family, other monogenetic diseases have been considered but apart from their homozygous LARS1 nonsense variant, there were no other pathogenic or likely pathogenic variants identified in these individuals by ES.
Apart from reduced aminoacylation activity, alteration of noncanonical functions of LARS1 could contribute to pathomechanism. An interaction of the C-terminal region of LARS1 with mTOR complex 1 (mTORC1) at the lysosomal membrane is known, directly stimulating the mTORC1 pathway inducing autophagy upon leucine deprivation, sensed by the leucine binding site of LARS1.30,31,32 We hypothesize that an altered C-terminal structure of LARS1 may stimulate the mTORC1 pathway and thus autophagy. As autophagy is pivotal to neurologic development and cell survival,33 this mechanism could be linked to neurodevelopmental delay in ILFS1. More functional studies are needed to understand the role of type and localization of LARS1 variants on aminoacylation activity, possibly autophagy, and ultimately clinical phenotype. To address these issues, aminoacylation activity and autophagy markers could be analyzed in cellular models after introduction of pathogenic LARS1 variants affecting different parts of the gene product (e.g., via CRISPR/Cas9 studies). Zebrafish with biallelic LARS1 variants affecting the catalytic domain show a clear hepatic phenotype.34 Further animal models harboring different LARS1 variants, including variants located in the catalytic domain but also models with variants affecting the C-terminal part of the gene, such as the UNE-L domain, will help to explore genotype–phenotype correlations in ILFS1.
Recurrent acute elevation of liver transaminases and encephalopathic episodes triggered by febrile illness are the most prominent findings in our cohort, and variants in LARS1 should be considered for differential diagnosis in both clinical conditions. Interestingly, MRI during encephalopathic episodes might disclose not only cortical, postictal changes, but also changes of deep gray matter suggesting metabolic stroke. Knowing the underlying cause can guide management and development of therapeutic strategies. The OMIM term “ILFS1” considers the prominent hepatic presentation disregarding the independent occurrence of encephalopathic episodes and the involvement of further organ systems. Additionally, “ILFS2” due to variants in NBAS is mechanistically not related to ARS deficiencies, making this nomenclature misleading. We suggest using a nomenclature separating ARS deficiencies from disorders of intracellular trafficking leading to a hepatic phenotype (such as CALFAN syndrome due to variants in SCYL1, NBAS deficiency, and RINT1 deficiency5,6,7).
Therapeutic options for patients with ILFS1 are limited. Leucine levels are normal in patients’ plasma; however, Casey et al. recommended a high daily protein intake of 2.5 g/kg/day10 and there is the anecdotal report of preference for a high protein diet in ILFS1 patients. However, data are lacking to judge the effect of leucine supplementation or high protein intake on the disease course of ILFS1 patients and future studies to quantify and critically analyze this potential dietary approach to ILFS1 are needed. As febrile illness triggered life-threatening episodes and increased temperature decreased aminoacylation in vitro, early antipyretic treatment should be considered. To prevent infectious triggers, the vaccination plan should carefully be followed with the option to vaccinate against influenza infections yearly, as these were the most noted pathogen triggering life-threatening decompensations with two fatal outcomes. Liver transplantation may be considered as a therapeutic option as there were no further liver decompensations or encephalopathic episodes, and anemia had resolved and albumin normalized in the single case where experience has been gathered. However, despite liver transplantation within the first months of life, there was poor weight gain, short stature, and more important pronounced developmental delay with necessity of special education and epilepsy (follow-up time 7 years), pointing at the multisystem character of the disease and the difficulty of transplant decision.
In conclusion, recurrent elevation of liver transaminases up to liver failure and/or encephalopathic episodes with or without metabolic stroke triggered by febrile illness in the first two years of life, even more if together with growth retardation, microcytic anemia, muscular hypotonia, and developmental delay, should prompt genetic testing for LARS1 or consideration for ES.
References
Squires RH Jr, Shneider BL, Bucuvalas J, et al. Acute liver failure in children: the first 348 patients in the pediatric acute liver failure study group. J Pediatr. 2006;148:652–658.
Kathemann S, Bechmann LP, Sowa JP, et al. Etiology, outcome and prognostic factors of childhood acute liver failure in a German Single Center. Ann Hepatol. 2015;14:722–728.
Jansen JC, Cirak S, van Scherpenzeel M, et al. CCDC115 deficiency causes a disorder of Golgi homeostasis with abnormal protein glycosylation. Am J Hum Genet. 2016;98:310–321.
Haack TB, Staufner C, Kopke MG, et al. Biallelic mutations in NBAS cause recurrent acute liver failure with onset in infancy. Am J Hum Genet. 2015;97:163–169.
Staufner C, Haack TB, Kopke MG, et al. Recurrent acute liver failure due to NBAS deficiency: phenotypic spectrum, disease mechanisms, and therapeutic concepts. J Inherit Metab Dis. 2016;39:3–16.
Lenz D, McClean P, Kansu A, et al. SCYL1 variants cause a syndrome with lowγ-glutamyl-transferase cholestasis, acute liver failure, and neurodegeneration(CALFAN). Genet Med. 2018;20:1255–1265.
Cousin MA, Conboy E, Wang JS, et al. RINT1 bi-allelic variations cause infantile-onset recurrent acute liver failure and skeletal abnormalities. Am J Hum Genet. 2019;105:108–121.
Staufner C, Peters B, Wagner M, et al. Defining clinical subgroups and genotype–phenotype correlations in NBAS-associated disease across 110 patients. Genet Med. 2020;22:610–621.
Fuchs SA, Schene IF, Kok G, et al. Aminoacyl-tRNA synthetase deficiencies in search of common themes. Genet Med. 2019;21:319–330.
Casey JP, Slattery S, Cotter M, et al. Clinical and genetic characterisation of infantile liver failure syndrome type 1, due to recessive mutations in LARS. J Inherit Metab Dis. 2015;38:1085–1092.
Kopajtich R, Murayama K, Janecke AR, et al. Biallelic IARS mutations cause growth retardation with prenatal onset, intellectual disability, muscular hypotonia, and infantile hepatopathy. Am J Hum Genet. 2016;99:414–422.
Nowaczyk MJ, Huang L, Tarnopolsky M, et al. A novel multisystem disease associated with recessive mutations in the tyrosyl-tRNA synthetase (YARS) gene. Am J Med Genet A. 2017;173:126–134.
van Meel E, Wegner DJ, Cliften P, et al. Rare recessive loss-of-function methionyl-tRNA synthetase mutations presenting as a multi-organ phenotype. BMC Med Genet. 2013;14:106.
Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158.
Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30:569–574.
Yao P, Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med. 2013;5:332–343.
Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genomics Hum Genet. 2008;9:87–107.
Diodato D, Ghezzi D, Tiranti V. The mitochondrial aminoacyl tRNA synthetases: genes and syndromes. Int J Cell Biol. 2014;2014:787956.
Casey JP, McGettigan P, Lynam-Lennon N, et al. Identification of a mutation in LARS as a novel cause of infantile hepatopathy. Mol Genet Metab. 2012;106:351–358.
El-Gharbawy A, Sebastian J, Ghaloul-Gonzalez L, et al. LARS mutations in non-Irish Travellers: an under-recognized multi-system disorder characterized by infantile hepatopathy during physiological stress. Mitochondrion. 2015;24:S40–S41.
Lin W, Zheng Q, Guo L, Cheng Y, Song YZ. First non-Caucasian case of infantile liver failure syndrome type I: clinical characteristics and molecular diagnosis. Chin J Contemp Pediatr. 2017;19:913–920.
El-Gharbawy A, Ranganathan S, Ghaloul-Gonzales L, et al. Management of two patients with Infantile liver failure syndrome type I due to LARS mutations: an under recognized reversible hepatopathy. J Inherit Metab Dis. 2018;41 (Suppl 1):S37–S219.
Husain RA, Dieckmann A, Betz B, et al. Abstracts of the 33rd Annual Conference of the Arbeitsgemeinschaft für Pädiatrische Stoffwechselstörungen (APS) (Association for Pediatric Metabolic Disorders). Monatsschr Kinderheilkd. 2019;167:359–376.
Peroutka C, Salas J, Britton J, et al. Severe neonatal manifestations of infantile liver failure syndrome type 1 caused by cytosolic leucine-tRNA synthetase deficiency. JIMD Rep. 2019;45:71–76.
Hebebrand M, Huffmeier U, Trollmann R, et al. The mutational and phenotypic spectrum of TUBA1A-associated tubulinopathy. Orphanet J Rare Dis. 2019;14:38.
McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122.
Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11:668–674.
Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809.
Scheper GC, Van Der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–723.
Han JM, Jeong SJ, Park MC, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424.
He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Ann Rev Genet. 2009;43:67–93.
Kim JH, Lee C, Lee M, et al. Control of leucine-dependent mTORC1 pathway through chemical intervention of leucyl-tRNA synthetase and RagD interaction. Nat Commun. 2017;8:732.
Kulkarni A, Chen J, Maday S. Neuronal autophagy and intercellular regulation of homeostasis in the brain. Curr Opin Neurobiol. 2018;51:29–36.
Wang Z, Song J, Luo L, Ma J. Loss of Leucyl-tRNA synthetase b leads to ILFS1-like symptoms in zebrafish. Biochem Biophys Res Commun. 2018;505:378–384.
Acknowledgements
We thank all the individuals involved in the study for their participation. We thank Selina Wächter for excellent technical assistance as well as Ina Ellrichmann and Martina Kohl for supporting data collection. C.S. is supported by the Dietmar Hopp Foundation, St. Leon-Rot, Germany (grant number 23011235). D.L. is supported by the Deutsche Leberstiftung (grant number S163/10052/2018). Additionally, this work was supported by the German Federal Ministry of Education and Research (BMBF) through the E-Rare project GENOMIT (01GM1603 to H.P.). T.B.H. was supported by the German Bundesministerium für Bildung und Forschung (BMBF) through the Juniorverbund in der Systemmedizin “mitOmics” (FKZ 01ZX1405C) and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, grant number 418081722). Furthermore, this work was supported in part by the National Institutes of Health, National Institute of Neurologic Disorders and Stroke (R35 NS105078) and a jointly funded National Human Genome Research Institute (NHGRI) and National Heart, Lung, and Blood Institute (NHLBI) grant to the Baylor‐Hopkins Center for Mendelian Genomics (UM1 HG006542) to J.R.L. J.E.P. is supported by NHGRI (K08 HG008986).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals and Novartis, and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting, and is on the Scientific Advisory Board of Baylor Genetics (BG). The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical genomic sequencing offered at BG (http://bmgl.com). The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lenz, D., Smith, D.E.C., Crushell, E. et al. Genotypic diversity and phenotypic spectrum of infantile liver failure syndrome type 1 due to variants in LARS1. Genet Med 22, 1863–1873 (2020). https://doi.org/10.1038/s41436-020-0904-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0904-4
Keywords
This article is cited by
-
Goldilocks principle and recessive disease
European Journal of Human Genetics (2024)
-
Familial Infantile Liver Failure Syndrome 1: Novel LARS1 Gene Mutation
Indian Journal of Pediatrics (2022)
-
Leucyl-tRNA synthetase deficiency systemically induces excessive autophagy in zebrafish
Scientific Reports (2021)