Abstract
Purpose
Abnormality of the corpus callosum (AbnCC) is etiologically a heterogeneous condition and the prognosis in prenatally diagnosed cases is difficult to predict. The purpose of our research was to establish the diagnostic yield using chromosomal microarray (CMA) and exome sequencing (ES) in cases with prenatally diagnosed isolated (iAbnCC) and nonisolated AbnCC (niAbnCC).
Methods
CMA and prenatal trio ES (pES) were done on 65 fetuses with iAbnCC and niAbnCC. Only pathogenic gene variants known to be associated with AbnCC and/or intellectual disability were considered.
Results
pES results were available within a median of 21.5 days (9–53 days). A pathogenic single-nucleotide variant (SNV) was identified in 12 cases (18%) and a pathogenic CNV was identified in 3 cases (4.5%). Thus, the genetic etiology was determined in 23% of cases. In all diagnosed cases, the results provided sufficient information regarding the neurodevelopmental prognosis and helped the parents to make an informed decision regarding the outcome of the pregnancy.
Conclusion
Our results show the significant diagnostic and prognostic contribution of CMA and pES in cases with prenatally diagnosed AbnCC. Further prospective cohort studies with long-term follow-up of the born children will be needed to provide accurate prenatal counseling after a negative pES result.
Similar content being viewed by others
INTRODUCTION
The corpus callosum (CC) is the largest brain commissure. It contains about 180 million axons connecting both hemispheres with a fundamental role in the integration of sensory, visuomotor, and cognitive processes. Abnormality of the corpus callosum (AbnCC) includes partial or complete agenesis or anomaly of its size and shape. The estimated incidence is about 1.8 per 100,000 in the general population and 230–600 per 100,000 in individuals with neurodevelopmental disorders.1,2,3 The intellectual abilities of people with AbnCC are extremely variable, ranging from normal development to profound intellectual disability (ID).
Prenatal diagnosis of AbnCC is usually performed on the 2nd trimester ultrasound (US).4,5 The parents’ primary concern is the neurodevelopmental prognosis, which is uncertain and mainly related to whether the AbnCC appears isolated (iAbnCC) (i.e., without other anomalies on further imaging including fetal magnetic resonance image [MRI]). In iAbnCC, data suggest that the neurodevelopment is within the normal range in 80% whereas 20% of the children will develop mild to severe ID.6,7,8 The parents therefore make their decision to continue with or terminate the pregnancy based on these probabilities. Non-isolated AbnCC (niAbnCC) is usually considered to be associated with poor developmental outcome. According to the French law, couples can request pregnancy termination at any time, regardless of the isolated or associated nature of the AbnCC.
As in most congenital malformations, the prognosis of AbnCC is linked to its etiological diagnosis. Up to now in France, the genetic investigations performed in prenatal care were fetal karyotyping and chromosomal microarray (CMA). In postnatal series including patients with AbnCC and developmental delay/ID, chromosomal anomalies account for only 10–15% of genetic causes, while about 40% are due to monogenic causes, representing most AbnCC etiologies.9,10,11 Recent studies have investigated the utility of prenatal exome sequencing (pES) in cases of various fetal anomalies during pregnancy12,13,14,15 but no series focusing on AbnCC have been reported so far.
In this study, we aimed to evaluate the feasibility of pES analysis (including copy-number variant [CNV] analysis) in prenatally diagnosed AbnCC and its contribution to parental counseling.
MATERIALS AND METHODS
Pregnant women with fetal isolated or nonisolated AbnCC who opted for invasive testing (amniocentesis) and consented for participation in the study were included.
AbnCC was identified at second and third trimester screening ultrasound (US) examinations. A second line US examination was performed to confirm AbnCC and to investigate other associated fetal anomalies. The pregnant women had three consecutive consultations in a referral center for prenatal diagnosis: with a fetal medicine specialist, a pediatric neurologist and a clinical geneticist. As part of the routine management, amniocentesis was offered and performed for fetal karyotyping and CMA. pES was offered as part as this protocol and parents both consented to blood sampling (for trio analysis) and genetic studies.
Fetal AbnCC comprised either complete or partial agenesis or short CC. Two groups were defined at inclusion time:
-
1.
Apparently isolated AbnCC (iAbnCC), defined as the absence of other malformations or presence of not more than only one minor anomaly (i.e., US finding that brings little or no significant prenatal or postnatal functional impact).16
-
2.
Nonisolated AbnCC (niAbnCC) defined as the presence of at least one additional major malformation (cerebral or extracerebral), or at least two minor anomalies.
Parental blood samples were collected for DNA extraction and fetal DNA was obtained from amniotic fluid. CMA and pES were carried out in parallel. We performed trio ES on a NextSeq 500 Sequencing System (Illumina, San Diego, CA), with a 2 × 150 bp high output sequencing kit after a 12-plex enrichment with SeqCap EZ MedExome kit (Roche, Basel, Switzerland), according to manufacturer’s specifications. Sequence quality was assessed with FastQC 0.11.5, then the reads were mapped using BWA-MEM (version 0.7.13), sorted and indexed in a bam file (samtools 1.4.1), duplicates were flagged (sambamba 0.6.6), coverage was calculated (picard-tools 2.10.10). Variant calling was done with GATK 3.7 Haplotype Caller. Coverage for these samples was >93% at a 20× depth threshold. Variants were annotated with SnpEff 4.3, dbNSFP 2.9.3, gnomAD, ClinVar, HGMD, OMIM, AbnCC genes (Supplementary data), and an internal database. Filtering was performed with criteria based on the consequence on the protein and frequency in gnomAD. CNVs were called with CNVKit and Excavator2 and annotated using annovar. To shorten turnaround times and lower the costs, pES was processed with routine postnatal samples (2 runs per week).
Variants were classified according to the American College of Medical Genetics and Genomics (ACMG) guidelines.17 All candidate pathogenic variants were confirmed by Sanger sequencing.
In accordance with the French law (French Council of State, July 2018), secondary findings were not analyzed for the fetus or for the parents, and variants of unknown significance (VUS) were not reported, as required by French legislation about prenatal diagnosis. Also, only parental status for the causative variants found in the fetus were reported.
In all cases with a diagnosis, pES results were explained during a consultation with a clinical geneticist.
Pregnancy outcome was systematically collected. All participants gave written informed consent and the study was approved by the Personal Protection Committee (CPP, Paris, France). The clinical trial number of the study was NCT03600792.
RESULTS
We prospectively included 65 fetuses with AbnCC from September 2018 to January 2020. The median gestational age at inclusion was 24.5 weeks gestation (WG; 19–32 WG). The study flowchart is shown in Fig. 1.
A total of 28 (43%) fetuses had complete agenesis of the CC, 18 (28%) had partial agenesis, and 19 (29%) had a short but complete CC. At inclusion time, 54 (83%) fetuses had apparently iAbnCC and 11 (17%) fetuses had niAbnCC. In 8/54 cases (15%), AbnCC was classified as isolated at inclusion, but further prenatal imaging revealed other malformations or anomalies.
Associated malformations included interhemispheric cysts (n = 2), pericallous lipoma, cerebellar hypoplasia or dysgenesis (n = 5), brain stem hypoplasia, gyration anomaly (n = 2), cleft palate, craniosynostosis, ventricular septal defect (n = 2), intrauterine growth restriction (n = 2), and club feet. Two fetuses had at least two minor anomalies and were classified as niAbnCC (tethered cord, pelviectasis, and vertebral segmentation defect in one case, and increased nuchal translucency [7.5 mm] with right aortic arch in another case). Three fetuses had only one minor anomaly and were classified as iAbnCC (respectively pelviectasis, Blake’s pouch cyst, and mild polyhydramnios).
In all cases, pES was successfully achieved. The median delay for pES results was 21.5 days (9–59 days). In 63 cases (97%), the pES result was delivered to the parents during the pregnancy. In two cases, the results were available only after birth (case 9 due to premature delivery and case 33 due to late inclusion after late identification of AbnCC).
At least one pathogenic or probably pathogenic variant was identified in 12 fetuses (BRAT1, PPP2R1A, PTPN11, TUBA1A, KIF1A, ASXL3, SHROOM4, ARID1B, ZEB2, ZBTB20, and KANSL1) including one fetus with two variants in two different genes (BRPF1 and RTTN) (Table 1). Thus, pES provided additional genetic diagnosis in 18% of cases compared with fetal karyotyping and CMA alone. In 11/12 cases (92%), the diagnosis was found to be associated with a poor neurodevelopmental outcome, with highly penetrant ID, whereas one diagnosis (PTPN11) was characterized by variable development outcome. Moreover, three CNVs were identified by pES (4.5%) (6qter deletion, 17q21 deletion, and 2q22 deletion), all confirmed by CMA. No cases of aneuploidy were detected in this series (apart from Turner syndrome in case 44, which did not explain the AbnCC phenotype).
Among the diagnosed fetuses, 13/15 presented with apparently iAbnCC when included. Among them, fetal MRI or second line US examination showed another malformation in five cases (ventricular septal defect [2q22 deletion, ZEB2 variant], vermian hypoplasia [TUBA1A variant], club feet [KIF1A variant] and cerebellar dysgenesis [BRPF1 and RTTN variants]).
A VUS was identified in six cases in six different genes but their pathogenicity could not be established based on molecular data. Phenotypic data provided by US or fetal MRI were not helpful to reclassify these variants. These VUS were discussed between specialists and were not reported to the parents.
In all the cases in which the diagnosis was reported during the pregnancy, 11/14 (73%) couples chose to terminate the pregnancy (17q21 deletion, 2q22 deletion, PPP2R1A, SHROOM4, KIF1A, ASXL3, ARID1B, ZBTB20, ZEB2, BRPF1 and RTTN, and TUBA1A) while 3/14 (KANSL1, BRAT1, 6qter deletion) decided to continue with the pregnancy. In the case of the BRAT1 variant, this diagnosis allowed the parents to prepare for the poor prognosis and redirect the postnatal care to palliation.
When pES did not identify a diagnosis, 42/50 couples (84%) continued with the pregnancy and 7/50 couples opted for termination of pregnancy (including 4 AbnCC with associated malformations).
DISCUSSION
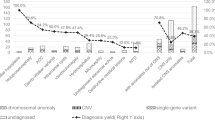
We report the first experience of pES in AbnCC, which is the most frequent cerebral malformation and is characterized by an uncertain prognosis. We found that pES was feasible with a result available during the pregnancy in most cases. Moreover, pES provided additional genetic diagnosis in up to 18% of cases compared with fetal karyotyping and CMA alone.
pES has already been reported in various fetal malformations with very variable diagnostic yields, from 8% to 57%.12,13,14,18 In the largest study (610 fetuses), the mean diagnostic yield was 8.5%, including broad phenotypic groups.12 In the group including brain anomalies, the diagnostic yield was less than 4%, but no details were available about the type of malformations. No study focusing on AbnCC has been reported so far.
In cases of AbnCC with unknown etiology, the prognosis mainly depends on the existence or lack of other major abnormalities. Gyration and sulci abnormalities can be detected in the second trimester. However, in view of the progressive brain development during the pregnancy, many migration disorders and late primary sulcation cannot be detected before the end of the second trimester and fetal MRI is often not offered until at least 30 WG.19
Current standard prenatal procedures in France are limited to the investigation of chromosomal anomalies by fetal karyotyping and CMA, although they represent a minority of AbnCC etiologies. However, the identification of the underlying etiology would enable the couples to make an informed decision to continue with or terminate the pregnancy.
By using pES during pregnancy, we identified pathogenic variants in 12 cases (18%) that were not detectable by current chromosomal diagnosis techniques. Moreover, three pathogenic CNVs were detected by pES, confirmed by CMA. Overall, the total diagnostic yield was 23%.
We were able to inform the parents of the result during the pregnancy in 97% of cases with a median delay of 21.5 days. Thus, pES in AbnCC is feasible during the pregnancy.
We found that the diagnostic yield was higher when AbnCC was nonisolated (42%) rather than when it was isolated (15%). This is in line with the higher rate of chromosomal anomalies found in niAbnCC11 and tends to confirm the poor prognosis of niAbnCC. In all iAbnCC fetuses with identified diagnosis during the pregnancy (six cases), pES result provided important prognostic information and shifted the prognosis of the fetus from a high probability of normal development to highly penetrant ID. These results significantly influenced the parental decision since 72% of the parents decided to terminate the pregnancy. Thus, impact of pES in cases with established diagnosis is evident.
In most iAbnCC cases without diagnosis (90%), parents decided to continue with the pregnancy. The negative pES results certainly influenced their decision to continue with the pregnancy although they were informed that a neurodevelopment disorder could not be excluded.
Our results led us to propose a new strategy for prenatal management of AbnCC: we suggest investigating the etiology of AbnCC by simultaneous pES and CMA as soon as the US diagnosis is confirmed (usually during the second trimester). If the etiology is identified, the fetal prognosis then depends on the cause. When CMA and pES are negative, the residual risk of disability may be less than 20% in case of iAbnCC. Long-term follow-up of born children identified prenatally with iAbnCC is of utmost importance to provide accurate prenatal counseling when both CMA and pES results show no detectable abnormalities.
References
Edwards TJ, Sherr EH, Barkovich AJ, Richards LJ. Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain. 2014;137:1579–1613.
Glass HC, Shaw GM, Ma C, Sherr EH. Agenesis of the corpus callosum in California 1983-2003: a population-based study. Am J Med Genet A. 2008;146A:2495–2500.
Schaefer GB, Bodensteiner JB. Radiological findings in developmental delay. Semin Pediatr Neurol. 1998;5:33–38.
Achiron R, Achiron A. Development of the human fetal corpus callosum: a high-resolution, cross-sectional sonographic study. Ultrasound Obstet Gynecol. 2001;18:343–347.
Santirocco M, Rodó C, Illescas T, et al. Accuracy of prenatal ultrasound in the diagnosis of corpus callosum anomalies. J Matern Fetal Neonatal Med. 2019. https://doi.org/10.1080/14767058.2019.1609931 [Epub ahead of print].
Moutard M-L, Kieffer V, Feingold J, et al. Isolated corpus callosum agenesis: a ten-year follow-up after prenatal diagnosis (how are the children without corpus callosum at 10 years of age?). Prenat Diagn. 2012;32:277–283.
Sotiriadis A, Makrydimas G. Neurodevelopment after prenatal diagnosis of isolated agenesis of the corpus callosum: an integrative review. Am J Obstet Gynecol. 2012;206:337.e1–337.e5.
des Portes V, Rolland A, Velazquez-Dominguez J, et al. Outcome of isolated agenesis of the corpus callosum: a population-based prospective study. Eur J Paediatr Neurol. 2018;22:82–92.
Bedeschi MF, Bonaglia MC, Grasso R, et al. Agenesis of the corpus callosum: clinical and genetic study in 63 young patients. Pediatr Neurol. 2006;34:186–193.
Schell-Apacik CC, Wagner K, Bihler M, et al. Agenesis and dysgenesis of the corpus callosum: clinical, genetic and neuroimaging findings in a series of 41 patients. Am J Med Genet A. 2008;146A:2501–2511.
Heide S, Keren B, Billette de Villemeur T, et al. Copy number variations found in patients with a corpus callosum abnormality and intellectual disability. J Pediatr. 2017;185:160–.e1.
Lord J, McMullan DJ, Eberhardt RY, et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet. 2019;393:747–757.
Chandler N, Best S, Hayward J, et al. Rapid prenatal diagnosis using targeted exome sequencing: a cohort study to assess feasibility and potential impact on prenatal counseling and pregnancy management. Genet Med. 2018;20:1430–1437.
Pangalos C, Hagnefelt B, Lilakos K, Konialis C. First applications of a targeted exome sequencing approach in fetuses with ultrasound abnormalities reveals an important fraction of cases with associated gene defects. PeerJ. 2016;4:e1955.
de Koning MA, Haak MC, Adama van Scheltema PN, et al. From diagnostic yield to clinical impact: a pilot study on the implementation of prenatal exome sequencing in routine care. Genet Med. 2019;21:2303–2310.
Rasmussen SA, Olney RS, Holmes LB, et al. Guidelines for case classification for the National Birth Defects Prevention Study. Birth Defects Res Part A Clin Mol Teratol. 2003;67:193–201.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424.
Abou Tayoun A, Mason-Suares H. Considerations for whole exome sequencing unique to prenatal care. Hum Genet. 2019. https://doi.org/10.1007/s00439-019-02085-7 [Epub ahead of print].
Malinger G, Zakut H. The corpus callosum: normal fetal development as shown by transvaginal sonography. AJR Am J Roentgenol. 1993;161:1041–1043.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Heide, S., Spentchian, M., Valence, S. et al. Prenatal exome sequencing in 65 fetuses with abnormality of the corpus callosum: contribution to further diagnostic delineation. Genet Med 22, 1887–1891 (2020). https://doi.org/10.1038/s41436-020-0872-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0872-8
Keywords
This article is cited by
-
Prenatal Genome-Wide Sequencing analysis (Exome or Genome) in detecting pathogenic Single Nucleotide Variants in fetal Central Nervous System Anomalies: systematic review and meta-analysis
European Journal of Human Genetics (2024)
-
NeuroSCORE is a genome-wide omics-based model that identifies candidate disease genes of the central nervous system
Scientific Reports (2022)