Abstract
Purpose
Spinal muscular atrophy (SMA) was added to the Recommended Uniform Screening Panel (RUSP) in July 2018, following FDA approval of the first effective SMA treatment, and demonstration of feasibility of high-throughput newborn screening using a primary molecular assay. SMA newborn screening was implemented in New York State (NYS) on 1 October 2018.
Methods
Screening was conducted using DNA extracted from dried blood spots with a multiplex real-time quantitative polymerase chain reaction (qPCR) assay targeting the recurrent SMN1 exon 7 gene deletion.
Results
During the first year, 225,093 infants were tested. Eight screened positive, were referred for follow-up, and confirmed to be homozygous for the deletion. Infants with two or three copies of the SMN2 gene, predicting more severe, earlier-onset SMA, were treated with antisense oligonucleotide and/or gene therapy. One infant with ≥4 copies SMN2 also received gene therapy.
Conclusion
Newborn screening permits presymptomatic SMA diagnosis, when treatment initiation is most beneficial. At 1 in 28,137 (95% confidence interval [CI]: 1 in 14,259 to 55,525), the NYS SMA incidence is 2.6- to 4.7-fold lower than expected. The low SMA incidence is likely attributable to imprecise and biased estimates, coupled with increased awareness, access to and uptake of carrier screening, genetic counseling, cascade testing, prenatal diagnosis, and advanced reproductive technologies.
Similar content being viewed by others
INTRODUCTION
Spinal muscular atrophy (SMA; OMIM *600354) is a neuromuscular disorder characterized by muscle weakness and atrophy, resulting from progressive degeneration and loss of the anterior horn cells in the spinal cord and brain stem nuclei.1 The pediatric SMA phenotype is variable with respect to age at onset, symptoms, and severity, with three major subtypes defined by the highest motor function achieved.1 Prior to the advent of SMA-specific treatment, infants were classified with SMA type 1 because of their inability to sit unassisted. The phenotype was characterized by a lack of motor development, extreme weakness, and impairments in sucking, swallowing, and breathing that typically began within six months of age. Without ventilatory support, affected infants typically died by two years of age from pneumonia or respiratory insufficiency. In SMA type 2, symptoms usually began between 6 and 18 months of age. By definition, SMA type 2 patients sat and some even stood at some point, but they never walked independently. Survival was normal or near normal and complications were primarily musculoskeletal in nature (scoliosis and contractures). SMA type 3 patients presented after age 18 months and were able to walk independently, though some lost the ability to walk, especially if presenting with symptoms under age three years (type 3a).2 SMA type 4, which presented in adulthood, was the least common and least severe. A rare congenital form, type 0, presented with profound weakness and early respiratory failure.
SMA is currently the most common genetic cause of death in infants and children. Incidence estimates range from 1 in 6000 to 1 in 11,000 births.3,4,5 The condition is autosomal recessive, resulting from mutation of the survival of motor neuron 1 gene, SMN1, most commonly, a deletion of exon 7. The number of genomic copies of survival of motor neuron 2, SMN2, is the major determinant of SMA severity, although other modifiers exist.1 SMN1 and SMN2 genes code for an identical protein; however, one of few nucleotide differences between the two genes results in only 5–10% properly spliced SMN2 transcripts, which can partially compensate for the loss of SMN1. In general, affected individuals with more genomic copies of SMN2 tend to have milder disease, while those with fewer copies have more severe, earlier-onset disease. In affected individuals, the number of SMN2 copies ranges from one to five,1 or rarely, more.
Several major barriers to newborn screening for SMA were overcome between 2016 and 2019. Historically, SMA was diagnosed after symptom onset, subsequent to irreversible motor neuron degeneration and loss,1 or presymptomatically via genetic testing for those with a family history. One systematic review estimated a mean diagnostic delay of 3.6, 14.3, and 43.6 months for SMA types 1, 2, and 3, respectively.6 Identification at birth prior to symptom onset via newborn screening was not previously feasible because there is no protein biomarker for SMA in blood. With implementation of the first primary molecular newborn screening test for severe combined immunodeficiency (SCID), screening for the common SMN1 exon 7 deletion via quantitative polymerase chain reaction (qPCR) became more feasible.7
The New York State (NYS) program initiated a pilot study for SMA newborn screening in January 2016.8 The goals of the study were to demonstrate feasibility of screening, and to assess whether parents wanted their infants screened for a disorder lacking an FDA-approved treatment. Testing was offered via informed consent at four New York City (NYC) hospitals. Among 16,712 infants tested over 32 months, one screened positive and was found to have two copies of SMN2, predicting severe, early onset SMA type 1.
Treatment of SMA was previously limited to supportive care, such as respiratory and nutritional support and physical therapy, until the FDA approved nusinersen (Spinraza) in December 2016, following successful clinical trials in affected and presymptomatic individuals.9 The family of the single affected infant identified via the NYS SMA pilot8 enrolled her in a presymptomatic nusinersen clinical trial,10 and as of November 2019, at nearly three years of age, she meets all motor milestones appropriate for age, including sitting, standing, and walking, which would never be achieved by an infant with severe SMA. Gene therapy (onasemnogene abeparvovec-xioi; Zolgensma)11 was subsequently approved by the FDA in May 2019. Outcomes are better when treatments are initiated prior to symptoms onset, justifying universal newborn screening.
Based on demonstration of feasibility of high-throughput molecular SMA testing in a public health laboratory setting with a high opt-in rate for screening (>93%);8 prospective identification and treatment of an asymptomatic infant with SMA who continues to meet motor milestones;8 recommendation that SMA be added to the Recommended Uniform Screening Panel (RUSP) by the Advisory Committee on Heritable Disorders in Newborns and Children (ACHDNC) in March 2018, with endorsement by the US Secretary of Health in July 2018; and availability of an FDA-approved treatment,9 SMA was added to the NYS newborn screening panel. Results from the first year of universal, population-based screening are presented.
MATERIALS AND METHODS
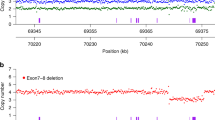
Screening for the common SMA exon 7 deletion was accomplished using a modified version of the real-time TaqMan qPCR assay used in the NYS pilot SMA screen.8 Briefly, for each infant, DNA was extracted from one 3-mm dried blood spot punch in a 96-well format. SMN1 primer/probe mix was multiplexed with the NYS SCID assay,12 modified to include three fluorescently labeled targets: T-cell receptor excision circles (TRECs [molecular SCID biomarker]; FAM-labeled), the recurrent SMN1 exon 7 deletion (VIC-labeled), and a fragment of RPPH1 as an internal control (ABY-labeled). Primer/probe mixes were purchased from Thermo Fisher Scientific (Waltham, MA), and the assay was run in a 384-well format using Quanta PerfeCTa Multiplex qPCR ToughMix (Beverly, MA) on a QuantStudio 12 K Flex Real-Time PCR System. Infants with SMN1 Ct ≥30 and RPPH1 Ct <30 were considered screen positive. Heterozygous carriers of the exon 7 deletion were not identified. SMN2 copy number was determined in infants who screened positive using a TaqMan assay targeting SMN2 exon 7 and RPPH1, and provided at the time of referral. The New York State Dept. of Health ruled that the work described in this manuscript was exempt, that this work was carried out in performance of public health activities.
RESULTS
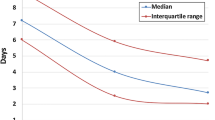
Universal newborn screening for SMA was implemented in NYS on 1 October 2018. During the first year, 225,093 infants were screened. Eight were homozygous for the SMN1 exon 7 deletion (Table 1). Three had two copies of SMN2, three had three copies, and two had at least four copies. Infants were referred to NYS Neuromuscular Specialty Care Centers (SCCs) for follow-up at a median of 7.5 days of life (DOL; range 5 to 12), facilitating rapid diagnostic confirmation and discussion of treatment options. All were evaluated within 13 DOL and each was confirmed to be homozygous for the SMN1 deletion, yielding a positive predictive value (PPV) of 100%. None reported a family history of SMA. One family reported having prenatal carrier screening (type and results not available) but stated they were unaware of the possibility of having a baby with SMA. Two were initially treated with nusinersen at four and six weeks of age, and then gene therapy at five and six months of age, with discontinuation of nusinersen at the parents’ request. Four infants with two or three copies of SMN2 underwent gene therapy only, all by age 38 days. All six infants with two or three copies of SMN2 were asymptomatic at the time of treatment initiation and at last follow-up. Patient 2 (Table 1), referred at seven DOL, had subclinical signs of absent deep tendon reflexes and tongue fasciculations at eight DOL, but was still clinically asymptomatic at last follow-up at eight months of age, meeting motor milestones for age with no complaints or concerns from parents or physicians. Among the two infants predicted to have later onset SMA based on SMN2 copy number of at least four, one has not received treatment and is being carefully monitored long-term for signs predictive of disease onset via routine SCC visits that include monitoring of compound muscle action potentials (CMAP). The second infant with at least four copies of SMN2 received gene therapy at six months of age in another state. The baby was asymptomatic at that time.
DISCUSSION
Based on the 225,093 infants tested during the first year, 20–38 screen positive infants with SMA were expected, corresponding to the often-cited incidence of 1 in 6000 to 1 in 11,000 live births per year (Table 2). However, only eight affected infants were identified, corresponding to an incidence of 1 in 28,137 (95% confidence interval [CI]: 1 in 14,259 to 55,525), 2.6- to 4.7-fold lower than expected. We are not aware of any cases born during this period who were missed by screening. Extending the data set through the end of February 2020, the NYS incidence is closer to expected, at approximately 1 in 21,000 (15 cases among approximately 314,000 screened), but still low. Point estimates will become more accurate as data accumulate from longer-term universal SMA screening.
SMA newborn screening is rapidly expanding. As of March 2020, NYS and 18 other states in the United States universally screen SMA in all births, and others have pending legislation or offer screening via pilot programs.13 None have published data on SMA incidence to date. Only a few are known to report SMN2 copy number at the time of referral, which we believe is important because it allows the specialist to rapidly assess (and convey to families) the need for urgent, presymptomatic treatment initiation without delay, for example, in infants with two SMN2 copies who may be undergoing rapid denervation.14
Approximately 95–98% of SMA results from homozygous SMN1 exon 7 deletion, the pathogenic genetic variant targeted by all screening programs. The remaining 2–5% of SMA patients will be missed by newborn screening because they are (1) compound heterozygous for another pathogenic SMN1 variant and the common deletion, (2) homozygous or compound heterozygous for two different SMN1 variants, or (3) rarely, due to variants in another gene (non-5q SMA).1 The target condition on the RUSP is SMA “due to homozygous deletion of exon 7 in SMN1.” The imperfect clinical sensitivity of the screen is a known limitation; however, the low SMA incidence we observed cannot alone be attributed to rare SMN1 variants. The low incidence is also not due to poor sensitivity of the screening assay, because rigorous validation studies demonstrated 100% sensitivity for the homozygous deletion. For routine testing, each 96-well assay plate included at least one homozygous SMN1 exon 7 deletion control and no template controls to monitor well contamination.
We hypothesize that there are two primary contributors to the low SMA incidence observed. First, there is increased awareness, access, and uptake of carrier screening, genetic counseling, cascade testing, prenatal diagnosis, and advanced reproductive technologies such as preimplantation genetic diagnosis. Although such data are not tracked systematically for this purpose, we are aware of two NYS cases confirmed prenatally that did not reach term following termination of pregnancy during the first year of universal screening. The American College of Medical Genetics and Genomics (ACMG) first recommended offering SMA carrier testing to all couples regardless of race/ethnicity in 2008,15 but the American College of Obstetricians and Gynecologists (ACOG) did not recommend carrier screening in women currently pregnant or considering pregnancy until 2017.16 Among infants identified as carriers via the NYS pilot study, at least 43% of families already knew at least one parent was a carrier, supporting high uptake of SMA carrier screening in pilot study hospitals in NYC. The pilot study also revealed a lower than expected SMA carrier frequency at 1 in 65.8 The frequency differed by race/ethnicity and by hospital, which differ in ancestry of the communities served. There was no difference in SMA incidence comparing NYC, Long Island and New Jersey births to all other NYS births (i.e., NYC vs. upstate) during the first year of universal screening, although the sample size is small. Extrapolating using published NYS maternal race/ethnicity proportions from 2017 birth certificates,17 SMA incidence is low in NYS Non-Hispanic/Latinx, Caucasian/White (approximately 1 in 21,900); Non-Hispanic/Latinx, Asian (1 in 24,900); Non-Hispanic/Latinx, African American/Black (1 in 33,400); and Hispanic/Latinx (1 in 52,000) groups. The exon 7 deletion carrier frequency is also known to be higher in the Caucasian and Ashkenazi Jewish populations, and lower in Asian, African American, and, especially, Hispanic populations.18 Furthermore, the carrier detection rate is also lower in race/ethnic groups with a higher frequency of the SMN1 2 + 0 genotype,1 that is, two copies of SMN1 in cis on one allele (duplication from one parent), and no copies of SMN1 (deletion from other parent) in trans on the other allele, which results in a false negative carrier screen because two copies of SMN1 are detected in the carrier without knowledge of phase. Approximately 2% of SMA cases occur as a result of de novo SMN1 variants and would also not be detected via carrier screening.1
Biased or imprecise historical3,4,5,19,20,21 or current incidence estimates likely play a role in lower than expected NYS SMA incidence. Due to a lack of population-based surveillance or comprehensive disease registries, most published estimates were derived based on small European subpopulations that are probably not representative of the general United States population. Most were conducted prior to identification of SMN1 gene variants,5,19 and estimates may have been inflated with cases diagnosed based on clinical presentation without genetic confirmation. Changes and differences in case and subtype definition and classification, and sample and ascertainment bias may also contribute. Exclusion of two screen positive infants with at least four copies of SMN2 predicted to have later onset/milder disease would yield an even lower NYS incidence (1 in 37,516), although it may be more comparable with earlier estimates using case definitions that could have excluded such cases. As an example of differences in predicted versus actual disease incidence, NYS was the first to screen Krabbe disease.22 The incidence was reportedly 1 in 100,000, with the severe, infantile-onset form (≤12 months) representing 90% of cases. More than 3.2 million NYS infants have been screened between August 2006 and February 2020, but only five cases of infantile-onset Krabbe disease have been identified (excludes one recently identified infant pending diagnostic confirmation, but very likely to present as infantile disease based on biochemical and genetic data). Many more have screened positive with confirmed low enzyme activity via diagnostic testing, likely representing later onset disease or subclinical enzyme deficiencies in otherwise healthy individuals. On the other hand, implementation of population-based screening may reveal a higher frequency of subclinical individuals, and later onset or mild forms of conditions that may have previously been misdiagnosed or gone unrecognized, compared with clinic-based estimates of prevalence based on symptomatic individuals with classic forms of disease. Population-based ascertainment of SMA cases identified at birth will permit more accurate estimates of the true current incidence in United States and other23,24,25,26 population groups, which may still be confounded by geographical differences in prenatal care and other socioeconomic factors.
All infants identified during the first year of SMA screening in NYS with two or three copies of SMN2 have received gene therapy, including two initially treated with nusinersen. Some families favored gene therapy due to the burden of repeated lumbar puncture and sedation required for oligonucleotide therapy. Other than mild liver enzyme abnormalities that subsequently resolved in one infant, adverse events have not been reported for NYS cases treated with gene therapy. Considering infants with two or three copies of SMN2, the median age at initial treatment with nusinersen or onasemnogene was 36 days (range 19–40) for six infants in NYS, compared with 24 days (15–39) for ten infants identified in the German pilot program,25 and 26.5 days (16–37) for eight infants identified in the Australian SMA pilot (Table 2).24 In NYS, treatment was initiated within one month of age for only two of the six infants with two or three copies of SMN2. For one of the six, gene therapy was delayed for five days due to a stop in production by the company for an undisclosed reason. Others were delayed due to insurance authorization requirements. We suspect that the time from diagnosis to treatment will decrease as insurers become aware of the need for prompt treatment and authorizations become standard, thereby reducing the number of lengthy appeals. The ability for all SCCs to administer both FDA-approved treatments without referral to a second center will further reduce delays.
Although long-term follow-up data are not yet available, antisense oligonucleotide and gene therapies have substantially altered the SMA phenotype.9,11 With presymptomatic therapeutic intervention, achievement of motor milestones will no longer be the optimal method of phenotype classification. Standardized clinical assessments and outcome measures for infants and children must be developed or refined based on the new natural history of treated SMA. A carefully designed national registry with key data elements will facilitate systematic prospective data collection. Importantly, a registry with a high participation rate can inform new standards of care that define when to initiate treatment(s), especially for individuals with at least four SMN2 copies. The first guidelines for infants identified by newborn screening with SMA and four or more copies of SMN2 recommended treatment initiation based on clinical judgment and the patient and/or family’s wishes.14 After review of new clinical data and the burden and benefit of early treatment, the group revised the guidelines, recommending immediate treatment for those with four SMN2 copies identified via newborn screening, and watchful waiting for infants with five copies of SMN2.27 Most clinical labs do not currently distinguish four from five or more copies of SMN2, and should now work toward utilization of technology that permits more precise determination of higher copy SMN2 number.
In summary, biased incidence estimates and informed reproductive decisions in the age of genomic medicine have likely contributed to a lower than expected incidence of SMA. Data from other screening programs will shed light on the true incidence in the United States. Long-term follow-up of infants treated with nusinersen and onasemnogene is still limited; however, at minimum, the quality of life for infants identified via newborn screening will likely be improved because they were diagnosed at birth. SMA newborn screening ensures equity in diagnosis of a common genetic condition in infants, such that affected children of families who cannot or choose not to undergo carrier screening, or who are not offered testing, are universally afforded the same benefit of new, life-saving treatments that are most effective when identified prior to symptom onset.
References
Prior TW, Leach ME. Finanger E. Spinal muscular atrophy. In: Adam MP, Ardinger HH, Pagon RA et al., editors. GeneReviews. Seattle, WA: University of Washington; 1993–2019.
Zerres K, Rudnik-Schöneborn S. Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch Neurol. 1995;52:518–523.
Sugarman EA, Nagan N, Zhu H, et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72 400 specimens. Eur J Hum Genet. 2012;20:27–32.
Ogino S, Leonard DGB, Rennert H, Ewens WJ, Wilson RB. Genetic risk assessment in carrier testing for spinal muscular atrophy. Am J Med Genet. 2002;110:301–307.
Emery AEH. Population frequencies of inherited neuromuscular diseases—a world survey. Neuromuscul Disord. 1991;1:19–29.
Lin CW, Kalb SJ, Yeh WS. Delay in diagnosis of spinal muscular atrophy: a systematic literature review. Pediatr Neurol. 2015;53:293–300.
Pyatt RE, Prior TW. A feasibility study for the newborn screening of spinal muscular atrophy. Genet Med. 2006;8:428–437.
Kraszewski JN, Kay DM, Stevens CF, et al. Pilot study of population-based newborn screening for spinal muscular atrophy in New York state. Genet Med. 2018;20:608–613.
Chiriboga CA. Nusinersen for the treatment of spinal muscular atrophy. Expert Rev Neurother. 2017;17:955–962.
De Vivo DC, Bertini E, Swoboda KJ, et al. Nusinersen initiated in infants during the presymptomatic stage of spinal muscular atrophy: Interim efficacy and safety results from the Phase 2 NURTURE study. Neuromuscul Disord. 2019;29:842–856.
Mendell JR, Al-Zaidy S, Shell R, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med. 2017;377:1713–1722.
Vogel BH, Bonagura V, Weinberg GA, et al. Newborn screening for SCID in New York State: experience from the first two years. J Clin Immunol. 2014;34:289–303.
Cure SMA. Newborn screening for SMA. https://www.curesma.org/newborn-screening-for-sma/. Accessed 5 March 2020.
Glascock J, Sampson J, Haidet-Phillips A, et al. Treatment algorithm for infants diagnosed with spinal muscular atrophy through newborn screening. J Neuromuscul Dis. 2018;5:145–158.
Prior T. Carrier screening for spinal muscular atrophy. Genet Med. 2008;10:840–842.
Committee on Genetics. Committee opinion no. 691: carrier screening for genetic conditions. Obstet Gynecol. 2017;129:e41–e55.
Martin JA, Hamilton BE, Osterman MJK, Driscoll AK, Drake P. Births: final data for 2017. Natl Vital Stat Rep. 2018;67:1–50.
Hendrickson BC, Donohoe C, Akmaev VR, et al. Differences in SMN1 allele frequencies among ethnic groups within North America. J Med Genet. 2009;46:641–644.
Verhaart IEC, Robertson A, Wilson IJ, et al. Prevalence, incidence and carrier frequency of 5q-linked spinal muscular atrophy—a literature review. Orphanet J Rare Dis. 2017;12:1–15.
Wilson RB, Ogino S. Carrier frequency of spinal muscular atrophy. Lancet. 2008;372:1542.
Ogino S, Wilson RB, Gold B. New insights on the evolution of the SMN1 and SMN2 region: simulation and meta-analysis for allele and haplotype frequency calculations. Eur J Hum Genet. 2004;12:1015–1023.
Orsini JJ, Kay DM, Saavedra-Matiz CA, et al. Newborn screening for Krabbe disease in New York State: the first eight years’ experience. Genet Med. 2016;18:239–248.
Chien YH, Chiang SC, Weng WC, et al. Presymptomatic diagnosis of spinal muscular atrophy through newborn screening. J Pediatr. 2017;190:124–129.e1.
Kariyawasam DST, Russell JS, Wiley V, Alexander IE, Farrar MA. The implementation of newborn screening for spinal muscular atrophy: the Australian experience. Genet Med. 2020;22:557–565.
Vill K, Kölbel H, Schwartz O, et al. One year of newborn screening for SMA—results of a German pilot project. J Neuromuscul Dis. 2019;6:503–515.
Czibere L, Burggraf S, Fleige T, et al. High-throughput genetic newborn screening for spinal muscular atrophy by rapid nucleic acid extraction from dried blood spots and 384-well qPCR. Eur J Hum Genet. 2020;28:23–30.
Glascock J, Sampson J, Connolly A, et al. Revised recommendations for the treatment of infants diagnosed with spinal muscular atrophy via newborn screening who have 4 copies of SMN2. J Neuromuscul Dis. 2020;7:97–100
Acknowledgements
Funding for universal screening was provided by the Centers for Disease Control and Prevention (CDC) (6 NU88EH001319–01–01) and the New York State Department of Health. Funding for the SMA pilot study and validation of the assay for universal screening was provided by Biogen, Idec to Columbia University Medical Center and contracted to the Wadsworth Center. The funding bodies had no role in the design of the study, collection and analysis of data or decision to publish. The authors thank New York State Newborn Screening Program staff, especially Jason Isabelle, Allison Madole, Robert Sicko, and Erin Hughes for technical support and data collection; Neuromuscular Disease Specialty Care Center staff, especially Katherine Hogan, Natasha Kleiman, Jacqueline Gomez, Dr. Angela Pena, Chelsea Kois, Dr. Chin-To Fong, Theresa Harte, Debra Guntrum, and Dawn Dawson; and Erin Collins and Joan Mountain for administrative support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
D.M.K.'s work has been funded by the CDC and Biogen, Idec. C.F.S.'s work has been funded by the CDC and Biogen, Idec. W.K.C.'s work has been funded by the NIH, Simons Foundation, JPB Foundation, and Biogen, Idec. She is on the scientific advisory board for the Regeneron Genetics Center. C.A.C. has received funding from Avexis, Biogen and Roche. She is a consultant (advisory board) for Cytokinetics, Avexis, Genentech, and Roche, and she is an educational speaker for Biogen. E.C. has received personal compensation for serving on advisory boards and/or as a consultant for Avexis, Biogen, Medscape, Pfizer, PTC Therapeutics, Sarepta Therapeutics, Ra Pharma, Wave, and Strongbridge Biopharma. She has received personal compensation for serving on a speaker’s bureau for Biogen and has received research and/or grant support from the CDC, CureSMA, Muscular Dystrophy Association, National Institutes of Health, the Patient-Centered Outcomes Research Institute, Parent Project Muscular Dystrophy, PTC Therapeutics, Santhera, Sarepta Therapeutics, Orphazyme, and the US Food and Drug Administration. She has received royalties from Oxford University Press and compensation from Medlink for editorial duties. B.H.L. receives research support from Sanofi Genzyme. A.S. has received compensation for advisory board consultation for Avexis, Biogen, PTC Therapeutics, and BioMarin. M.C.'s work has been funded by the CDC and Biogen, Idec. The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kay, D.M., Stevens, C.F., Parker, A. et al. Implementation of population-based newborn screening reveals low incidence of spinal muscular atrophy. Genet Med 22, 1296–1302 (2020). https://doi.org/10.1038/s41436-020-0824-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0824-3
Key words
This article is cited by
-
Cost-Effectiveness of Newborn Screening for Spinal Muscular Atrophy in England
Neurology and Therapy (2023)
-
Advances and limitations for the treatment of spinal muscular atrophy
BMC Pediatrics (2022)
-
Spinal muscular atrophy
Nature Reviews Disease Primers (2022)
-
Systematic Literature Review to Assess the Cost and Resource Use Associated with Spinal Muscular Atrophy Management
PharmacoEconomics (2022)
-
A Window of Opportunity for Newborn Screening
Molecular Diagnosis & Therapy (2022)