Abstract
Purpose
Plasma cell-free DNA (cfDNA) variant analysis is commonly used in many cancer subtypes. Cell-free methylated DNA immunoprecipitation sequencing (cfMeDIP-seq) has shown high sensitivity for cancer detection. To date, studies have not compared the sensitivity of both methods in a single cancer subtype.
Methods
cfDNA from 40 metastatic RCC (mRCC) patients was subjected to targeted panel variant analysis. For 34 of 40, cfMeDIP-seq was also performed. A separate cohort of 38 mRCC patients were used in cfMeDIP-seq analysis to train an RCC classifier.
Results
cfDNA variant analysis detected 21 candidate variants in 11 of 40 mRCC patients (28%), after exclusion of 2 germline variants and 6 variants reflecting clonal hematopoiesis. Among 23 patients with parallel tumor sequencing, cfDNA analysis alone identified variants in 9 patients (39%), while cfDNA analysis focused on tumor sequencing variant findings improved the sensitivity to 52%. In 34 mRCC patients undergoing cfMeDIP-seq, cfDNA variant analysis identified variants in 7 (21%), while cfMeDIP-seq detected all mRCC cases (100% sensitivity) with 88% specificity in 34 control subjects. In 5 patients with cfDNA variants and serial samples, variant frequency correlated with response to therapy.
Conclusion
cfMeDIP-seq is significantly more sensitive for mRCC detection than cfDNA variant analysis. However, cfDNA variant analysis may be useful for monitoring response to therapy.
Similar content being viewed by others
INTRODUCTION
The presence of fragmented DNA in the bloodstream has been observed for decades. In recent years, it has been recognized that cancer cells contribute to plasma cell-free DNA (cfDNA), enabling so-called liquid biopsies to detect actionable tumor variants and determine mechanisms of resistance to targeted therapy.
Renal cell carcinoma (RCC) accounts for 15,000 deaths in the United States per year.1 Few reports have described cfDNA variant analysis in RCC. One study using hybrid capture detected genomic alterations in 79% of metastatic RCC (mRCC) patients (n = 220) with TP53 the most commonly mutated gene (35%), followed by VHL (23%).2 Other studies have shown that circulating tumor DNA (ctDNA) was detectable in more than 75% of patients with advanced tumors of various origins, but was much less common in advanced RCC (40%).3,4
One issue with cfDNA variant analysis is lack of reproducibility; one study demonstrated a 60% concordance rate between two commercial cfDNA sequencing platforms.5 In addition, clonal hematopoiesis (CH), a process in which growth-promoting somatic variants drive expansion of clonal hematopoietic cell populations,6 is recognized as an important confounding factor in detection of cfDNA variant analysis.7 Prior cfDNA analyses in RCC have not accounted for CH,2 likel y inflating the detection rate with variant calls due to CH and not due to RCC-derived somatic variants.
An alternative approach to the detection of tumor derived DNA in the circulation is cell-free methylated DNA immunoprecipitation sequencing (cfMeDIP-seq). The principle of this method is that tumor cells acquire recurrent aberrant DNA methylation that is distinct from blood and other normal tissue cells, enabling sensitive and specific detection of tumor cfDNA. cfMeDIP-seq has shown high sensitivity and accuracy for detecting and classifying several tumor types, including RCC.8
The relative performance of cfDNA variant analysis versus cfMeDIP-seq profiling has not been evaluated systematically for any cancer subtype. Herein, we present a direct comparison between these two methods for 34 patients with mRCC. We also show improvement in cfDNA variant detection when guided by tumor sequencing, we show that CH is common in mRCC, and we assess the utility of cfDNA variant analysis in monitoring response to therapy in patients with serial samples.
MATERIALS AND METHODS
Study design and patient cohort
Forty patients with mRCC were seen at Brigham and Women’s Hospital, Dana-Farber Cancer Institute (DFCI), and/or Beth Israel Deaconess Medical Center, and provided signed informed consent for this study between May 2016 and November 2017. Twenty-three of these patients also had tumor panel sequencing analysis performed. An additional independent cohort of 38 patients with RCC and 34 cancer-free controls were used to develop a cell-free methylated DNA-based classifier for RCC detection; samples were collected between February 2005 and May 2015. The study was approved by the Institutional Review Board of DFCI (Gelb Center proposal 16-13).
Sample processing
Plasma cfDNA was isolated by standard methods (Supplementary Methods). Buffy coats were used for leukocyte DNA isolation. The same cfDNA aliquots were subjected to cfDNA targeted sequencing and cfMeDIP-seq for all subjects in whom there was sufficient cfDNA.
cfDNA variant analysis
A custom bait set was designed covering the exonic regions of 27 genes mutated at a significant rate in any of the three major types of RCC.9,10,11 cfDNA was subjected to targeted exon capture and sequencing at the DFCI Center for Cancer Genome Discovery. The mean, median, and range of mean depth of read coverage for the 40 RCC samples were 989×, 1012×, and 456–1520×, respectively.
A custom analysis pipeline was employed to detect low frequency variants. Candidate single-nucleotide variants (SNVs) were identified using the following criteria: observed in >3 reads, including at least 1 read in each orientation; allele frequency (AF) of ≥0.5%; probable functional effect; and AF at least three times that of the second highest allele frequency for the same variant within a sequencing batch of 8–12 samples. Candidate insertion and deletion (indel) variants were identified as having AF ≥ 0.2%; observed in at least two reads; and a read count three times that of the second highest AF within a sequencing batch. All candidate variants were reviewed using Integrative Genomics Viewer12 to exclude artifacts. SNVs and small indels observed at any frequency in gnomAD13 were excluded. Intronic variants within 15 bp of exon boundaries were retained, while others were excluded. In addition, variants in genes that were not significantly mutated in the subtype of mRCC for each individual patient were not considered further.
Validation of cfDNA and tumor variant calls
Candidate SNVs and indels were validated by Sanger sequencing for variants with AF ≥ 20%, and amplicon massive parallel sequencing (aMPS) for variants with <20% AF, as described previously. A read depth of 13,366–3,048,392, median = 148,077×, average = 122,035× was achieved in aMPS. Variant AFs on control DNA samples were 0–0.04% for indels (median 0%, mean 0.005%), and 0–0.20% for SNVs (median 0.02%, mean 0.03%) by this method.
Detection of differentially methylated regions
cfMeDIP-seq was performed using previously published methods (Supplementary Methods).8 An independent cohort of 38 RCC samples (stages I–IV) and 34 cancer-free controls were used as a training set to generate a model to classify samples as RCC or controls. Differentially methylated regions (DMRs) of size 300 bp were detected by first separating the genome into 300-bp bins. Bins for which the average read count was ≥0.2 across all samples were tested for differential read counts between RCC and control samples using limma-voom14 on TMM-normalized counts.15 We identified 29,786 DMRs between cases and controls at a false discovery rate (FDR) < 0.05. A set of 300 DMRs was obtained by selecting the top 150 DMRs with gain in RCC (all FDR < 9.45 × 10−6) and the top 150 DMRs with loss in RCC (all FDR < 4.11 × 10−10) (Supplementary Figure S1).
Prediction of sample status using the DMR signature
Three hundred DMRs were detected as described above using the independent cohort of 38 RCC samples and a random selection of 26 of the 34 cancer-free controls. This was repeated 100 times to generate 100 bootstrap samples of 300 DMRs, each using a different set of 26 control samples and holding out 8 to be classified. The 100 test sets, consisting of 34 of the 40 mRCC cfDNA samples subject to variant analysis and 8 random withheld control samples, were TMM-normalized using the training set as a reference. Log-transformed TMM-normalized values were then used to calculate methylation scores (with 95% confidence interval [CI]) for each sample across the 100 training test sets (Supplementary Methods).
Tumor targeted sequencing
Tumor targeted sequencing was performed using an institutional analytic platform, OncoPanel, as described previously.16,17
Statistical analysis
Statistical analysis was performed using GraphPad Prism (7.0) and JMP Pro 13. The Kaplan–Meier method with log-rank test and Cox regression analyses were performed to assess overall survival. All tests were two-sided and considered statistically significant at p < 0.05.
Radiological assessment
Tumor burden scores were assessed by a radiologist (A.B.S.) blinded to clinical and molecular data. Quantitative assessment was performed by measuring the maximum diameter of each metastatic lesion on axial images from computed tomography (CT) scans of the chest, abdomen, and pelvis as well as extremity and brain CT or magnetic resonance image (MRI) whenever metastases were identified in those organ systems. The tumor burden score was calculated as follows: the score for each organ (lymph nodes, liver, lung, peritoneum, soft tissue, bone, brain) was the sum of the maximum diameter of each metastatic lesion.
RESULTS
Patient characteristics
Forty RCC patients—31 clear cell and 9 nonclear cell—were studied (Tables 1, S1.1). The median age at diagnosis was 62 years (range 24–77); the male-to-female ratio was 2.6:1. Median follow-up for the entire cohort was 48 months (range 4–143 months). At enrollment, 20 patients (50%) had lung metastases, 13 (33%) had liver metastases, and 12 (30%) had bone metastases. Twelve of 40 (30%) had serial samples (Table S1.1).
cfDNA variant analysis, variant identification, and validation
Using targeted variant analysis of cfDNA, 35 candidate variants were found in 19 of 40 (48%) patients (Table S1.2). Variant allele fraction (VAF) ranged from 0.26% to 48%. Twenty-nine of the 35 (83%) candidate variants were validated by aMPS in 15 patients (38%). Among the 29, the genes most commonly mutated were TP53, SETD2, PBRM1, ATM, and VHL (10, 3, 3, 2, and 2 variants respectively). Comparison between the VAF observed in the initial cfDNA sequencing and in validation is shown in Figure S2.
Clonal hematopoiesis and false positivity in cfDNA
Recognizing that CH could lead to false positive variant calls, all cfDNA variants identified were assessed in matched patient leukocyte DNA. Eight of the 29 validated variants were observed in leukocyte DNA as well as cfDNA (Table S1.3). Two of the 8 variants were likely germline heterozygous alleles (in ATM and BAP1, Table S1.3), and may have predisposed these individuals to RCC development. The other six variants occurred at AFs similar to those seen in cfDNA (Table S1.3). Four of six variants (in four patients) occurred in TP53, which is commonly affected in CH.6,18 Two variants were identified in genes not known to be commonly mutated in CH, MTOR and KDM6A, and may reflect either CH or generalized mosaicism. Overall, after excluding germline and CH variants, cfDNA variant analysis identified variants in known RCC genes in 11 of 40 (28%) patients.
Comparison between tumor variant analysis and cfDNA findings
Given the allele frequency detection limit of our cfDNA sequencing method, we hypothesized that cfDNA variant analysis focused on variants identified by tumor sequencing might enhance the detection rate—an individualized or bespoke approach. Twenty-three of 40 subjects studied by cfDNA analysis had prior tumor sequencing (OncoPanel/PROFILE, Table S1.4).
Forty-seven variants in 27 genes were identified in 20 (87%) of the 23 patients by tumor sequencing (Table S1.5). Sixteen (34%) of 47 tumor sequencing findings had been independently identified in the initial cfDNA sequencing in nine patients (Table S1.2, S1.4). Of the 16 common findings, 1 was a possible CH variant (found in KDM6A), 2 were germline, and the remaining 13 (81%) were seen in cfDNA, but not leukocyte DNA. Thirty-one (66%) of the tumor sequencing findings had minimal or no supporting evidence (<3 reads for SNVs, <2 reads for indels) in the initial targeted sequencing of cfDNA. However, using aMPS (see “Materials and Methods”), 9 of 31 (29%) variants identified by tumor sequencing were validated in cfDNA at allele frequencies as low as 0.11% (Table S1.4). Overall, 25 of 47 (53%) variants identified by tumor sequencing were validated in the cfDNA of 12 of 23 (52%) patients, including one CH variant and two germline variants. The most frequently mutated genes in the cfDNA of this subset of patients were SETD2, TP53, PBRM1, and VHL (4, 3, 3, 3 variants respectively). Five variants found and validated in the independent cfDNA sequencing were not seen in tumor sequencing, of which one was a TP53 CH variant (Table S1.6A).
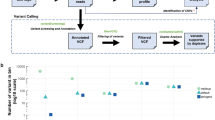
For the 17 patients for whom paired tumor sequencing was not available, targeted cfDNA variant analysis led to identification of 11 variants, of which 4 were validated in two patients and 4 others (in four patients) validated as probable or definite CH variants (Table S1.6B). Overall, after removal of CH and germline variants, 30 somatic variants were identified in cfDNA at AFs of 0.11–32% in 14 of 40 (35%) patients. The most frequently mutated genes were TP53, PBRM1, SETD2, and VHL (6, 5, 5, and 4 variants respectively, Fig. 1, Table S1.2, 1.4).
All cfDNA variants are shown, including those found in the primary cfDNA analysis and those found after comparison with tumor sequencing. Germline and clonal hematopoiesis (CH) findings are not shown. Renal cell carcinoma (RCC) subtype, availability of tumor variant data, and variants found are indicated.
cfMeDNA analysis and comparison with cfDNA variant analysis
Of 40 mRCC patients subject to cfDNA variant analysis, 34 had sufficient cfDNA available (>1.5 ng) to perform cfMeDIP-seq. A separate training cohort of 38 RCC patients and 34 controls were used first to develop a classifier of RCC (Table S1.7). Using a methylation score threshold of 0.375, 34 of 34 (100%) mRCC samples were accurately classified as RCC with a specificity of 88% (Fig. 2). The mean methylation score for the 34 RCC samples was 0.685 (range 0.465–0.857). In comparison, the mean methylation score of the 34 control samples was 0.160 (range 0.005–0.637, Fig. 2, Table S1.8). Although the methylation score for four outlier control samples was relatively high (0.460–0.637), there was a clear distinction between the remaining 30 control samples (all < 0.313) and the RCC samples (all > 0.465). This resulted in a mean area under the receiver operating curve of 0.983 for accurately distinguishing RCC cases from cancer-free controls. cfDNA variant analysis detected tumor variants in only 7 of these 34 patients (21%).
Association between variant allele frequency in cfDNA and clinical features
We examined whether cfDNA variant analysis might be utilized as a prognostic biomarker. Patients with any cfDNA variant finding had a significantly shorter overall survival compared with those without any cfDNA finding (log-rank test p < 0.001, Fig. 3). However, a higher tumor burden score (as a continuous variable) was also significantly associated with shorter overall survival. For every one point increase in the tumor burden score, a 3% rise in the hazard ratio was observed (hazard ratio [HR] = 1.03, 95% CI = 1.01–1.06, p = 0.006). Further, a positive cfDNA variant finding was strongly associated with a higher tumor burden score (Figure S3), as expected. Hence the association between cfDNA finding and survival may reflect the influence of tumor burden. However, the tumor burden score did not correlate with VAF in those subjects with a cfDNA variant finding (Spearman's Rho = -0.07, p = 0.8, n = 14, Supplementary Figure S4). We did not have sufficient subjects in this study to permit formal multivariable analysis to determine which factor was most important. Interestingly, age at metastatic disease, and International Metastatic RCC Database Consortium (IMDC) risk score were not associated with overall survival (p = 0.95, and p = 0.26, respectively).
Serial cfDNA analyses
cfDNA variant analysis has been useful, in some studies, to monitor therapeutic treatment responses longitudinally.19,20,21,22,23 We assessed this possibility in five subjects for whom serial (≥2) cfDNA samples were available (Fig. 4, Table S1.9). Although the number of subjects was small, there was a correlation between the level of VAF in cfDNA and response to therapy, assessed by RECIST v1.1. In particular, the VAF for seven variants in two subjects was either markedly reduced or 0 in subsequent cfDNA samples at the time partial response was achieved (Fig. 4).
Variant allele frequency and tumor burden score are plotted on the same graph for each of five renal cell carcinoma (RCC) patients (Panels a–e), with time from the first cfDNA sample shown on the x-axis. VAF is shown on the left axis; tumor burden score (dotted line) is shown on the right axis. Boxes indicate treatment intervals. Horizontal arrows start at the time when PR was assessed and extend as long as PR was maintained. Vertical arrows point to the time of progression of disease on therapy. ADC antibody-dependent cytotoxic agent, ICI immune checkpoint inhibitor TKI tyrosine kinase inhibitor PR Partial response.
DISCUSSION
cfDNA variant analysis is now a standard clinical tool used in multiple clinical cancer settings.24,25 To date, cfDNA studies in mRCC are limited.2 Moreover, until now, head-to-head comparisons between cfDNA variant analysis and cfMeDIP-seq have not been performed using the same input DNA. Our findings lead to the following conclusions.
First, cfDNA variant analysis has limited sensitivity for disease detection in patients with mRCC at a VAF of 0.5% (11 of 40, 28%) similar to previous reports.4 Second, cfDNA variant analysis is confounded by CH, which we found to be relatively common in mRCC (6 of 40 patients, 15%). Compared with one prior study,2 in which a 79% cfDNA variant detection rate was reported, our detection rate was relatively low. However, that study did not consider CH, and many genes in which they found variants have no known role in RCC development.2
Third, cfDNA variant analysis informed by prior tumor variant analysis, the bespoke approach, improves sensitivity (12 of 23, 52%) compared with cfDNA variant analysis alone (9 of 23, 39%). Nonetheless, of the variants identified in the 23 tumor samples and not seen in cfDNA, the median VAF obtained by amplicon sequencing was 0.01%, with many tumor variants being seen in none of 18,000–100,000 reads (Table S1.4), suggesting that there was a complete absence of tumor contribution to the cfDNA. On the other hand, we identified four non-CH variants in cfDNA from three subjects, in PTEN, SETD2, PBRM1, and TP53, that were not seen in tumor variant analysis. These variants may have been subclonal in the original cancer, and not present in the tumor sample analyzed, a well-known phenomenon in RCC.26 Alternatively, they may have developed in the time between tumor biopsy and plasma cfDNA analysis.
Fourth, cfMeDIP-seq has markedly improved sensitivity (100%) for detecting patients with mRCC compared with cfDNA variant analysis (21%). The enhanced sensitivity of the cfMeDIP-seq method likely relates to the analysis of multiple genomic regions by computational means, compared with a single site in variant analysis. In addition, DNA methylation changes are more numerous in RCC than are variants, with more than ten times as many regions demonstrating aberrant DNA methylation as variants in the Cancer Genome Atlas (TCGA) analysis.9,10,11 However, we also note that 4 of 34 (12%) normal controls had methylation scores that were similar to the RCC samples, and are false positives by the cfMeDIP-seq analysis.
Fifth, a positive finding by cfDNA variant analysis is associated with a worse overall survival (Fig. 3), as seen previously in melanoma and lung cancer.27,28 However, this may be due to the strong association between tumor burden and presence of cfDNA variants. Sixth, cfDNA variant analysis may be a useful molecular tool in monitoring treatment response. Analysis of serial plasma samples revealed that there were dynamic changes in the cfDNA VAFs that correlated with response to treatment and reduction of tumor burden as assessed by imaging (Fig. 4), as seen previously in chronic lymphocytic leukemia,23 pancreatic cancer29 and melanoma,27 but not in all prior studies.30,31
Several limitations of the current study must be noted. First, our sample size was limited. Second, tumor biopsies for sequencing and cfDNA analyses were not performed concurrently, as often happens in routine clinical care. Third, only one tumor area was used for sequencing. Fourth, we limited our analysis to the 27 genes known to be involved in the different types of RCC. Fifth, it is likely that a higher sensitivity assay, capable of detecting variants at 0.1% VAF would have led to detection of cfDNA variants in a higher fraction of subjects. Sixth, although cfMeDIP-seq analysis showed 100% sensitivity and 88% specificity in identification of RCC samples in this cohort, the false positive classification of 4 of 34 normal controls as having a high methylation of RCC is of concern, and this approach requires validation in independent cohorts.
In conclusion, we show that cfMeDIP-seq is significantly more sensitive for RCC detection in patients with metastatic disease than cfDNA variant analysis, with a 100% detection rate in our cohort at 88% specificity. Further study is needed to assess the potential sensitivity of cfMeDIP-seq for early stage RCC. Nonetheless, cfDNA variant analysis may be useful clinically in RCC patients to enable disease monitoring in response to treatment. cfDNA analysis may also detect variants not seen in earlier tumor samples due to tumor heterogeneity or clonal evolution. Last, we highlight CH as an important confounder that contributes to false positive findings in cfDNA variant analysis and emphasize the importance of concurrent leukocyte variant analysis whenever cfDNA variant analysis is done.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
Pal SK, Sonpavde G, Agarwal N, et al. Evolution of circulating tumor DNA profile from first-line to subsequent therapy in metastatic renal cell carcinoma. Eur Urol. 2017;72:557–564.
Zill OA, Banks KC, Fairclough SR, et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res. 2018;24:3528–3538.
Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224.
Torga G, Pienta KJ. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol. 2018;4:868–870.
Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498.
Hu Y, Ulrich BC, Supplee J, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24:4437–4443.
Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579–583.
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49.
Cancer Genome Atlas Research N, Linehan WM, Spellman PT, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. 2016;374:135–145.
Davis CF, Ricketts CJ, Wang M, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330.
Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291.
Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25.
Nassar AH, Umeton R, Kim J. et al. Mutational analysis of 472 urothelial carcinoma across grades and anatomic sites. Clin Cancer Res. 2018;15:2458–2470.
Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1:e87062.
Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376:536–547.
Christensen E, Birkenkamp-Demtroder K, Sethi H, et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J Clin Oncol. 2019;37:1547–1557.
Gray ES, Rizos H, Reid AL, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6:42008–42018.
Herbreteau G, Vallee A, Knol AC, et al. Quantitative monitoring of circulating tumor DNA predicts response of cutaneous metastatic melanoma to anti-PD1 immunotherapy. Oncotarget. 2018;9:25265–25276.
Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20:1698–1705.
Yeh P, Hunter T, Sinha D, et al. Circulating tumour DNA reflects treatment response and clonal evolution in chronic lymphocytic leukaemia. Nat Commun. 2017;8:14756.
Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8:1324.
Manier S, Park J, Capelletti M, et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nat Commun. 2018;9:1691.
Turajlic S, Xu H, Litchfield K, et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx Renal. Cell. 2018;173:595–610 e511.
Santiago-Walker A, Gagnon R, Mazumdar J, et al. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res. 2016;22:567–574.
Wang S, An T, Wang J, et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2010;16:1324–1330.
Wei T, Zhang Q, Li X. et al. Monitoring tumor burden in response to FOLFIRINOX chemotherapy via profiling circulating cell-free DNA in pancreatic cancer. Mol Cancer Ther. 2018;18:196–2013.
Kis O, Kaedbey R, Chow S, et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat Commun. 2017;8:15086.
Dizman N, Bergerot PG, Bergerot CD, et al. Association between mutation variant allele fractions (mVAFs) and response to immunotherapy (IO) in metastatic renal cell carcinoma (mRCC). J Clin Oncol. 2018;36:e16575–e16575.
Acknowledgements
We thank all the patients who participated in this study and their providers.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
D.A.B. reported nonfinancial support from Bristol-Myers Squibb, and personal fees from Octane Global, Defined Health, Dedham Group, Adept Field Solutions, Slingshot Insights, Blueprint Partnerships, Charles River Associates, Trinity Group, and Insight Strategy, outside of the submitted work. T.K.C. reports research (institutional and personal) support from AstraZeneca, Alexion, Bayer, Bristol-Myers Squibb/ER Squibb and sons LLC, Cerulean, Eisai, Foundation Medicine Inc., Exelixis, Ipsen, Tracon, Genentech, Roche, Roche Products Limited, F. Hoffmann-La Roche, GlaxoSmithKline, Lilly, Merck, Novartis, Peloton, Pfizer, Prometheus Labs, Corvus, Calithera, Analysis Group, Sanofi/Aventis, Takeda, as well as honoraria from AstraZeneca, Alexion, Sanofi/Aventis, Bayer, Bristol-Myers Squibb/ER Squibb and sons LLC, Cerulean, Eisai, Foundation Medicine Inc., Exelixis, Genentech, Roche, Roche Products Limited, F. Hoffmann-La Roche, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, EMD Serono, Prometheus Labs, Corvus, Ipsen, Up-to-Date, NCCN, Analysis Group, NCCN, Michael J. Hennessy (MJH) Associates, Inc (Healthcare Communications Company with several brands such as OnClive, PeerView and PER), Research to Practice, L-path, Kidney Cancer Journal, Clinical Care Options, Platform Q, Navinata Healthcare, Harborside Press, American Society of Medical Oncology, NEJM, Lancet Oncology, Heron Therapeutics, Lilly. T.K.C. has played a consulting or advisory role to the following: AstraZeneca, Alexion, Sanofi/Aventis, Bayer, Bristol-Myers Squibb/ER Squibb and sons LLC, Cerulean, Eisai, Foundation Medicine Inc., Exelixis, Genentech, Heron Therapeutics, Lilly, Roche, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, EMD Serono, Prometheus Labs, Corvus, Ipsen, Up-to-Date, NCCN, Analysis Group, Pionyr, Tempest, and has stock ownership in Pionyr and Tempest. T.K.C. has also held the following leadership roles: Director of GU Oncology Division at Dana-Farber and past President of Medical Staff at Dana-Farber, member of NCCN Kidney panel and the GU Steering Committee, past chairman of the Kidney Cancer Association Medical and Scientific Steering Committee. T.K.C. reports the following patents, royalties, or other intellectual properties: International Patent Application number PCT/US2018/12209, entitled “PBRM1 Biomarkers Predictive of Anti-Immune Checkpoint Response,” filed 3 January 2018, claiming priority to U.S. Provisional Patent Application No. 62/445,094, filed 11 January 2017; International Patent Application number PCT/US2018/058430, entitled “Biomarkers of Clinical Response and Benefit to Immune Checkpoint Inhibitor Therapy,” filed 31 October 2018, claiming priority to US Provisional Patent Application number 62/581,175, filed 3 November 2017. T.K.C. reports travel, accommodations, and expenses, in relation to consulting, advisory roles, or honoraria. T.K.C. will provide a CV upon request for scope of clinical practice and research. Medical writing and editorial assistance support may have been funded by communications companies funded by pharmaceutical companies (ClinicalThinking, Envision Pharma Group, Fishawack Group of Companies, Health Interactions, Parexel, Oxford PharmaGenesis, and others). The institution (Dana-Farber Cancer Institute) may have received additional independent funding of drug companies or/and royalties potentially involved in research around the subject matter. T.K.C. has mentored several non-US citizens on research projects with potential funding (in part) from non-US sources/foreign components: (1) Asmar Wood S.A.L. is a private company based in Beirut, Lebanon that will provide a total of $100,000 in salary support to Sarah Abou Alaiwi from 1 July 2018 to 1 July 2020 during her postdoctoral research fellowship at DFCI; (2) Fondation Arc Pour La Recherche Sur Le Cancer is a not-for-profit foundation based in Villejuif, France that provides 2561.04€ per month in salary support to Ronan Flippot during his clinical training at DFCI from 2 May 2018 to 4 November 2018. The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lasseter, K., Nassar, A.H., Hamieh, L. et al. Plasma cell-free DNA variant analysis compared with methylated DNA analysis in renal cell carcinoma. Genet Med 22, 1366–1373 (2020). https://doi.org/10.1038/s41436-020-0801-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0801-x
Keywords
This article is cited by
-
Diagnostic liquid biopsy biomarkers in renal cell cancer
Nature Reviews Urology (2024)
-
Liquid biopsy at the frontier in renal cell carcinoma: recent analysis of techniques and clinical application
Molecular Cancer (2023)
-
Longitudinal monitoring of cell-free DNA methylation in ALK-positive non-small cell lung cancer patients
Clinical Epigenetics (2022)
-
Tumor fractions deciphered from circulating cell-free DNA methylation for cancer early diagnosis
Nature Communications (2022)
-
Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes
Nature Medicine (2020)