Abstract
Purpose
Cytochrome P450 2D6 (CYP2D6) genotype–guided opioid prescribing is limited. The purpose of this type 2 hybrid implementation-effectiveness trial was to evaluate the feasibility of clinically implementing CYP2D6-guided postsurgical pain management and determine that such an approach did not worsen pain control.
Methods
Adults undergoing total joint arthroplasty were randomized 2:1 to genotype-guided or usual pain management. For participants in the genotype-guided arm with a CYP2D6 poor (PM), intermediate (IM), or ultrarapid (UM) metabolizer phenotype, recommendations were to avoid hydrocodone, tramadol, codeine, and oxycodone. The primary endpoints were feasibility metrics and opioid use; pain intensity was a secondary endpoint. Effectiveness outcomes were collected 2 weeks postsurgery.
Results
Of 282 patients approached, 260 (92%) agreed to participate. In the genotype-guided arm, 20% had a high-risk (IM/PM/UM) phenotype, of whom 72% received an alternative opioid versus 0% of usual care participants (p < 0.001). In an exploratory analysis, there was less opioid consumption (200 [104–280] vs. 230 [133–350] morphine milligram equivalents; p = 0.047) and similar pain intensity (2.6 ± 0.8 vs. 2.5 ± 0.7; p = 0.638) in the genotype-guided vs. usual care arm, respectively.
Conclusion
Implementing CYP2D6 to guide postoperative pain management is feasible and may lead to lower opioid use without compromising pain control.
Similar content being viewed by others
INTRODUCTION
More Americans suffer from pain than are affected by heart disease, cancer, and lung disease combined.1 Opioids are commonly used to treat pain and are among the most widely prescribed medications in the United States.2 Given the role of opioids in acute, postoperative pain management, it is no surprise that approximately 35% of all opioid prescriptions originate from surgeons.3 However, interindividual variability in analgesic response to opioids has been observed, which may compromise postoperative pain control.4,5,6,7 Considering factors underlying this variability in opioid response may allow surgeons to individualize treatments and optimize opioid prescribing.
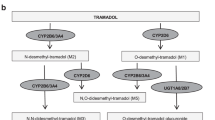
Hydrocodone, tramadol, codeine, and oxycodone are among the most prescribed opioids, and the cytochrome P450 2D6 (CYP2D6) enzyme is central to generation of highly potent metabolites for these opioids.8 Codeine and tramadol are dependent on bioactivation by the CYP2D6 enzyme to morphine and O-desmethyltramadol, respectively, which have 200-fold greater affinity for the µ-opioid receptor than their parent compounds.8,9 Tramadol also has nonopioid mechanisms of action, via reuptake inhibition of serotonin and norepinephrine, which do not require bioactivation, thus providing some nonopioid analgesic activity with the parent compound.10 Hydrocodone and oxycodone undergo similar metabolism via CYP2D6 to hydromorphone and oxymorphone, respectively, which have 10- to 40-fold higher receptor affinity than the parent compound.8,9,11
The CYP2D6 gene is highly polymorphic, with over 130 alleles defined (https://www.pharmvar.org/gene/CYP2D6). CYP2D6 variation affects opioid metabolism and contributes to the interindividual variability in opioid response.8 Clinically, CYP2D6 genotype is used to predict CYP2D6 enzyme activity. Approximately 5–10% of individuals are poor metabolizers (PMs), with no enzyme activity, and another 2–11% are intermediate metabolizers (IMs), with significantly reduced enzyme activity.8 Patients with a CYP2D6 PM or IM phenotype have a reduced capacity to biotransform codeine and tramadol to their active metabolites.8 Clinically, this may result in insufficient analgesia, and data suggest these drugs should be avoided in PMs and potentially in IMs.8,12 At the opposite extreme, approximately 1–2% of individuals are ultrarapid metabolizers (UMs) secondary to CYP2D6 copy-number variation. Patients with the UM phenotype are at increased risk for toxic concentrations of active opioid metabolites, with reports of life-threatening toxicity and death with codeine or tramadol.13,14,15 The data are less clear with hydrocodone and oxycodone. However, evidence that the effectiveness of hydrocodone is reduced with concomitant use of CYP2D6 inhibitors suggests that CYP2D6 genotype has implications for response to hydrocodone as well.16,17
Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines support CYP2D6 genotype–guided use of opioid analgesics,8 but this is rarely done in clinical practice. Given frequent postoperative opioid use and the role of CYP2D6 in opioid metabolism and response, this pharmacogene is uniquely poised to facilitate opioid prescribing decisions and individualize postoperative pain management. Therefore, the purpose of this study was to evaluate the feasibility of implementing a CYP2D6-guided postsurgical pain management paradigm and determine that such an approach did not worsen postoperative pain control. We specifically hypothesized that CYP2D6-guided management of postsurgical pain is feasible, with reduced use of codeine, tramadol, hydrocodone, and oxycodone in PMs/IMs/UMs. Additionally, we hypothesized that a CYP2D6-guided approach would not worsen pain control compared with an unguided approach.
MATERIALS AND METHODS
Study design
This study was a randomized, open label, type 2 hybrid implementation-effectiveness trial18 with the coprimary endpoints of feasibility of clinical implementation and opioid utilization. The study was conducted in 260 adult participants undergoing unilateral total joint arthroplasty, funded under a University of Florida (UF) Health–Clinical and Translational Science Institute Learning Health System Initiative, with additional support from UF Health Shands Hospital. According to the PRagmatic-Explanatory Continuum Indicator Summary (PRECIS)-2 tool, this trial was highly pragmatic as opposed to explanatory in six of nine domains assessed (Supplementary Table 1).19 Participants were enrolled between June 2018 and June 2019. This study initially recruited participants from a single UF orthopedic clinic and later expanded to include a second UF orthopedic clinic site. Five orthopedic physicians were involved in this study and referred patients for study participation.
Study participants
Eligible participants were adults ≥18 years of age scheduled for primary unilateral total hip or knee arthroplasty. Those scheduled to undergo a revision or bilateral procedure; with planned discharge to a rehabilitation facility; receiving long-term opioid therapy, defined as use of opioids on most days for more than three months; or with an allergy to opioids were excluded.
Randomization and baseline procedures
Prior to surgery, patients typically had two orthopedic clinic visits. The first was an initial evaluation visit when the decision for surgery was made. The second was a preoperative visit that occurred within one month of surgery, during which the postoperative pain management plan was developed, and a prescription for postoperative opioid therapy was provided. Clinical research coordinators approached patients about study participation at the initial evaluation visit after the decision for surgery was made. After providing written informed consent, a buccal sample and data on participant demographics and medical history were collected, and participants were randomized 2:1 to a CYP2D6-guided or usual postoperative pain management approach. Simple random allocation with block sizes of 6 or 12 was performed within each site. For participants in the CYP2D6-guided arm, buccal samples were batched each week and processed in the College of American Pathologists(CAP)/CLIA–licensed UF Health Pathology Laboratory, and CYP2D6 genotype and metabolizer phenotype results were reported to the electronic health record (EHR). This process allowed genotype results to be available in time to inform the postoperative pain management plan. Participants randomized to the usual care pain management arm had their DNA sample stored in the pathology laboratory until they completed their study participation, at which time the sample was processed, and CYP2D6 genotype and metabolizer phenotype were reported in the EHR for future use. Samples were not genotyped for participants in the usual care arm who did not complete study procedures.
CYP2D6 genotyping, phenotype assignment, and recommendations
CYP2D6 genotype was determined for all but four participants using a Luminex xTAG CYP2D6 Kit v3 (Luminex Corporation, Austin, TX, USA). In early June 2019, the pathology laboratory transitioned genotyping to the QuantStudio 12 K Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA), which was used to genotype the final four participants. Both platforms tested for the CYP2D6 *2, *3, *4, *5, *6, *7, *8, *9, *10, *17, *29, and *41 alleles and copy-number variation. The Luminex kit additionally tested for the *11, *15, and *35 alleles. The activity score (AS) method was used to assign CYP2D6 metabolizer phenotype based on genotype and concomitant use of strong or moderate CYP2D6 inhibitors. First, an AS was assigned for each allele based on CPIC guidance at the time of study enrollment.8 An activity value of 1 was assigned for each normal function allele (i.e., *1, *2, *35), 0.5 for each decreased function allele (i.e., *9, *10, *17, *29, *41), and 0 for each no function allele (i.e., *3–*8).8,20,21 The allele activity scores were summed to derive the total AS for the diplotype. Then, the total AS was multiplied by a factor of 0.5 for individuals taking a moderate inhibitor (i.e., duloxetine) or 0 for individuals taking a strong CYP2D6 inhibitor (i.e., bupropion, fluoxetine, paroxetine) to derive the clinical CYP2D6 activity score.8,20,22,23 Phenotypes were then assigned based on the clinical CYP2D6 activity score that accounted for CYP2D6 genotype and phenoconversion: 0, PM; >0–0.75, IM; 1–2, normal metabolizer (NM); and >2, UM. The genotyping assay could detect the presence of allele duplication, but could not determine which allele was duplicated or the number of allele copies, and thus, ranged phenotypes were possible (e.g., NM to UM).
After return of CYP2D6 genotype, a clinical pharmacist provided a standardized consult note in the EHR, as has been described,24 to communicate prescribing recommendations based on CYP2D6 phenotype. For participants in the genotype-guided arm with a CYP2D6 PM, IM, or UM clinical phenotype (high-risk phenotype), recommendations were to avoid tramadol, hydrocodone, codeine, and oxycodone and to use an alternative opioid (e.g., morphine, hydromorphone) or non-opioid (e.g., nonsteroidal anti-inflammatory drug [NSAID]) analgesic. Tramadol was recommended as the preferred opioid in NMs because of its opioid and non-opioid mechanisms and purported lower risk for misuse.25,26 Given the risks for toxicity in UMs, participants with the NM to UM range phenotype were treated as UM in regard to opioid recommendations.
Study surveys
At 1 week postsurgery, participants in both arms completed the Patient-Reported Outcomes Measurement Information System (PROMIS) pain intensity survey. At 2 weeks (±4 days) postsurgery, participants in both arms were asked to complete the PROMIS 43 and pain intensity surveys (www.nihpromis.org) and the Hip Disability and Osteoarthritis Outcome Score Short Form (HOOS-JR) or Knee Injury and Osteoarthritis Outcome Score Short Form (KOOS-JR) based on surgery indication (e.g., total hip arthroplasty [THA] vs. total knee arthroplasty [TKA]). At both postoperative follow-up timepoints, participants were also asked about their consumption of any opioid prescribed since discharge home from surgery. Pain intensity surveys were utilized to determine composite pain intensity, defined as the mean of current pain and worst and average pain over the past seven days.12 Participants rated their pain intensity for each of the three questions on a 5-point Likert scale: Had no pain = 1, Mild = 2, Moderate = 3, Severe = 4, and Very severe = 5. The PROMIS 43 survey assessed the domains of physical functioning, anxiety, depression, fatigue, sleep disturbance, ability to participate in social activities, pain interference, prescription pain medication misuse, and average pain intensity.27 The HOOS-JR and KOOS-JR surveys assessed the participant’s ability to complete usual activities and are part of the standard clinical postoperative follow-up assessment. Opioid consumption was evaluated by asking participants to report the strength and quantity of the prescribed opioid and the number of pills remaining in the bottle (Supplementary Fig. 1). A series of questions was also posed regarding medication refills to capture opioid use beyond what was prescribed at the preoperative appointment. If the participant had a follow-up clinic visit at the two-week timepoint, surveys and assessment of opioid consumption were completed in person with the study coordinator. Otherwise, based on participant preference, surveys and assessment of opioid consumption were completed by email, via a link provided in a text message, or by phone call with a pharmacist or pharmacy technician from the UF Center for Quality Medication Management.
Data analysis
Data are presented as mean ± standard deviation, median [interquartile range], or count (%). The primary outcomes were feasibility of implementing a genotype-guided opioid prescribing approach for participants undergoing an elective surgical procedure and opioid utilization. The following implementation metrics were assessed: percentage of patients who agreed to participate, percentage of participants in both arms with a clinical phenotype (based on CYP2D6 and phenoconversion) warranting alternative therapy, percentage of participants in the genotype-guided arm for whom a clinical phenotype-guided recommendation was accepted by the clinician, and specific opioids prescribed by CYP2D6 phenotype.
Consult recommendations were considered accepted for participants with a high-risk phenotype if an alternative opioid (e.g., hydromorphone, morphine) was prescribed. For CYP2D6 NMs, consult recommendations were accepted if tramadol was prescribed. Participants whose genotype resulted after the preoperative appointment were excluded from the analysis of acceptance of consult recommendations.
Opioid consumption was calculated as the differences between participant reported opioid pills prescribed at the preoperative appointment and opioid pills remaining at the 1-week and 2-week follow-up timepoints. This difference was calculated for each opioid and then expressed as morphine milligram equivalents (MME) using standard conversion factors and the medication strength of the prescribed opioid analgesic.28 If a participant was prescribed multiple opioids, MMEs were calculated for each opioid and then summed to give a total MME value.
Secondary outcomes included composite pain intensity, PROMIS-43 measures, and mobility as assessed by the HOOS-JR or KOOS-JR survey. Data on pain intensity were collected to document that there were not obvious trends of the CYP2D6-guided approach leading to worse pain control, which in turn could lead to worse postsurgical outcomes. While composite pain intensity and opioid consumption were collected at both 1 week and 2 weeks postsurgery, the 2-week timepoint was designated as the primary timepoint for data on pain control.
Study participant characteristics, composite pain intensity, and other survey results were compared between the CYP2D6-guided and usual care arms by chi-squared, Fisher’s exact, or two independent samples t-test where appropriate. Opioid consumption data were not normally distributed and were compared between study arms using the Mann–Whitney U-test. Main analyses were conducted in all participants in each study arm, with subset analyses conducted in IM/PMs and separately in NMs. Comparisons between the CYP2D6-guided and usual care groups for opioid consumption and composite pain intensity were also performed by analysis of covariance, adjusting for baseline differences between groups. Data analyses were performed using Python.29
This study was originally funded as a pilot project within our National Institutes of Health (NIH)-funded Learning Health System, with planned enrollment of 130 participants. In an a priori power calculation assuming 25% of participants in the CYP2D6-guided arm would have a PM, IM, or UM phenotype (based on genotype and drug interactions) and that codeine, tramadol, hydrocodone, and oxycodone would be avoided in these participants, whereas the remaining 75% of participants in the implementation arm would be prescribed tramadol and nearly all (i.e., 95%) of controls would be prescribed tramadol, codeine, hydrocodone, or oxycodone, including 130 patients was estimated to have >80% power, with an alpha of 0.05, to detect a difference in use of codeine, tramadol, hydrocodone, or oxycodone between arms. After the trial started, we received additional funding from our health system, which enabled expansion of our sample size to 260 participants. A post hoc power calculation based on the number of patients who completed the assessment of opioid consumption showed that including 126 participants in the CYP2D6-guided arm and 68 participants in the usual care arm, with an alpha of 0.05, provided 80% power to detect an effect size of 0.43, which is equivalent to a difference of 60 MME between arms.
RESULTS
Participant population
Of 282 participants approached about study participation, 260 (92%) agreed to participate and were randomized to the genotype-guided (n = 173) or usual care (n = 87) arm (Fig. 1). Prevailing reasons why patients declined participation included being uncomfortable with research (n = 5) or genotyping (n = 3), unwilling to take an opioid analgesic (n = 3) or complete follow-up surveys (n = 2), and content with postoperative analgesics used with prior surgery (n = 1). The remaining declinations were for other, unknown, or logistical reasons (n = 8). Of those who agreed to participate, total joint arthroplasty was performed in 90% (234/260) of participants, including 154 in the genotype-guided arm and 80 in the usual care arm. Study participant demographics and clinical variables for these 234 participants are summarized in Table 1. Clinical CYP2D6 phenotype and body mass index (BMI) varied between treatment arms. The CYP2D6 NM phenotype occurred less frequently in the usual care arm (56%) compared with the genotype-guided arm (77%; p = 0.017). Otherwise, participant characteristics were well balanced between the CYP2D6-guided and usual care arms.
Feasibility and implementation metrics
Clinical CYP2D6 genotyping was successful from the initial buccal swab in >99% (227/228) of participants in both study arms. The only indeterminate genotype result occurred in a usual care participant who was unable to be recontacted to obtain a second sample for repeat genotyping. CYP2D6 phenotype frequencies based on genotype alone and with consideration of phenoconversion are listed in Table 2. Based on CYP2D6 genotype, 6% of participants were PMs, 7% were IMs, and 16% (36/228) had a high-risk CYP2D6 phenotype that warranted a perioperative opioid other than codeine, tramadol, hydrocodone, or oxycodone. Once phenoconversion was considered in the overall study population, there was a 2.8-fold increase in the prevalence of the PM phenotype and the proportion of participants with a high-risk CYP2D6 phenotype increased to 27% (62/228). Of those who underwent surgery within the CYP2D6-guided arm, 94% (145/154) had genotype results returned prior to the preoperative appointment when the opioid prescription was provided, with a median genotype turnaround time of 8 (range: 2–23) days.
In the CYP2D6-guided arm, 20% (29/145) of those with genotype returned prior to the preoperative appointment had a high-risk (IM/PM/UM/NM-UM) CYP2D6 phenotype, of whom 72% (21/29) received an opioid other than codeine, tramadol, hydrocodone, or oxycodone. Hydromorphone was the most commonly prescribed alternative opioid in those with a high-risk phenotype (20/21; 95%, Fig. 2). None of the usual care participants with a high-risk phenotype received an alternative opioid (Fig. 2,p < 0.001 for comparison with the genotype-guided arm). Among those with a NM phenotype, 88% (101/115) of CYP2D6-guided and 82% (37/45) of usual care participants were prescribed a tramadol-based regimen (p = 0.355). In most cases (87% of those in genotype-guided arm and 78% in usual care arm) hydrocodone, or another opioid, was prescribed concomitantly with tramadol as is usual practice at the clinics where participants were enrolled. Tramadol was prescribed as the sole opioid to 1% of NMs in the genotype-guided arm and 4% of NMs in the usual care arm.
Opioid consumption and pain intensity
MME data were provided by 72% (111/154) and 82% (126/154) of CYP2D6-guided and 71% (57/80) and 85% (68/80) of usual care participants at the 1- and 2-week postoperative timepoints. One- and two-week postoperative pain intensity surveys were completed by 86% (133/154) and 89% (137/154) of CYP2D6-guided and 84% (67/80) and 93% (74/80) of usual care participants, respectively. In the trial population overall at 2 weeks postsurgery, opioid consumption was lower in the CYP2D6-guided group (200 mg [104 mg–280 mg]) compared with the usual care group (230 mg [133 mg–350 mg]; p = 0.047; Fig. 3a). However, composite pain intensity was similar between CYP2D6-guided (2.6 ± 0.8) and usual care (2.5 ± 0.7) arms (p = 0.638; Fig. 3b); most reported a mild to moderate level of pain intensity. Adjusting for BMI and clinical CYP2D6 phenotype did not influence results for pain intensity (p = 0.640) or MME consumption (p = 0.029). None of the participants with the UM phenotype completed study follow-up, and all participants with the NM-UM ranged phenotype were in the usual care arm. Subset analyses were conducted in those with the IM/PM and NM phenotypes. Within these subsets, MME use and pain intensity did not differ significantly between study arms (Supplementary Table 2). There were similar trends in the opioid consumption and composite pain intensity data at the 1-week postsurgery timepoint (Supplementary Table 3).
Two-week postoperative opioid consumption and pain intensity were greater following TKA, compared with THA in both the genotype-guided and usual care arms (Table 3). Opioid consumption did not differ between CYP2D6-guided and usual care arms for participants undergoing THA (p = 0.865). However, among TKA recipients, opioid consumption was lower in the CYP2D6-guided versus usual care arm (Table 3; p = 0.003). Composite pain intensity was similar between CYP2D6-guided and usual care arms when stratified by surgery indication (Table 3).
Other PROMIS measures and KOOS-JR/HOOS-JR interval scores were similar between CYP2D6-guided and usual care arms when considering all patients in the study arm and when limiting to PM/IMs and NMs (Supplementary Tables 4 and 5). Opioid-related adverse events were not systematically collected as part of the study. However, none of the participants required naloxone in the postoperative period, or throughout their hospitalization.
DISCUSSION
Data from the current investigation demonstrate the feasibility of implementing CYP2D6 genotyping to guide postoperative pain management, with most participants agreeing to testing and high provider acceptance of genotype-guided recommendations in those with a high-risk phenotype. Further, our data suggest a genotype-guided approach may lead to lower opioid use without compromising pain control. Similarities in composite pain intensity and HOOS/KOOS-JR interval scores between study arms suggests participants consumed opioids as needed to minimize postoperative pain, but those in the genotype-guided arm needed fewer opioids for similar pain control.
Results when stratified by surgery type imply that the impact of a genotype-guided approach on opioid consumption may be greatest in those undergoing more painful surgical procedures. Participants who underwent TKA had greater composite pain intensity scores and consumed more postoperative opioids than their THA counterparts, regardless of the postoperative pain management approach. The CYP2D6-guided approach was associated with decreased opioid consumption after TKA, but not THA, suggesting that overall results were driven by participants undergoing TKA.
While not directly assessing the opioid epidemic, our results may have implications in this regard in that the majority of individuals who misuse opioids report the pursuit of pain relief as a primary motivator.30 Similar pain intensity with less opioid consumption in the CYP2D6-guided arm indicates that this guided approach leads to better pain control,31 and as such, may potentially avert future opioid misuse. Indeed, postoperative opioid use is proposed to be a gateway to chronic opioid use, with evidence of new persistent opioid use three to six months after a surgical procedure in approximately 6% of individuals who were opioid naïve prior to surgery.32 Physicians in certain specialties, like orthopedic surgery, prescribe opioids at a higher rate than those of other specialties and primary care physicians.3,33 Opioid sparing measures for postoperative analgesia have thus become a priority for surgeons to expedite patient recovery and combat the opioid epidemic,32,34 and patients undergoing orthopedic surgery may especially benefit from measures, such as CYP2D6 genotyping, to guide opioid prescribing. Patients have also expressed a desire for more individualized information regarding pain management after hip and knee arthroplasty.35 Indeed, over 90% of patients approached about the current study agreed to participate. The elective surgical setting further lends itself well to a genotype-guided opioid approach. Even with a median genotype turnaround time of 8 days, 95% of genotypes resulted prior to the preoperative visit, demonstrating that a major logistical barrier to pharmacogenetic implementation, that is genotype turnaround time, can be largely overcome in the elective surgery setting. While this study primarily focused on utilizing CYP2D6 genotype to guide postoperative opioid prescribing, studies evaluating perioperative pharmacogenomic implementations are underway.36,37
Multiple studies have evaluated postoperative opioid effectiveness according to CYP2D6 genotype. In line with our findings, a longitudinal cohort study of patients undergoing abdominal or thoracic surgeries showed similar postoperative pain scores after tramadol administration across CYP2D6 phenotypes; however, opioid consumption was not assessed.38 Other studies evaluating tramadol effectiveness in relieving postoperative pain included both pain intensity and tramadol consumption as outcome measures. Among patients undergoing elective nephrectomy or major abdominal surgery, increased pain intensity, despite higher opioid consumption, was observed in those with CYP2D6 nonfunctional or reduced function genotype versus NMs.39,40 A study of patients undergoing knee arthroscopy showed that pain intensity, but not tramadol consumption, varied by CYP2D6 phenotype in the immediate postoperative period.41 The totality of these data, in addition to findings from the current study, support the importance of assessing both opioid consumption and pain intensity when evaluating postsurgical pain management approaches.
The current study also highlights the importance and prevalence of phenoconversion (i.e., use of moderate and strong CYP2D6 inhibitors) in clinical phenotype determination among patients undergoing total joint arthroplasty. After considering phenoconversion in this cohort of participants, the prevalence of high-risk CYP2D6 phenotypes increased from 16% to 27%. More specifically, the prevalent use of strong CYP2D6 inhibitors resulted in the PM phenotype nearly tripling. These results suggest future pain-related pharmacogenomics research should screen for the concomitant use of moderate and strong CYP2D6 inhibitors.
This study has limitations that deserve consideration. First, assessment of MME may be considered exploratory in that it was not the focus of our a priori power calculation. In addition, reliance on participant reported opioid consumption restricts MME analysis to those who successfully completed the two-week survey. Given this was a pragmatic study, participants could have received any number of analgesics at various doses. Therefore, reliance on participant responses was key to calculating MME at the follow-up timepoints, and incomplete participant-reported data reduces the available sample to assess postoperative opioid consumption. Second, CYP2D6 phenotype distributions were unequal between genotype-guided and usual care arms, with the usual care group displaying higher IM and PM percentages than expected. This was possibly the result of adding the second study site over the course of the trial that had a less ancestrally diverse population, but not initially adjusting the randomization approach to account for a second site (e.g., randomizing separately by site). Nonetheless, adjusting for CYP2D6 phenotype did not affect the overall results. Third, only five providers were involved in the perioperative care of these patients, and the high acceptance rate of genotype-guided recommendations observed in this study may not be generalizable across a larger provider group. However, surgeons have made opioid-sparing analgesic regimens a priority in clinical practice to reduce postoperative opioid use without compromising pain control. The overall study goals aligned with this clinical practice paradigm. Thus, surgeons were motivated to follow CYP2D6-guided recommendations, as evidenced by the high physician acceptance rate, which we expect may be true more generally. Fourth, differences in opioid consumption may be subject to a placebo effect since study participants were not blinded. To mitigate a placebo effect, CYP2D6 results were not actively provided to study participants. However, we cannot rule out that surgeons discussed results with patients or that CYP2D6 results were accessed through the patient portal. Finally, the practice of prescribing tramadol with another opioid, most commonly hydrocodone, suggests surgeons were not comfortable with prescribing tramadol as the sole postoperative opioid for NMs. Thus, further research to demonstrate whether use of tramadol alone in NMs results in sufficient pain control is warranted.
Future efforts will examine the effects of having CYP2D6 genotype on pain management and pain control. Findings from this study informed the design of a multicenter, National Human Genome Research Institute (NHGRI)-funded, clinical trial (ADOPT-PGx: A Depression and Opioid Pragmatic Trial in Pharmacogenomics; https://gmkb.org/) evaluating the efficacy of a CYP2D6-guided approach on postoperative pain management as part of the IGNITE Pragmatic Trials Network.
In summary, preemptive CYP2D6-guided opioid selection is feasible in an elective surgery setting, with high patient acceptance of genotyping and provider acceptance of genotype-guided recommendations in PMs/IMs/UMs. We also show that CYP2D6 can be obtained in the preoperative period after the decision for surgery is made so that results are available in time to guide postoperative opioid selection. Our results also suggest that a CYP2D6-guided approach may reduce postoperative opioid utilization without sacrificing pain control, with the greatest effect potentially in those undergoing more painful surgical procedures (e.g., TKA). Overall, utilizing a CYP2D6-guided approach to prescribe postoperative opioids is feasible and pain control may be achieved with less opioid use. In the current opioid climate, the need to develop safe and effective pain management strategies has never been more imperative, and precision medicine initiatives may prove invaluable.
Data availability
Data set and code available upon request.
References
Gereau, R. W. T. et al. A pain research agenda for the 21st century. J. Pain. 15, 1203–1214 (2014).
Hoots B. E. et al. 2018 Annual surveillance report of drug-related risks and outcomes-United States. Surveillance Special Report. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services. Published August 31, 2018. Accessed May 30, 2020.
Levy, B., Paulozzi, L., Mack, K. A. & Jones, C. M. Trends in opioid analgesic-prescribing rates by specialty, U.S., 2007-2012. Am. J. Prev. Med. 49, 409–413 (2015).
Aubrun, F., Langeron, O., Quesnel, C., Coriat, P. & Riou, B. Relationships between measurement of pain using visual analog score and morphine requirements during postoperative intravenous morphine titration. Anesthesiology. 98, 1415–1421 (2003).
Abdel Shaheed, C., Maher, C. G., Williams, K. A., Day, R. & McLachlan, A. J. Efficacy, tolerability, and dose-dependent effects of opioid analgesics for low back pain: a systematic review and meta-analysis. JAMA Intern. Med. 176, 958–968 (2016).
Kalso, E., Edwards, J. E., Moore, R. A. & McQuay, H. J. Opioids in chronic non-cancer pain: systematic review of efficacy and safety. Pain. 112, 372–380 (2004).
Lopes, G. S. et al. Sex differences in associations between CYP2D6 phenotypes and response to opioid analgesics. Pharmacogenomics Pers. Med. 13, 71–79 (2020).
Crews, K. R. et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 95, 376–382 (2014).
Volpe, D. A. et al. Uniform assessment and ranking of opioid mu receptor binding constants for selected opioid drugs. Regul Toxicol. Pharmacol. 59, 385–390 (2011).
Ultram (tramadol) [package insert]. Titusville, NJ: Janssen Pharmaceutical Companies; 2019.
Davis, M. P. et al. Normal-release and controlled-release oxycodone: pharmacokinetics, pharmacodynamics, and controversy. Support Care Cancer 11, 84–92 (2003).
Smith, D. M. et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet. Med. 21, 1842–1850 (2019).
Ciszkowski, C., Madadi, P., Phillips, M. S., Lauwers, A. E. & Koren, G. Codeine, ultrarapid-metabolism genotype, and postoperative death. N. Engl. J. Med. 361, 827–828 (2009).
Gasche, Y. et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N. Engl. J. Med. 351, 2827–2831 (2004).
Koren, G., Cairns, J., Chitayat, D., Gaedigk, A. & Leeder, S. J. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet. 368, 704 (2006).
Kapil, R. P. et al. Effects of paroxetine, a CYP2D6 inhibitor, on the pharmacokinetic properties of hydrocodone after coadministration with a single-entity, once-daily, extended-release hydrocodone tablet. Clin. Ther. 37, 2286–2296 (2015).
Monte, A. A. et al. The effect of CYP2D6 drug-drug interactions on hydrocodone effectiveness. Acad. Emerg. Med. 21, 879–885 (2014).
Curran, G. M., Bauer, M., Mittman, B., Pyne, J. M. & Stetler, C. Effectiveness-implementation hybrid designs: combining elements of clinical effectiveness and implementation research to enhance public health impact. Med. Care 50, 217–226 (2012).
Loudon, K. et al. The PRECIS-2 tool: designing trials that are fit for purpose. BMJ. 350, h2147 (2015).
Gaedigk, A. et al. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 83, 234–242 (2008).
Hicks, J. K., Swen, J. J. & Gaedigk, A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr. Drug. Metab. 15, 218–232 (2014).
Borges, S. et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J. Clin. Pharmacol. 50, 450–458 (2010).
Kim, K. M. et al. Pharmacogenetics and healthcare outcomes in patients with chronic heart failure. Eur. J. Clin. Pharmacol. 68, 1483–1491 (2012).
Mosley, S. A. et al. Design and rational for the precision medicine guided treatment for cancer pain pragmatic clinical trial. Contemp. Clin. Trials 68, 7–13 (2018).
Adams, E. H. et al. A comparison of the abuse liability of tramadol, NSAIDs, and hydrocodone in patients with chronic pain. J. Pain Symptom Manage. 31, 465–476 (2006).
Preston, K. L., Jasinski, D. R. & Testa, M. Abuse potential and pharmacological comparison of tramadol and morphine. Drug Alcohol Depend. 27, 7–17 (1991).
Cella, D. et al. The Patient-Reported Outcomes Measurement Information System (PROMIS): progress of an NIH Roadmap cooperative group during its first two years. Med. Care. 45(5 Suppl 1), S3–S11 (2007).
Dowell, D., Haegerich, T. M. & Chou, R. CDC guideline for prescribing opioids for chronic pain—United States, 2016. JAMA. 315, 1624–1645 (2016).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. https://doi.org/10.1038/s41592-019-0686-2 (2020).
Han, B. et al. Prescription opioid use, misuse, and use disorders in U.S. adults: 2015 National Survey on Drug Use and Health. Ann. Intern. Med. 167, 293–301 (2017).
Dai, F. et al. Integration of pain score and morphine consumption in analgesic clinical studies. J. Pain. 14, 767–777 (2013).
Brummett, C. M. et al. New persistent opioid use after minor and major surgical procedures in US adults. JAMA Surg. 152, e170504 (2017).
Ringwalt, C. et al. Differential prescribing of opioid analgesics according to physician specialty for Medicaid patients with chronic noncancer pain diagnoses. Pain Res. Manag. 19, 179–185 (2014).
Wick, E. C., Grant, M. C. & Wu, C. L. Postoperative Multimodal Analgesia Pain Management With Nonopioid Analgesics and Techniques: A Review. JAMA surgery 152, 691–697 (2017).
Sjoveian, A. K. H. & Leegaard, M. Hip and knee arthroplasty - patient’s experiences of pain and rehabilitation after discharge from hospital. Int J Orthop Trauma Nurs 27, 28–35 (2017).
Jhun, E. H. et al. Pharmacogenomic considerations for medications in the perioperative setting. Pharmacogenomics. 20, 813–827 (2019).
Truong, T. M. et al. The ImPreSS Trial: Implementation of Point-of-Care Pharmacogenomic Decision Support in Perioperative Care. Clin Pharmacol Ther 106, 1179–1183 (2019).
Seripa, D. et al. Role of CYP2D6 Polymorphisms in the Outcome of Postoperative Pain Treatment. Pain Med (Malden, Mass) 16, 2012–2023 (2015).
Dong, H. et al. Effect of the CYP2D6 gene polymorphism on postoperative analgesia of tramadol in Han nationality nephrectomy patients. Eur J Clin Pharmacol 71, 681–686 (2015).
Stamer, U. M. et al. Impact of CYP2D6 genotype on postoperative tramadol analgesia. Pain. 105, 231–238 (2003).
Slanar, O. et al. Tramadol efficacy in patients with postoperative pain in relation to CYP2D6 and MDR1 polymorphisms. Bratisl Lek Listy 113, 152–155 (2012).
Acknowledgements
This work was supported by the University of Florida Clinical and Translational Science Institute, which is supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427 and University of Florida Health Shands. Work by C.D.T. was supported by T32 HG008958. Research reported in this publication was also supported by NIH/NHGRI (U01 HG 007269). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Conceptualization: L.H.C., J.A.J., R.B.F., H.K.P., C.F.G., C.D.T. Data curation: C.D.T. Formal analysis: C.D.T., Y.G. Funding acquisition: J.A.J., L.H.C. Investigation: H.K.P., C.F.G., J.T.D., H.A.P., L.F.P., E.N.E., C.D.T. Methodology: L.H.C., J.A.J., R.B.F., H.K.P., C.F.G., P.S., Y.G. Project administration: A.R.E., E.N.E. Resources: J.T.D., C.F.G., H.A.P., H.K.P., L.F.P., P.S. Supervision: L.H.C., R.B.F., J.A.J., H.K.P. Validation: L.H.C., R.B.F., Y.G., J.A.J., H.K.P., C.D.T. Visualization: C.D.T. Writing—original draft: C.D.T., L.H.C., Y.G., J.A.J. Writing—review & editing: L.H.C., J.T.D., R.B.F., C.F.G., Y.G., J.A.J., H.A.P., H.K.P., L.F.P., P.S., C.D.T.
Corresponding author
Ethics declarations
Competing interests
Y.G., R.B.F., J.A.J., and L.H.C. have received research funding from the National Institutes of Health. P.S. receives support from University of Florida Health Pathology Laboratories. C.F.G. is a stockholder for ROMTech. The other authors declare no competing interests.
Ethics declaration
All study participants provided written, informed consent. The study was approved by the University of Florida Institutional Review Board, and all procedures were in accordance with the ethical standards of the Declaration of Helsinki and registered in ClinicalTrials.gov (NCT03534063).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Thomas, C.D., Parvataneni, H.K., Gray, C.F. et al. A hybrid implementation-effectiveness randomized trial of CYP2D6-guided postoperative pain management. Genet Med 23, 621–628 (2021). https://doi.org/10.1038/s41436-020-01050-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-01050-4