Abstract
Purpose
Neurodevelopmental disorders (NDD) caused by protein phosphatase 2A (PP2A) dysfunction have mainly been associated with de novo variants in PPP2R5D and PPP2CA, and more rarely in PPP2R1A. Here, we aimed to better understand the latter by characterizing 30 individuals with de novo and often recurrent variants in this PP2A scaffolding Aα subunit.
Methods
Most cases were identified through routine clinical diagnostics. Variants were biochemically characterized for phosphatase activity and interaction with other PP2A subunits.
Results
We describe 30 individuals with 16 different variants in PPP2R1A, 21 of whom had variants not previously reported. The severity of developmental delay ranged from mild learning problems to severe intellectual disability (ID) with or without epilepsy. Common features were language delay, hypotonia, and hypermobile joints. Macrocephaly was only seen in individuals without B55α subunit-binding deficit, and these patients had less severe ID and no seizures. Biochemically more disruptive variants with impaired B55α but increased striatin binding were associated with profound ID, epilepsy, corpus callosum hypoplasia, and sometimes microcephaly.
Conclusion
We significantly expand the phenotypic spectrum of PPP2R1A-related NDD, revealing a broader clinical presentation of the patients and that the functional consequences of the variants are more diverse than previously reported.
Similar content being viewed by others
INTRODUCTION
Recent advances in exome sequencing have shown that de novo pathogenic variants are the most important cause of intellectual disability (ID) in outbred populations,1,2 and this has led to the identification of many new genetic conditions. Among the new causes discovered are de novo, often recurrent, pathogenic variants in subunits of the protein phosphatase type 2A (PP2A), the major serine/threonine phosphatase in the human body.1,2,3,4,5,6 Since PP2A counterbalances Ser/Thr-specific protein kinase activity, this phosphatase plays an essential regulatory role in cellular signaling and physiology.7,8 PP2A phosphatases are holoenzymes, comprising three subunits: a catalytic C, a scaffolding A and a regulatory B-type subunit, encoded by 19 different genes in total (2 for C, 2 for A, and 15 for B subunits).8 The B subunits determine substrate specificity and regulation of the complex,9,10,11 and thus are the main determinants of PP2A functions in cells and tissues.
By far the largest number of reported PP2A mutated cases harbor de novo pathogenic variants in PPP2R5D, encoding the regulatory B56δ subunit (25 patients).4,5,12,13 All but one (p.P53S) of the B56δ variants that were biochemically characterized (p.E198K, p.E200K, p.P210R, and p.W207R) showed decreased A and C binding, suggesting a dominant-negative mode of action.4 PPP2R5C (encoding B56γ; one case) and PPP2R5B (encoding B56β; one case) are two additional PP2A genes, pathogenic variants of which were associated with ID and overgrowth.5 Although not yet biochemically characterized, the pathogenic variants affect the very same, highly conserved acidic loop in B56 isoforms that is recurrently mutated in PPP2R5D-affected individuals. In PPP2R5B, this loop supposedly serves as a substrate specifying binding site.14 Seven patients (five variants) were reported with de novo pathogenic variants in PPP2R1A, encoding the scaffolding Aα subunit.4,15,16 Four of these variants (p.P179L, p.R182W, p.R183Q, and p.R258H) also occurred somatically in tumor tissue, and had defective binding of specific B-type subunits, except B56δ.4,17 However, mutant Aα-B56δ complexes proved catalytically impaired, potentially due to increased binding of TIPRL1, an endogenous inhibitor of PP2A.17 Consistent with a dominant-negative mode of action, ectopic expression of the p.R183Q Aα variant in wild-type (WT) PPP2R1A endometrial cancer cells increased anchorage-independent growth and tumor formation, and triggered hyperphosphorylation of oncogenic PP2A substrates in GSK-3β, Akt, and mTOR/p70 S6K signaling pathways.17 Most recently, 16 individuals were reported with 15 variants in PPP2CA, encoding the PP2A Cα subunit. Functional characterization also revealed losses of function, consistent with decreased PP2A-B56(δ) functionalities in most cases—either by a dominant-negative mechanism, or by haploinsufficiency.6 Finally, a non-sense variant in BOD1 (encoding a cellular inhibitor of PP2A-B56 complexes) was reported to cosegregate with ID in a consanguineous family,18 and a familial reciprocal translocation (4;6)(p16.1;q22) disrupting PPP2R2C (encoding the PP2A-B55γ subunit) was associated with mild ID, epilepsy, and behavioral problems.19
In the current study, we report on 30 additional cases (16 variants) with a de novo pathogenic variants in PPP2R1A. We find that the clinical presentation of the patients is much broader and the functional consequences of the variants are more diverse than previously reported, expanding the spectrum of this PP2A-related neurodevelopmental disorder.
MATERIALS AND METHODS
Ethics statement
All participants who were ascertained via research studies gave consent to their research participation. Those individual studies received approval from an institutional review board (IRB) or research ethics committee. Participants who were ascertained via clinical diagnostics procedures gave clinical consent for testing, and their permission for inclusion in this case collection, including photographs, was obtained using standard forms used locally at each site. The overall case collection study reported here did not require separate ethics approval, based on the institutional policies of all involved sites.
Generation of PPP2R1A mutants
The coding region of WT Aα complementary DNA (cDNA) (NM_014225.6) was cloned into an N-terminal HA-tag eukaryotic expression vector (pMB001) using XbaI/BamHI sites. Mutated Aα constructs were directly generated from this plasmid by polymerase chain reaction (PCR)–based site-directed mutagenesis (Stratagene) with Pwo polymerase (Roche Applied Science) and cDNA oligonucleotides (Sigma Genosys) containing the desired point mutations (Table S1). All variants were verified by Sanger sequencing (LGC Genomics).
Cellular PP2A binding assays
HEK293T cells (ATCC, characterized by short tandem repeat profiling, mycoplasma-free and used at passage number <15) were transfected with PEI transfection reagent using standard protocol. All GFP-B-type subunit expression vectors are previously described.4,17,20 Forty-eight hours post-transfection, cells were rinsed with PBS, lysed in 150 μl NET buffer (50 mM Tris.HCl pH 7.4, 150 mM NaCl, 15 mM EDTA, and 1% Nonidet P-40) containing protease and phosphatase inhibitor cocktail (Roche Applied Science) and centrifuged for 15 minutes at 13,000g. For binding assays with GFP-STRN3, no phosphatase inhibitors were added. If the experiment required measuring phosphatase activity, TBS was used instead of PBS and phosphatase inhibitors were omitted.
For pulldown experiments, lysates were incubated at 4 °C for 1 hour with 800 µl NENT100 buffer (20 mM Tris.HCl pH 7.4, 1 mM EDTA, 0.1% Nonidet P-40, 25% glycerol, 100 mM NaCl) containing 1 mg/ml bovine serum albumin, and 20 μl anti-HA-Agarose beads (Sigma-Aldrich, for HA pull down), or 20 μl GFP-trap-A beads (ChromoTek, for GFP trapping). Beads were washed three times with 1 ml NENT300 (containing 300 mM NaCl), and two times with 1 ml NENT150 (containing 150 mM NaCl). Bound proteins were eluted in 2× NuPage sample buffer (Invitrogen) and boiled for subsequent analysis by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 4–12% (w/v) Bis-Tris gels (BioRad) and western blotting. Membranes were blocked in 5% milk in TBS/0.1% Tween-20 for 1 hour at room temperature and incubated with the primary antibody overnight at 4 °C. Primary mouse monoclonal antibodies were anti-HA (clone HA-7, Sigma-Aldrich), anti-GFP (clone 9F9.F9, Abcam), anti-PP2A-C subunit (clone F2.6A10, generously supplied by Dr. S. Dilworth, Middlesex University, London, UK). After washing in TBS/0.1% Tween-20, membranes were incubated with horseradish peroxidase–conjugated secondary antibodies (Dako) and developed using Western Bright ECL (Advansta) on the ImageQuant LAS4000 scanner (GE Healthcare). All densitometric quantifications were done with Image Studio Lite software (version 5.2).
PP2A activity assays
After HA pulldown, beads were washed with 50 mM Tris-HCl pH 8.0 supplemented with 1 mM DTT, and resuspended in 75 µl of the same buffer. Assays were performed with 35 µl of this phosphatase suspension, and 10 µl of 2mM R-K-(p)T-I-R-R phosphopeptide or pRb phosphopeptide (I-N-G-S-P-R-(p)T-P-R-R-G-Q-N-R) for 10 and 20 minutes at 30 °C (in the linear range of the assay). Released phosphate was determined by addition of Biomol®Green (#BML-AK111-0250, Enzo) and absorbance measurement at 620 nm in a multichannel spectrophotometer. Specific phosphatase activity was obtained by correcting the measured absorbance for the input of HA-tagged Aα as determined and quantified by immunoblotting.
Functional studies with p.Ser152Phe variant
Mouse experiments were performed according to protocols approved by University of Tours and INSERM (Project 01456.03, French Ministry of Research). Hippocampal neurons from embryonic day 17 embryos were prepared and plated at a density of 450 cells/mm2 as described.21 Mature primary hippocampal mouse neurons (13 days in vitro) were transfected with pEGFP (vector only), Aα WT or Aα p.Ser152Phe-GFP fusion expression vector (three independent transfections). Forty-eight hours later, confocal microscopy (Leica SP8) images of fixed neurons were analyzed, and the number of dendritic spines in 10-µm sections determined (pEGFP: 134 sections, 19 cells; Aα-GFP: 190 sections, 18 cells; Aα Ser152Phe-GFP: 231 sections, 18 cells).
Statistics
Statistical analysis of biochemical data was assessed with one-sample Student t-tests using GraphPad Prism 8.4.2 software—data were compared with WT values that were set at 100% in each experimental replicate. At least three replicates were analyzed for each measurement; p values below 0.05 were considered significant (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001). Statistical comparative analysis of the counted number of spines was performed with GraphPad Prism 6 software and a Kruskal–Wallis test followed by Dunn multiple comparisons test.
RESULTS
Mutational spectrum is diverse and suggests impaired PPP2R1A subunit functioning
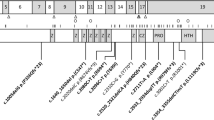
In total, 16 different de novo variants (of which 6 were recurrent) were identified in 30 individuals, in most cases through routine clinical diagnostic procedures, including exome sequencing. Individuals 5, 7, 10, 17, 19, and 25 were identified through the Deciphering Developmental Disorders (DDD) Study.3 Individual 23 was identified via a research study that provided trio exome sequencing. All variants resulted in nonidentical, single amino acid substitutions (Fig. 1a). Four variants, p.Pro179Leu (1 case), p.Arg182Trp (3 cases), p.Ser219Leu (3 cases), and p.Arg258His (2 cases) had previously been reported;4,16 hence, 12 variants were novel.
(a) De novo pathogenic variants in PPP2R1A identified in individuals with intellectual disability and (neuro)developmental delay are shown with respect to their exonic and Aα protein localization respectively, with exon notation by Roman numbering (I–XV) and indications of the 15 HEAT repeats (HRs) within the protein structure. (b) Detailed amino acid sequence of the affected HRs (HR1, HR4, HR5, HR6, and HR7), with the intrarepeat loop indicated in purple, and the affected amino acids underlined and in bold. (c) More schematic representation of the antiparallel helical structures of the HRs with the B subunit binding intrarepeat loops. The red crosses indicate the approximate locations of the mutated amino acids found in the affected individuals.
Structurally, the Aα subunit consists of 15 HEAT repeats (HRs; Huntingtin - Elongation factor - A subunit - TOR), organized into a horseshoe-shaped molecule.22 Each HR is constituted of two antiparallel helices that are connected by an intrahelical loop (Fig. 1b, c). These intrahelical loops form a continuous ridge at one side of the molecule that provides the direct binding contacts with other PP2A subunits: in particular, the loops from HR1 to HR7 provide the contacts with different B-type subunits (Fig. 1a, c),23,24,25,26,27 while the loops from HR 11 to HR15 bind to the catalytic C subunit (Fig. 1a).28 Interestingly, seven of the novel variants, p.Thr178Asn, p.Thr178Ser, p.Met180Val, p.Met180Thr, p.Met180Lys, p.Met180Arg, and p.Arg183Trp, reside immediately adjacent to or affect the same amino acid as the previously reported Aα HR5 loop variants (p.Pro179Leu,4 p.Arg182Trp,4 and p.Arg183Gln15) (Fig. 1a–c). The novel variant p.Val220Met resides immediately adjacent to the reported p.Ser219Leu16 in the loop of HR6, and another novel variant, p.Arg258Ser, affects the same amino acid as the previously reported one (p.Arg258His4) in the loop of HR7. The novel p.Phe141Ile variant resides in the intrarepeat loop of HR4 (Fig. 1a–c), a loop not previously reported to be affected. Thus, the large majority of Aα variants affect amino acids predicted to be involved in B-type subunit binding and reside in the intrarepeat loops of HR4, HR5, HR6, and HR7. In contrast, the novel p.Ile32Met and p.Ser152Phe variants reside in one of both helical structures of HR1 or HR4, suggesting they might not be directly involved in B-type subunit binding (Fig. 1a–c).
Clinical spectrum associated with PPP2R1A variants
The core clinical information on all 30 PPP2R1A-affected patients is summarized in Table 1; more details can be found in Table S2. Facial features of 12 individuals are displayed in Fig. S1. Individuals were aged 10 months to 27 years, and all except one had developmental delay of a severity that was certainly or likely within the ID range. The one exceptional individual, with a de novo p.Ser152Phe variant (individual 3, Table 1), had an autism spectrum disorder (ASD) and a full scale IQ of 86. In the others with ID, most were moderate to severe, but two could have mild ID only. The presence of epilepsy or brain malformations (corpus callosum hypoplasia, ventriculomegaly, and/or periventricular leukomalacia) was found in individuals with moderate to severe ID. Table 2 provides an overview of the main phenotypic features and their prevalence. A subgroup of 11 individuals had macrocephaly; the rest either normocephaly or microcephaly (n = 7) (Table 2). Uniform features were neurodevelopmental delay with prolonged hypotonia and language delay. The subgroup with macrocephaly did not have epilepsy, and all cases with ptosis (n = 5) and hearing loss or microtia (n = 3) belonged to this group (Table 2). The facial features were variable, from fairly normal to clearly dysmorphic (Fig. S1). Frontal bossing or a tall forehead was typical for the macrocephalic group (individuals 2, 9, 13, 14, Fig. S1), and these features could also be reminiscent of Sotos syndrome (individual 2) or Weaver syndrome (individual 14), but body lengths were usually normal (Table S2). Of note, digital malpositioning was seen in the severely p.Arg182Trp affected cases (Fig. S1). Affection of brain function and morphology is the main clinical problem caused by PPP2R1A dysfunction, but as a clue to diagnosis, mild facial dysmorphism (e.g., frontal bossing, tall forehead, downslanting or upslanting palpebral fissures) can be seen in some (Table 1, Fig. S1). As mentioned, head circumference was either normal (38%), macrocephalic (38%) or microcephalic (24%), representing an unusual distribution across the full range. Head size did not show a clear correlation with the degree of ID or the presence of epilepsy, but the macrocephalic individuals mostly had moderate ID (8/11). Malformations are not common in this cohort, and the persistent ductus arteriosus in 3/30 individuals could be a random association—or a mild susceptibility. It is also noteworthy that no individuals, except one, had been diagnosed with a malignancy: individual 22 was operated for a paratesticular lipoblastoma at age 1 year. Tumor nodules were composed of mature adipocytes and vacuolated lipoblasts showing varying degrees of differentiation.
To correlate clinical with biochemical findings, a crude clinical severity score was calculated based on degree of developmental delay (1–4 points), epilepsy (+1), macro/microcephaly (+1), malformations (+1), corpus callosum hypoplasia (+1), short stature (+1), and other major findings (+1) for a maximum score of 10 (Tables 1, S3). There was a correlation between variant type and severity, e.g., individuals with p.Met180Thr and p.Met180Val usually had moderate ID and all individuals with p.Arg182Trp had severe ID, with some degree of variation for a given variant (Table 1).
PPP2R1A variants show altered PP2A B-type subunit binding in a variant-specific manner
We assessed binding of 13 Aα variants (p.Phe141Leu, p.Ser152Phe, p.Thr178Asn, p.Pro179Leu, p.Met180Thr, p.Met180Val, p.Met180Lys, p.Arg182Trp, p.Arg183Trp, p.Ser219Leu, p.Val220Met, p.Arg258His, and p.Arg258Ser) to at least one representative of the regulatory B/B55, B’/B56, B”/PR72 and B”’/striatin subunit classes. To this end, N-terminally GFP-tagged B55α (encoded by PPP2R2A), B56α to ε (encoded by PPP2R5A-R5E), B”/PR72 (encoded by PPP2R3A), and B”’/STRN3 (encoded by PPP2R6B) (Figs. 2, S2–S9) were coexpressed with HA-tagged WT or mutant Aα. The retrieved Aα proteins in GFP traps were subsequently visualized by anti-HA immunoblotting. As previously reported,4 we confirmed near complete loss of binding of the B55α subunit to the p.Pro179Leu, p.Arg182Trp, and p.Arg258His variants (Figs. 2a, S2), while the binding behavior of not previously biochemically tested variants appeared in part identical (for variants p.Arg183Trp, p.Ser219Leu, p.Val220Met, and p.Arg258Ser), and in part completely opposite to this observation. Binding of B55α was indeed fully retained in p.Phe141Ile, p.Ser152Phe, p.Thr178Asn, p.Met180Thr, and p.Met180Val, and was 50% decreased in p.Met180Lys (Figs. 2a, S2). This binding behavior was consistent with results from PP2A activity assays using a PP2A-B55 holoenzyme-specific substrate, derived from the pRb protein.29 In these assays, significant PP2A activity was only found in the variants that still significantly bound B55α (Fig. 2b).
(a, c–i). GFP-tagged B55α (a), B”’/STRN3 (c), B56δ (d), B56α (e), B56β (f), B56γ1 (g), B56ε (h), B”/PR72 (i) or GFP alone (as a control) was coexpressed with HA-tagged wild-type (WT) or variant Aα proteins in HEK293T cells. The presence of Aα variants was subsequently assessed in GFP traps by anti-HA immunoblotting. Representative blots can be found in Figs. S2–S9. Shown is the average value +/- SD of the ratios of the quantified anti-HA signal versus the quantified anti-GFP signal for a given Aα variant, relative to those of WT Aα (set at 100% in each experiment, dotted line), as determined in at least three independent binding experiments (n ≥ 3). A one-sample t-test (compared with 100%) was used to assess statistical significance (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). (b) PP2A activity measurements on a PP2A-B55-specific phosphopeptide. HA-tagged WT and variant Aα proteins were purified from transfected HEK293T cells by HA pulldown, and absolute PP2A activity was determined on the I-N-G-S-P-R-(p)T-P-R-R-G-Q-N-R phosphopeptide substrate using Biomol®Green. Specific PP2A activities were calculated by correcting the measured activities for actual Aα inputs, determined by anti-HA immunoblotting. Results represent the average specific activity +/- SD for a given Aα variant, relative to the specific activity of WT Aα (set at 100% in each experiment), as determined in at least three independent experiments (n ≥ 3). A one-sample t-test (compared with 100%) was used to assess statistical significance (*p < 0.05; **p < 0.01; ****p < 0.0001).
We next performed binding assays with STRN3, a representative member of the PP2A B”’ subunit family. Interestingly, no significant differences in binding compared with WT Aα were observed with Aα variants exhibiting retained B55α binding (i.e., p.Phe141Ile, p.Ser152Phe, p.Thr178Asn, p.Met180Thr, p.Met180Val), while a significant (or trend toward) gain of binding of STRN3 was observed for the variants with lost or decreased B55α binding (p.Met180Lys, p.Arg182Trp, p.Arg183Trp, p.Ser219Leu, p.Val220Met, p.Arg258His, and p.Arg258Ser), with p.Pro179Leu being the only exception (Figs. 2c, S3).
Within the B56 family of B subunits, isoform-specific binding dysfunctions were observed for different Aα variants. Consistent with previous observations,4 binding of B56δ was retained in all variants (Figs. 2d, S4). For B56α, loss of or significantly decreased binding was seen to Aα variants p.Phe141Ile, p.Thr178Asn, p.Pro179Leu, p.Met180Lys/Thr/Val, p.Arg182Trp, and p.Arg183Trp, while binding was retained in all other Aα variants tested (Figs. 2e, S5). For B56β, the same pattern of binding deficiencies was observed, in addition to decreased binding to p.Ser219Leu and p.Val220Met (Figs. 2f, S6). B56γ1 and B56ε showed a near identical pattern of binding deficiencies, with significantly impaired or lost binding to all Aα variants, except p.Ser152Phe, p.Val220Met, Arg258His and p.Arg258Ser (Figs. 2g, h, S7–S8).
For the B”/PR72 subunit, another unique binding deficiency pattern was found, with lost or significantly decreased binding to Aα variants p.Phe141Ile, p.Met180Thr/Val, p.Arg182Trp, p.Arg183Trp, and p.Arg258His/Ser (Figs. 2i, S9).
Some PPP2R1A variants show altered C subunit binding and impaired overall PP2A activity
We next assessed binding of HA-tagged Aα variants to endogenous PP2A C subunit (Fig. 3a, b). Whereas, compared with WT Aα, no obvious impairment of C subunit binding could be observed for Aα variants p.Ser152Phe, p.Thr178Asn, and p.Met180Lys/Thr/Val, C subunit binding was significantly affected by about 50% or more for p.Phe141Ile, p.Pro179Leu, p.Arg182Trp, p.Arg183Trp, p.Ser219Leu, p.Val220Met, and p.Arg258His/Ser (Fig. 3a, b). PP2A activity measurements using a nonspecific artificial phosphopeptide (i.e., one that is equally well dephosphorylated by all PP2A holoenzymes), showed significantly decreased activities by 48–70% for p.Thr178Asn, p.Pro179Leu, p.Arg182Trp, and p.Arg258His/Ser variants (Fig. 3c).
(a) C subunit binding. HA-tagged wild-type (WT) Aα and Aα variants were purified from transfected HEK293T cells by HA pulldown, and the presence of endogenous C subunit in the complexes determined by anti-A immunoblotting. (b) Quantified C subunit binding values. Results represent the average value +/- SD of the ratios of the quantified anti-C signal versus the quantified anti-HA signal for a given Aα variant, relative to those of WT Aα (set at 100% in each experiment), as determined in at least three independent experiments (n ≥ 3). A one-sample t-test (compare to 100%) was used to assess statistical significance (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). (c) Associated phosphatase activity of Aα variants, measured on a nonspecific PP2A substrate. HA-tagged WT Aα and Aα variants were purified from transfected HEK293T cells by HA pulldown, and PP2A activity was determined on the artificial, holoenzyme nonspecific K-R-(p)T-I-R-R phosphopeptide substrate using Biomol®Green. Specific PP2A activities were calculated by correcting the measured activities for actual Aα inputs, determined by anti-HA immunoblotting. Results represent the average specific activity +/- SD for a given Aα variant, relative to the specific activity of WT Aα (set at 100% in each experiment), as determined in at least three independent experiments (n ≥ 3). A one-sample t-test (compared with 100%) was used to assess statistical significance (*p < 0.05; **p < 0.01; ***p < 0.001). (d, e) Dendritic spine analysis in hippocampal neurons expressing Aα variant p.Ser152Phe. Confocal images of fixed primary hippocampal neurons (DIV 13) 48 hours after transfection with pEGFP (negative control), WT Aα–GFP or Aα Ser152Phe-GFP (d). Quantification of the number of dendritic spines for the transiently expressed GFP, WT Aα, and Aα variant p.Ser152Phe (e). For each neuron analyzed, the number of spines in 10-μm sections was counted: pEGFP (3 transfections; 19 cells, 134 sections), Aα–GFP (3 transfections; 18 cells, 190 sections), Aα Ser152Phe-GFP (3 transfections; 18 cells, 231 sections). Each plot represents the mean value of sections analyzed in one neuron. Statistical comparative analysis was performed using a nonparametric Kruskal–Wallis test followed by Dunn multiple comparisons tests. ****p < 0.0001, ns not significant.
Decreased number of dendritic spines in hippocampal neurons expressing p.Ser152Phe
So far, only the p.Ser152Phe variant did not show any functional impairments in our biochemical assays. To provide further insights into its potential pathogenicity, we expressed it as a GFP fusion protein in mature primary hippocampal neurons. Compared with GFP or WT Aα-GFP expressing neurons, a significantly decreased number of dendritic spines was found in these neurons (Fig. 3d, e), suggesting p.Ser152Phe may indeed be pathogenic. Of note, the PP2A Aα subunit is part of the postsynaptic density,30 and PP2A function could have importance for regulating dendritic spine number during neuronal development.
DISCUSSION
Although incompletely understood, PP2A phosphatases play important roles in mammalian brain development and function,8,31,32 e.g., in regulating signaling33,34,35 and preventing degeneration in neurons.36,37 In 2015, PP2A dysfunction was reported as a new cause of syndromic intellectual disability and (neuro)developmental delay, involving de novo loss-of-function pathogenic variants in PPP2R1A and PPP2R5D,4 and likely also in PPP2R5C and PPP2R5B, although the latter has not yet been functionally confirmed.5 Later, the set of affected genes was extended to PPP2CA,6 and the overlapping syndromes, all characterized by severe PP2A dysfunction, were proposed to be called “PP2A-related neurodevelopmental disorders.” Here, we significantly expand the spectrum of PPP2R1A variants and phenotypes through analysis of 30 additional patients. While all previously reported PPP2R1A-affected cases represented individuals with severe ID, often associated with epilepsy and microcephaly, more diversity is seen in the current cohort, with several cases being phenotypically milder, and at least one without ID but with only learning problems and autistic features. Interestingly, this wider phenotypic spectrum correlates well with the more diverse biochemical dysfunctions found in this broader cohort, with the most pronounced difference seen in the binding behavior of Aα variants to the B-type subunits, B55α and STRN3. Indeed, patients with less severe ID, no seizures, and sometimes macrocephaly typically showed normal B55α and STRN3 binding, while patients with more severe ID, seizures, and sometimes microcephaly typically lost B55α binding and showed increased STRN3 binding. Notably, these functionally more disruptive variants were also found in the Catalogue Of Somatic Mutations In Cancer (COSMIC), or were affected in the same amino acid as cancer-associated PPP2R1A variants (Table S3). Reported functional studies of these cancer-associated Aα variants confirmed their increased oncogenic potential in cancer cell growth, migration, and drug resistance.17,38,39,40 Only for p.Pro179Leu was the inverse correlation between loss of B55α binding and gain of STRN3 binding not found, which could be explained by the broader structural disruptive nature of this variant, as shown before for the cancer-associated p.Pro179Arg variant.39 It is noteworthy that one individual (22, p.Ser219Leu, present in COSMIC) was affected at young age by a relatively rare tumor (paratesticular lipoblastoma). However, since this is the first PPP2R1A case reported with a malignancy, it is unclear whether this is mainly coincidental or part of the clinical phenotype of the more severely affected PPP2R1A subgroup.
With respect to B56 isoform binding, all new ID-associated Aα variants, without exception, retained binding to B56δ, as reported previously4 (Table S3). Binding to all other B56 isoforms was significantly decreased or completely lost for Aα variants affected in the intrarepeat loops of HR4 and HR5, while Aα variants affected in the loop of HR7 fully retained binding. Aα variants affected in the loop of HR6 showed a more variable B56 isoform-specific pattern of binding deficiencies. In contrast, B”/PR72 binding was significantly decreased or lost to Aα variants affected in the intrarepeat loops of HR4 and HR7, while it was completely retained with Aα variants affected in the loop of HR6. A more variable B”/PR72 binding deficiency pattern was found with Aα variants affected in the loop of HR5. Thus, the binding requirements of different B-type subunit families to each of the Aα HEAT repeats 4–7 appeared in part overlapping, in part clearly different (Fig. S10).
Structural data from different PP2A trimers, each harboring a single representative of the four PP2A B-type subunit classes, further underscore the presumed functional importance of most affected Aα residues in B-type subunit binding. Indeed, structural data from crystallized Aα-B56γ1-C trimers highlighted the importance of Aα Trp140 and Phe141 residues in making multiple van der Waals interactions with hydrophobic residues in B56γ1,23,24 while Arg183 donates a pair of charge-stabilized hydrogen bonds to Glu214 of B56γ1.23 Other Aα residues involved in direct contacts with B56γ1 are Asp177, Thr178, Pro179, and Met180.24 A smaller contact area between Aα and B56γ1 involved the ridge of Aα HR7, where Trp257 hydrogen bonds to a main chain carbonyl group of B56γ1.23 Six Aα residues make direct contacts with the B55α subunit: Trp140, Phe141, Thr178, Arg183, Asp218, and Trp257,25 while Trp140, Met180, Arg183, and Arg258 are part of an extensive binding surface between Aα and the B”/PR70 subunit.26 In contrast, truncation of HR1–HR3 of Aα sufficed to abolish interaction with the coiled coil domain of the B”’/striatins, suggesting that Aα-striatin contacts are largely mediated by HR1 to HR3.27 However, despite the availability of these structural data, the pattern of specific B-type subunit-binding defects to the various Aα variants, experimentally determined here, appeared largely, if not completely, unpredictable. For instance, despite the fact that Phe141, Thr178, and Arg183 were reported to make direct contacts with B55α, the p.Phe141Ile and p.Thr178Asn variants clearly fully retained B55α binding, while p.Arg183Trp completely lost B55α binding. Likewise, while Met180 was reported to be part of the binding surface with B”/PR70, only Met180Thr/Val substitutions significantly affected B”/PR72 binding, while p.Met180Lys bound normally. This testifies to complex regulation and precise fine-tuning of amino acid contacts between Aα and the diverse B-type subunits.
In accordance with previous studies,4,17 eight of the tested ID-associated Aα variants bound significantly less to the PP2A catalytic subunit, despite the C subunit-binding domain residing in HR10-HR15.28 However, this decrease in C binding did not seem to correlate well with decreased Aα-associated PP2A activity, which was only observed for three of these affected variants (p.Pro179Leu and p.Arg258His/Ser). Thus, in the latter cases, a dominant-negative mechanism could be in place, as these Aα variants still form trimeric complexes with respectively, B56δ and PR72 (for p.Pro179Leu), and all B56 isoforms (for p.Arg258His/Ser), but show impaired phosphatase activity. Conversely, p.Thr178Asn showed decreased associated PP2A activity, while having normal C subunit binding, perhaps hinting at a different substrate specificity for this variant.
Finally, of all PPP2R1A variants functionally examined here, only p.Ser152Phe appeared biochemically normal in our assays (Table S3). This is both in concordance with the mild phenotype of the affected individual (individual 3), as well as with the localization of Ser152 outside of the intrahelical loop, but within one of the helices, of HR4 (Fig. 1). Nevertheless, neurons expressing this variant presented with fewer dendritic spines, testifying of an altered functionality, compared with WT Aα (Fig. 3d, e). These findings thus expand the PPP2R1A phenotype to include ASD without ID.
In summary, the extended number of de novo PPP2R1A variants reported here revealed a wider phenotypic spectrum than previously anticipated, which correlated well with the more diverse biochemical dysfunctions found. Apart from p.Ser152Phe, all functionally tested variants were impaired in binding to a specific set of B-type subunits, except for binding to B56δ (which was normal in all cases), and for binding to STRN3 (which was increased in some cases). Combined with significantly decreased PP2A activity found in a number of variants, these binding deficiencies result in a complicated pattern of PP2A dysfunctions, which may affect specific trimeric holoenzymes both by dominant-negative as well as haploinsufficiency mechanisms. One subgroup of variants (p.Phe141Ile, p.Thr178Asn, p.Met180Val/Thr/Lys; absent from COSMIC) presented overall with a milder biochemical and clinical phenotype, while another subgroup (p.Pro179Arg, p.Arg182Trp, p.Arg183Trp, p.Ser291Leu, p.Val220Met, p.Arg258His/Ser; all with affected amino acids present in COSMIC) was overall more severely affected, both biochemically and phenotypically. These findings have major consequences for diagnosis, clinical management, and follow-up of PPP2R1A-affected patients, and provide additional rationales for functional follow-up studies in cell and mouse models.
References
Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347.
de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929.
Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228.
Houge G, Haesen D, Vissers LE, et al. B56δ-related protein phosphatase 2A dysfunction identified in patients with intellectual disability. J Clin Invest. 2015;125:3051–3062.
Loveday C, Tatton-Brown K, Clarke M, et al. Mutations in the PP2A regulatory subunit B family genes PPP2R5B, PPP2R5C and PPP2R5D cause human overgrowth. Hum Mol Genet. 2015;24:4775–4779.
Reynhout S, Jansen S, Haesen D, et al. De novo mutations affecting the catalytic Cα subunit of PP2A, PPP2CA, cause syndromic intellectual disability resembling other PP2A-related neurodevelopmental disorders. Am J Hum Genet. 2019;104:139–156.
Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439.
Reynhout S, Janssens V. Physiologic functions of PP2A: Lessons from genetically modified mice. Biochim Biophys Acta Mol Cell Res. 2019;1866:31–50.
Haesen D, Sents W, Lemaire K, Hoorne Y, Janssens V. The basic biology of PP2A in hematologic cells and malignancies. Front Oncol. 2014;4:347.
Hertz EPT, Kruse T, Davey NE, et al. A conserved motif provides binding specificity to the PP2A-B56 phosphatase. Mol Cell. 2016;63:686–695.
Cundell MJ, Hutter LH, Nunes Bastos R, et al. A PP2A-B55 recognition signal controls substrate dephosphorylation kinetics during mitotic exit. J Cell Biol. 2016;214:539–554.
Shang L, Henderson LB, Cho MT, et al. De novo missense variants in PPP2R5D are associated with intellectual disability, macrocephaly, hypotonia, and autism. Neurogenetics. 2016;17:43–49.
Yeung KS, Tso WWY, Ip JJK, et al. Identification of mutations in the PI3K-AKT-mTOR signalling pathway in patients with macrocephaly and developmental delay and/or autism. Mol Autism. 2017;8:66.
Saraf A, Oberg EA, Strack S. Molecular determinants for PP2A substrate specificity: charged residues mediate dephosphorylation of tyrosine hydroxylase by the PP2A/B’ regulatory subunit. Biochemistry. 2010;49:986–995.
Wallace A, Caruso P, Karaa A. A newborn with severe ventriculomegaly: expanding the PPP2R1A gene mutation phenotype. J Pediatr Genet. 2019;8:240–243.
Zhang Y, Li H, Wang H, Jia Z, Xi H, Mao X. A de novo variant identified in the PPP2R1A gene in an infant induces neurodevelopmental abnormalities. Neurosci Bull. 2020;36:179–182.
Haesen D, Abbasi Asbagh L, Derua R, et al. Recurrent PPP2R1A mutations in uterine cancer act through a dominant-negative mechanism to promote malignant cell growth. Cancer Res. 2016;76:5719–5731.
Esmaeeli-Nieh S, Fenckova M, Porter IM, et al. BOD1 is required for cognitive function in humans and Drosophila. PLoS Genet. 2016;12:e1006022.
Backx L, Vermeesch J, Pijkels E, de Ravel T, Seuntjens E, Van Esch H. PPP2R2C, a gene disrupted in autosomal dominant intellectual disability. Eur J Med Genet. 2010;53:239–243.
Janssens V, Jordens J, Stevens I, et al. Identification and functional analysis of two Ca2+-binding EF-hand motifs in the B”/PR72 subunit of protein phosphatase 2A. J Biol Chem. 2013;278:10697–10706.
Ung DC, Iacono G, Méziane H, et al. Ptchd1 deficiency induces excitatory synaptic and cognitive dysfunctions in mouse. Mol Psychiatry. 2018;23:1356–1367.
Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96:99–110.
Xu Y, Xing Y, Chen Y, et al. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251.
Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57.
Xu Y, Chen Y, Zhang P, Jeffrey PD, Shi Y. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol Cell. 2008;31:873–885.
Wlodarchak N, Guo F, Satyshur KA, et al. Structure of the Ca2+-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation. Cell Res. 2013;23:931–946.
Chen C, Shi Z, Zhang W, et al. Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2:2 heterotetrameric core of striatin-interacting phosphatase and kinase (STRIPAK) complex. J Biol Chem. 2014;289:9651–9661.
Xing Y, Xu Y, Chen Y, et al. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell. 2006;127:341–353.
Agostinis P, Derua R, Sarno S, Goris J, Merlevede W. Specificity of the polycation-stimulated (type-2A) and ATP,Mg-dependent (type-1) protein phosphatases toward substrates phosphorylated by P34cdc2 kinase. Eur J Biochem. 1992;205:241–248.
Laumonnier F, Cuthbert PC, Grant SG. The role of neuronal complexes in human X-linked brain diseases. Am J Hum Genet. 2007;80:205–220.
Liu B, Sun LH, Huang YF, Guo LJ, Luo LS. Protein phosphatase 2ACalpha gene knock-out results in cortical atrophy through activating hippo cascade in neuronal progenitor cells. Int J Biochem Cell Biol. 2018;95:53–62.
Panicker N, Coutman M, Lawlor-O’Neill C, Kahl RGS, Roselli S, Verrills NM. Ppp2r2a knockout mice reveal that protein phosphatase 2A regulatory subunit, PP2A-B55α, is an essential regulator of neuronal and epidermal embryonic development. Front Cell Dev Biol. 2020;8:358.
Kapfhamer D, Berger KH, Hopf FW, et al. Protein phosphatase 2a and glycogen synthase kinase 3 signaling modulate prepulse inhibition of the acoustic startle response by altering cortical M-type potassium channel activity. J Neurosci. 2010;30:8830–8840.
Louis JV, Martens E, Borghgraef P, et al. Mice lacking phosphatase PP2A subunit PR61/B′delta (Ppp2r5d) develop spatially restricted tauopathy by deregulation of CDK5 and GSK3beta. Proc Natl Acad Sci USA. 2011;108:6957–6962.
Leslie SN, Nairn AC. cAMP regulation of protein phosphatases PP1 and PP2A in brain. Biochim Biophys Acta Mol Cell Res. 2019;1866:64–73.
Taleski G, Sontag E. Protein phosphatase 2A and tau: an orchestrated ‘pas de deux’. FEBS Lett. 2018;592:1079–1095.
Taymans JM, Baekelandt V. Phosphatases of α-synuclein, LRRK2, and tau: important players in the phosphorylation-dependent pathology of Parkinsonism. Front Genet. 2014;5:382.
Jeong AL, Han S, Lee S, et al. Patient derived mutation W257G of PPP2R1A enhances cancer cell migration through SRC-JNK-c-Jun pathway. Sci Rep. 2016;6:27391.
Taylor SE, O’Connor CM, Wang Z, et al. The highly recurrent PP2A Aα subunit mutation P179R alters protein structure and impairs PP2A enzyme function to promote endometrial tumorigenesis. Cancer Res. 2019;79:4242–4257.
O’Connor CM, Leonard D, Wiredja D, et al. Inactivation of PP2A by a recurrent mutation drives resistance to MEK inhibitors. Oncogene. 2020;39:703–717.
Acknowledgements
The authors thank the individuals and their parents for participating in the study. V.J. was funded by Jordan’s Guardian Angels Foundation; F.L. by Association pour le Développement de la Neurogénétique (INSERM) and French Ministry of Health (NCT01770548); A.R. by Swiss National Science Foundation. S.R. received an FWO-SB fellowship (Research Foundation-Flanders). Patient 23 was ascertained in the Duke Genome Sequencing Clinic as participant in a research study (Duke Protocol 00032301). Sequencing for this patient was performed at the Institute for Genomic Medicine, Columbia University (New York, NY, USA). Funding for the Duke Genome Sequencing Clinic is supported by Duke University Health System. The DDD Study presents independent research commissioned by the Health Innovation Challenge Fund (grant HICF-1009-003). This study makes use of DECIPHER (http://decipher.sanger.ac.uk/), which is funded by Wellcome. A. McTague is supported by National Institute for Health Research (NIHR) Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the author(s) and not necessarily those of the NHS, NIHR, or the UK Department of Health.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. If you remix, transform, or build upon this article or a part thereof, you must distribute your contributions under the same license as the original. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/.
About this article
Cite this article
Lenaerts, L., Reynhout, S., Verbinnen, I. et al. The broad phenotypic spectrum of PPP2R1A-related neurodevelopmental disorders correlates with the degree of biochemical dysfunction. Genet Med 23, 352–362 (2021). https://doi.org/10.1038/s41436-020-00981-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-00981-2
Key words
This article is cited by
-
A pigmentary manifestation associated with PPP2R5D-related neurodevelopmental disorder: a case report and review of literature
Bulletin of the National Research Centre (2023)
-
Genomic regulation of transcription and RNA processing by the multitasking Integrator complex
Nature Reviews Molecular Cell Biology (2023)