Abstract
Purpose
Genomic testing is routinely utilized across clinical settings and can have significant variant interpretation challenges. The extent of genetic counselor (GC) engagement in variant interpretation in clinical practice is unknown. This study aimed to explore clinical GCs’ variant interpretation practice across specialties, understand outcomes of this practice, and identify resource and educational needs.
Methods
An online survey was administered to National Society of Genetic Counselors members providing clinical counseling.
Results
Respondents (n = 239) represented all major clinical specialties. The majority (68%) reported reviewing evidence documented by the laboratory for most (>60%) variants reported; 45.5% report seeking additional evidence. Prenatal GCs were less likely to independently assess reported evidence. Most respondents (67%) report having reached a different conclusion about a variant’s classification than the testing laboratory, though infrequently. Time was the most commonly reported barrier (72%) to performing variant interpretation, though the majority (97%) indicated that this practice had an important impact on patient care. When presented with three hypothetical scenarios, evidence typically used for variant interpretation was generally applied correctly.
Conclusion
This study is the first to document variant interpretation practice broadly across clinical GC specialties. Our results suggest that variant interpretation should be considered a practice-based competency for GCs.
Similar content being viewed by others
INTRODUCTION
Broad genomic testing options, such as next-generation sequencing (genome/exome and multigene panels) and chromosomal microarray, have been incorporated into clinical care across many specialties, including prenatal, pediatric, hereditary cancer, cardiovascular, general genetics, and other subspecialties.1,2,3,4,5 This approach has improved the diagnostic process by reducing time to diagnosis, eliminating a tiered testing approach, and avoiding phenotype-based ascertainment bias.1,2,3,4,5 However, these tests also identify a significant number of genomic variants that are not easily interpreted. This challenge has resulted in the development of new variant interpretation guidelines, databases, and other resources.6,7,8 Traditionally, the practice of variant interpretation has been the purview of clinical genetics laboratories; however, the laboratory genetic counselor role has grown steadily over the past two decades.9,10,11 While laboratory genetic counselor roles can vary by setting, formal variant interpretation and variant data curation skills have been described, and it has been suggested that these skills are applicable and transferrable to patient-facing clinical practice.9,10,12
The extent to which genetic counselors in patient-facing clinical roles, subsequently referred to as clinical genetic counselors, are incorporating variant interpretation activities into clinical practice is not known, though an understanding of variant interpretation has been acknowledged as relevant in clinical settings.12,13,14,15 Thus far, studies exploring variant interpretation within clinical genetic counselors’ scope of practice have focused on a single clinical specialty or setting and have differed in terms of study objectives and outcomes.14,16,17,18 Reuter et al. surveyed clinical cardiovascular genetic counselors (n = 46) and found that 96% were evaluating information relevant to variant interpretation beyond what was provided in the laboratory report and 81% were assigning a classification term, such as “pathogenic,” to reflect their own assessment. This was done in a team setting with other health-care professionals, though the genetic counselor typically led this process. A survey of clinical hereditary cancer genetic counselors (n = 224) identified similar findings with 96% of respondents conducting data searches to inform or confirm a variant interpretation.18 Two studies have described variant interpretation activities in pediatric clinical settings, but broad, multi-institution assessments of this practice have not been done within pediatric or prenatal settings.13,17 Establishing the extent to which clinical genetic counselors are involved in variant interpretation activities is necessary to define the practice and ensure it is recognized in the genetic counselor's scope of practice. This is also needed to inform graduate training, board certification examinations, and postcertification educational curricula.
Documentation of the outcomes of clinical genetic counselor variant interpretation activities is also limited. The data available suggest this practice impacts medical management, decisions to test at-risk relatives, and approaches for discussing test results with a patient or family.12,14,16,18 Variant interpretation discrepancies have been encountered by clinical genetic counselors as observations of interpretation discrepancies between laboratories and as discrepancies between the laboratory and the clinical team.13,14,16,17,18 In the hereditary cancer setting, 93% of respondents reported having identified a variant interpretation discrepancy between laboratories, typically via public databases like ClinVar; however, this study did not assess the outcome of the genetic counselor’s own variant interpretation process.18,19 Variant interpretation discrepancies between the genetic counselor–led clinical team and the laboratory have been estimated to occur for 18–19% of reported variants in the cardiovascular genetics setting.14,16 These genetic counselor–laboratory variant interpretation discrepancies have been reported to impact patient management decisions, cascade testing decisions, and counseling.12,13,14,18
The perspectives of clinical providers, including medical geneticists, genetic counselors, and specialized care teams, are increasingly recognized as an important component of variant interpretation and large-scale variant curation efforts.13,17,20 Baldridge et al.13 described the medical geneticist’s role in providing clinical perspective, post-test evaluations, and subsequent literature searches in variant interpretation and reported that post-test clinical correlation resulted in diagnostic reclassification for 21 of 155 (14%) patients who had exome sequencing. The impact of variant interpretation by genetic counselors in a transdisciplinary pediatric setting was further explored by some members of our group, describing genetic counselor–led variant interpretation and a large-scale ClinVar submission of 303 clinically obtained genomic variants.17 Through this practice, genetic counselor–laboratory interpretation discrepancies were identified for 21% of copy-number variants (32 of 155).17 These discrepancies were typically related to older reports that utilized outdated data and classification terms; 31% (10 of 32) resulted in the genetic counselor downgrading the variant to an uncertain classification.17 Additionally, genomic variant data sharing among clinical specialized care centers has been successful in reducing variant interpretation discrepancies for cardiovascular disorders.20
Recognizing the important role that clinical providers have in variant interpretation, and the impact that these skills could have on clinical practice, the Clinical Genome Resource (ClinGen) Education, Coordination, and Training Working Group (https://clinicalgenome.org/working-groups/ect/) developed a Variant Interpretation Education Subgroup to address educational and training needs related to variant interpretation within ClinGen and the greater genomics communities. The aims of this study were to (1) explore variant interpretation practice by clinical genetic counselors across specialties, (2) understand the outcomes of this practice, and (3) assess educational and resource needs to support such activities.
MATERIALS AND METHODS
Human subjects and informed consent
This study was approved by the Geisinger Institutional Review Board (2016–0426). Written consent requirements were waived; consent was indicated by participation.
Participants
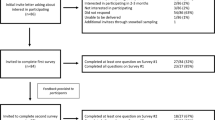
An invitation to participate in this study was emailed to National Society of Genetic Counselors (NSGC) members at the beginning of January 2018 and data were collected through that month. Participants were eligible if they provided part- or full-time clinical genetic counseling, either in-person or via telemedicine. Participants could opt to respond anonymously.
Data collection and analysis
A survey was developed and piloted by the ClinGen Variant Interpretation Education Subgroup members and was administered via SurveyMonkey (SurveyMonkey, San Mateo, CA). It comprised five sections (listed below); included yes/no questions, Likert-scale responses, and multiresponse items; and took approximately 15–20 minutes to complete (Supplemental Materials and Methods). Survey topics included:
Demographics and clinical practice information
Genetic counselor variant interpretation practice
Results of variant interpretation activities
Genomic resources used in variant interpretation
Application of variant interpretation knowledge via clinical scenarios
Survey data were analyzed using SPSS version 25.0 (IBM Corporation, Armonk, NY) and descriptive statistics are provided. Logistic regression was performed to identify significant correlations in frequency of variant interpretation practice (using collapsed categories) and rate of genetic counselor–laboratory interpretation discrepancies across clinical specialties, and to identify any potential influence of a previous or current laboratory/research role on this practice. Respondents reporting laboratory/research roles were asked if that role included variant interpretation. The significance of correlations with independent clinical specialties were verified through additional logistic regression analyses that excluded participants reporting more than one specialty. All analyses were two-sided, and results were considered significant if the p value was less than 0.05. Free text responses were reviewed (K.E.W.) and counted based on response category/type.
RESULTS
Demographics and clinical practice
There were 239 survey respondents. According to the 2016 NSGC Professional Status Survey (PSS), 69% of genetic counselors reported direct patient care;21 at the time of recruitment, there were 3616 NSGC members (NSGC direct communication). Thus, we estimate a 9.6% (239/2495) response rate of eligible NSGC members (2495 of 3616). Demographics are generally consistent with NSGC membership (Table S1), except for a higher proportion of participants in the 25–29 years age range (41% of respondents vs. 25% from PSS) and a lower rate of participants reporting prenatal practice (24% of respondents vs. 41% from PSS).21,22 All major clinical specialties were represented and 77 participants (32%) reported multiple specialties (Table S1). Common examples of other specialties included infertility (n = 6), neurogenetics (n = 3), metabolic (n = 3), and ophthalmology (n = 3).
Genetic counselor variant interpretation practice
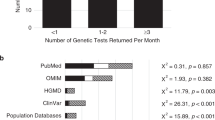
Across all clinical specialties, 68% (n = 153) of participants reported assessing the evidence documented by the clinical laboratory in the clinical report for most variants (≥60%). Forty-six percent of respondents (n = 101) reported searching for additional evidence, beyond what was documented in the report, for most variants (≥60%). As shown in Tables 1 and 2, genetic counselors reporting prenatal and other specialties were less likely to review the evidence in the laboratory report compared with other specialties (p = 0.03 and p = 0.035, respectively) but were not significantly different from other specialties with regard to searching for additional evidence beyond the report. No other significant differences were identified between specialties. Having a previous or current research or laboratory role did not correlate with increased review of evidence provided on a laboratory report or seeking additional evidence beyond the report (Tables 1 and 2). Years of experience also did not correlate with these activities. Respondents reported performing variant interpretation independently (n = 98, 43%) and in a variety of team settings, including taking a lead role and discussing with colleagues (n = 70, 31%), formal case conferences (n = 69, 31%), and researching information for other team members to assess (n = 66, 29%). A laboratory classification of variant of uncertain significance (VUS) was the factor most often noted to influence the decision to independently evaluate a variant (n = 167, 76% of respondents) (Fig. 1).
Lack of time was the most commonly reported barrier to performing variant interpretation (n = 156, 72%), followed by lack of familiarity with resources (n = 103, 48%) and lack of basic knowledge (n = 64, 30%). Lack of interest was noted by only 9% (n = 20). Lack of familiarity with resources, lack of basic knowledge, and lack of interest were found to significantly correlate with lower reported frequency of performing variant interpretation activities, but lack of time did not.
Free text responses were provided by 112 respondents to indicate resources that could support variant interpretation activities. Suggestions included additional training and resource guides (n = 41, 37%), improved laboratory reports (n = 13, 12%), institutional access to literature and/or licensed databases and tools (n = 13, 12%), and increased laboratory submissions to a single public database, such as ClinVar (n = 13, 12%). Requests for additional training and resources were often general, but specific examples included “webinar training refresher,” “better understanding on how to use various databases,” and “a short comprehensive guide book.” Comments about improved laboratory reports generally pertained to improved standardization, clarity, and transparency: “various labs provide different explanations and evidence” and “having all the primary data on the variant provided by the laboratory so I don’t have to look the data up.” The need for a single, public database was acknowledged as important for optimal curation and to simplify workflow: “having one centralized source to search the variant and get up-to-date information” and “consolidated variant database that is properly curated and updated.”
Results of variant interpretation activities
Ninety-seven percent of respondents (n = 213) indicated a perceived impact of genetic counselor–driven variant interpretation on clinical practice, including how they counsel/educate the patient (n = 189, 86%), how the patient is managed (n = 132, 60%), whether familial testing is offered (n = 121, 55%), improvement in the clinical team’s understanding of the result (n = 114, 52%), and provision of psychosocial support (n = 79, 36%). Some participants provided additional comments: counseling/educating the patient (“Adjust clinical counseling time,” “Allows me to provide more information for the patient,” “For my understanding of the variant so I am better able to communicate results to the patient”); clinical management (“The lab is often forced to call a variant ‘VUS’ by ACMG criteria while we (and often they) feel that it is causative of the patient’s phenotype”); and offering familial testing (“I try to look at VUS in light of the clinical symptoms and see if there is sufficient concern for us to ask for more clinical information or a family study”).
Although 67% of participants (n = 149) reported having a variant interpretation discrepancy with the laboratory at some point in their career, this was not a frequent occurrence. The majority indicated that discrepancies between their clinical variant interpretation and the laboratory’s were rare (<15% of variants; n = 113, 78% of respondents) or infrequent (15–29% of variants, n = 27, 19% of respondents). Genetic counselor–laboratory discrepancies were reported across all clinical specialties. As shown in Table 3, experiencing a genetic counselor–laboratory discrepancy at some point was reported by more genetic counselors in pediatric (n = 52, 84%, p = 0.002), cardiology (n = 22, 92%, p = 0.017), and other (n = 21, 88%, p = 0.037) settings and by fewer in prenatal (n = 21, 38%, p < 0.001) and cancer (n = 70, 61%, p = 0.05) settings. Having a previous or current research or laboratory role was not found to influence the frequency of genetic counselor–laboratory discrepancies.
The factors most often noted to influence a genetic counselor–laboratory discrepancy included identifying discrepant assertions between laboratories in ClinVar (n = 90, 62%), clinical correlation at the time of the report (n = 89, 61%), and identifying subsequent literature after the report date (n = 66, 46%). The most common type of interpretation discrepancy that prompted follow-up was an uncertain versus likely pathogenic or pathogenic classification (n = 97, 66%). Reported follow-up for a genetic counselor–laboratory discrepancy included discussion with the laboratory (n = 122, 82%), discussion with the medical team/colleagues (n = 124, 77%), discussion with the patient/family (n = 91, 56%), and/or requesting a new report (n = 23, 16%). Only 1% of respondents (n = 2) indicated no follow-up when a genetic counselor–laboratory discrepancy occurred.
Genomic resources used in variant interpretation
Respondents were asked to indicate their frequency of use and comfort with 16 resources relevant to variant interpretation that were identified by the authors (Fig. 2). Of all resources assessed, ClinVar was used most frequently (n = 150, 73% “use often”) and, correspondingly, respondents had the highest reported comfort using this resource (n = 114, 61% “very comfortable”).19 Population databases for allele frequency data (with the Exome Aggregation Consortium provided as an example) were the second most frequently used resource (n = 48, 24% “use often”), followed by published sequence variant interpretation guidelines (n = 41, 20% “use often”).7,23 Reported comfort using a resource correlated generally with frequency of use (Fig. 2), and no significant differences were identified based on clinical specialty, laboratory/research role, or years of experience. Several publicly available resources and databases, including ClinGen resources and DECIPHER, were not as familiar to participants and were used less frequently.6,24
Application of variant interpretation knowledge via clinical scenarios
Three brief scenarios were created to represent a variety of clinical settings, inheritance patterns, and variant interpretation challenges. These included (1) preconception counseling regarding carrier status for an autosomal recessive, neonatal-onset disorder; (2) an unaffected adult with a family history of a VUS identified in a relative with early-onset cancer; and (3) a de novo VUS identified by exome sequencing in a child with epilepsy and developmental delay (Supplemental Materials and Methods). For each scenario, respondents were provided with different examples of evidence representing population, computational, segregation, variant type, gene-level constraint, and functional data. Respondents were asked to assess if the evidence indicated that the variant in the scenario was benign or pathogenic, if it was neutral, or if the respondent didn’t know. Generally, variant interpretation evidence was applied correctly, particularly for allele frequency data from population databases, de novo/segregation data, and consideration of previously published cases. Self-reported uncertainty (Don’t Know responses) was greatest for gene-level data (e.g., constraint metrics) from population databases (n = 75, 38% uncertain, scenario 3) which can be used to infer whether genomic variation in a gene is tolerated or not, and thus, whether variants in a gene of uncertain significance might have a clinical impact.23 The implications of a specific variant type in a given disease context and considerations of potential mechanisms of disease were also associated with some reported uncertainty across scenarios: a missense variant (n = 28, 14% Don’t Know, scenario 1), a premature termination (nonsense variant) (n = 8, 4% Don’t Know, scenario 2), or a variant with an expected splicing impact (n = 32, 16% Don’t Know, scenario 3).
DISCUSSION
This study is the first to assess variant interpretation practice among clinical genetic counselors across specialties and to document this practice across pediatric and prenatal settings. With most genetic counselors in this study (68%) engaging in variant interpretation activities for most reported variants (>60%), our findings indicate that variant interpretation activities are prevalent across clinical settings. The reported rates of variant interpretation activities by cardiovascular and hereditary cancer genetic counselors in our study are notably lower than those reported previously (58% vs. 96% and 66% vs. 96%, respectively).14,18 This could be due to survey design and recruitment strategies given that the previous studies recruited participants through the NSGC Cardiovascular and Cancer Special Interest Groups, respectively, as opposed to the general NSGC membership.
Data across clinical specialties indicate that variant interpretation is a routine component of clinical genetic counseling practice throughout the profession and is not limited to particular subspecialties. In general, we did not identify significant associations between clinical specialty and variant interpretation practice, with the exception of genetic counselors reporting prenatal and other specialties who were less likely to review evidence provided in the laboratory report. However, interestingly, these genetic counselors were as likely to report reviewing evidence beyond what was provided in the laboratory report and did not differ in terms of perceived barriers to variant interpretation. It is possible that genomic testing and reporting approaches utilized in prenatal settings are sufficiently unique from those in other settings to cause this observed difference. Laboratories may use more conservative reporting criteria for prenatal cases, such as larger size thresholds for chromosomal microarray to reduce the reported VUS rate, and broad sequencing tests, such as exome sequencing, may not be as frequently utilized in the prenatal setting.25 Additionally, carrier screening test reports typically only include pathogenic and likely pathogenic variants.26 Prenatal laboratory reports may also provide more detailed evidence, reducing the need for independent assessment. There are likely fewer opportunities for clinical correlation for variants identified in a fetus, which may also be a factor. Almost all respondents across specialties (97%) indicated a positive impact of variant interpretation activities on clinical practice. Given the frequency of variant interpretation and the direct impact that it has across genetic counseling practice, it is reasonable to consider basic knowledge of variant interpretation as a core competency for genetic counselors in patient-facing clinical roles.
Understanding how variant interpretation activities have been incorporated into clinical practice can inform genetic counselor training, continued competence, and current practice improvements more broadly. Our results indicate that variant interpretation activities are conducted by genetic counselors in both an independent and a team-based manner. A team approach was particularly common when a genetic counselor–laboratory discrepancy was identified, with 77% of participants discussing such cases with the medical team or other colleagues. This ability to incorporate variant interpretation into clinical practice in both independent and team-based manners will likely prove necessary as more genetic counselors join team-based models with non-genetics providers.12,14,17
Previous studies have estimated genetic counselor/clinician–laboratory interpretation discrepancies for 14–21% of variants, though these studies differ in terms of clinical setting and available data on factors that may influence these discrepancies.13,14,16,17 Although the majority of participants in this study reported having a discrepancy with a laboratory’s interpretation at some time, this was overall infrequent, with only 19% of participants reporting discrepancies that often. Discrepancies were reported more frequently in the pediatric and cardiovascular settings. This warrants further study, particularly to determine if this could be due to additional opportunities for phenotypic evaluation. Importantly, our results provide information regarding how such discrepancies are addressed in clinical practice; 82% of respondents discussed genetic counselor–laboratory discrepancies with the clinical testing laboratory. While this indicates that clinical genetic counselors consider the laboratory to be part of the overall clinical care team and are open to collaborative interpretation, we would advocate that all discrepancies be discussed in this way to promote resolution and mutual understanding.27,28 Our findings also indicate that improvements to laboratory reports, such as complete literature summaries and clear outlines of criteria evaluated and applied for variant classification, could facilitate this communication.
Despite the reported frequency and value of variant interpretation in clinical genetic counseling practice, we identified significant barriers to completing these activities. Most notably, lack of time was identified as a barrier by 72% of respondents, though this response was not associated with a decreased likelihood to engage in variant interpretation activities. This finding further supports the importance of these activities in genetic counseling practice but indicates a need for additional employer and institutional support. Participant suggestions for this type of support include support staff for clerical or administrative tasks and improved access to licensed databases and medical literature. Variant interpretation tasks, like other case preparation tasks, should be included in workload requirement calculations for genetic counseling staffing purposes. Improvements to laboratory reports could reduce the time needed for clinical genetic counselors to manually obtain references or details that are not explicitly provided in a report.
A perceived lack of basic knowledge and lack of familiarity with and comfort using available resources were also frequently reported barriers to variant interpretation in clinical practice. These barriers correlated with reduced variant interpretation practice in our study, and additional training opportunities and resource guides were the most common suggestions for improvement. Given the general frequency and the high reported value of variant interpretation activities in this study, barriers that significantly deter this practice are concerning. Continuing education opportunities have been available at national genomics conferences and online educational materials are available through ClinGen and other sources, yet these efforts may not be meeting the needs of the genetic counseling community. Most participants indicated some lack of comfort using most of the variant interpretation resources included in this survey, with the exception of ClinVar. This finding illustrates the need for new educational strategies and/or variant interpretation resources that are easier to utilize or aggregate into a single source.
Despite this perceived lack of familiarity, participants generally applied variant interpretation knowledge correctly in all three of the clinical scenarios provided. Self-reported uncertainty was highest for gene-level topics, such as the use of constraint metrics from population databases and predictions of a variant’s impact based on disease mechanism. This result supports the argument that genetic counselors have a strong educational foundation for variant interpretation and indicates that focused education may be most needed, in addition to improving how variant interpretation resources are made available.12,14
Improvements in applied variant interpretation education by graduate training programs could be promoted if variant interpretation skills in clinical practice were specifically acknowledged as practice-based competencies by the Accreditation Council for Genetic Counseling. Variant interpretation considerations are currently included in the content outline for the certification examination administered by the American Board of Genetic Counseling, and training programs are required to offer laboratory-based educational opportunities, though these can vary widely in terms of format, length, and content. Ad hoc laboratory rotations, available for a subset of genetic counseling students, can provide more in-depth exposure and some training programs are focusing on variant interpretation education more purposely.29 However, formal acknowledgement of the importance of variant interpretation skills in genetic counseling practice is needed to ensure competency and will reinforce the profession’s position as broad genomic testing approaches are adopted across expanding clinical settings.
Study limitations
This study did not specifically assess how clinical genetic counselor variant interpretations differ from laboratory interpretations when a discrepancy occurred. Thus, we could not assess the frequency of discrepancies that are often considered most clinically significant (i.e., VUS vs. likely pathogenic/pathogenic).30 We also did not assess how interpretation discrepancies were documented in patient medical records. While study participants generally represented the NSGC membership, our cohort included a higher proportion of genetic counselors in the 25–29 year age range, which could introduce bias, particularly if they were more likely to receive variant interpretation training in graduate school. There could also be a selection bias toward genetic counselors with an interest in variant interpretation. Prenatal genetic counselors were somewhat underrepresented and additional studies are needed.
Practice implications and research recommendations
The findings from this study strongly indicate that variant interpretation knowledge and skills are actively incorporated into patient-facing clinical genetic counseling practice across all major clinical specialties on a routine basis. These clinical activities have positive impacts on several components of genetic counseling practice. However, important practical and educational needs may interfere with a genetic counselor’s ability to develop and implement these skills. We recommend that variant interpretation knowledge and skills be formally acknowledged in the practice-based competencies for genetic counselors to recognize the frequency and value of this work. Future research to build on our results will continue to define this practice and its impact on clinical care and will inform optimal continuing education strategies to meet the needs of the genetic counseling workforce.
References
Farwell KD, Shahmirzadi L, El-Khechen D, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2015;17:578–86.
Hershberger RE, Givertz MM, Ho CY, et al. Genetic evaluation of cardiomyopathy—a Heart Failure Society of America practice guideline. J Card Fail. 2018;24:281–302.
Hooker GW, Clemens KR, Quillin J, et al. Cancer genetic counseling and testing in an era of rapid change. J Genet Couns. 2017;26:1244–53.
Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64.
Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704.
Rehm HL, Berg JS, Brooks LD, et al. ClinGen—the Clinical Genome Resource. N Engl J Med. 2015;372:2235–42.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Kearney HM, Thorland EC, Brown KB, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–5.
Christian S, Lilley M, Hume S, Scott P, Somerville M. Defining the role of laboratory genetic counselor. J Genet Couns. 2012;21:605–11.
Waltman L, Runke C, Balcom J, et al. Further defining the role of the laboratory genetic counselor. J Genet Couns. 2016;25:786–98.
Zetzsche LH, Kotzer KE, Wain KE. Looking back and moving forward: an historical perspective from laboratory genetic counselors. J Genet Couns. 2014;23:363–70.
Wain K. A commentary on opportunities for the genetic counseling profession through genomic variant interpretation: reflections from an ex-lab rat. J Genet Couns. 2018;27:747–50.
Baldridge D, Heeley J, Vineyard M, et al. The Exome Clinic and the role of the medical genetics expertise in the interpretation of exome sequencing results. Genet Med. 2017;19:1040–8.
Reuter C, Grove ME, Orland D, Spoonamore K, Caleshu C. Clinical cardiovascular genetic counselors take a leading role in team-based variant classification. J Genet Couns. 2018;27:751–60.
Scherr CL, Lindor NM, Malo TL, Couch FJ, Vadaparampil ST. A preliminary investigation of genetic counselors’ information needs when receiving a variant of uncertain significance result: a mixed methods study. Genet Med. 2015;17:739–46.
Bland A, Harrington EA, Dunn K, et al. Clinically impactful differences in variant interpretation between clinicians and testing laboratories: a single-center experience. Genet Med. 2017;20:369–73.
Wain KE, Palen E, Savatt JM, et al. The value of genomic variant ClinVar submissions from clinical providers: beyond the addition of novel variants. Hum Mutat. 2018;39:1660–7.
Zirkelbach E, Hashmi S, Ramdaney A, et al. Managing variant interpretation discrepancies in hereditary cancer: clinical practice, concerns, and desired resources. J Genet Couns. 2017;27:761–9.
Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D7.
Furqan A, Arscott P, Girolami F, et al. Care in specialized centers and data sharing increase agreement in hypertrophic cardiomyopathy genetic test interpretation. Circ Cardiovasc Genet. 2017;10:e001700.
National Society of Genetic Counselors. Professional status survey. 2016. https://www.nsgc.org/p/do/si/topic=562. Accessed 4 May 2018.
National Society of Genetic Counselors. Professional status survey. 2018. https://www.nsgc.org/p/do/sd/sid=7524. Accessed 4 May 2018.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Firth HV, Richards SM, Bevan AP, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84:524–33.
Wou K, Chung WK, Wapner RJ. Laboratory considerations for prenatal genetic testing. Semin Perinatol. 2018;42:307–13.
Edwards JG, Feldman G, Goldberg J, et al. Expanded carrier screening in reproductive medicine-points to consider: a joint statement of the American College of Medical Genetics and Genomics, American College of Obstetricians and Gynecologists, National Society of Genetic Counselors, Perinatal Quality Foundation, and Society for Maternal-Fetal Medicine. Obstet Gynecol. 2015;125:653–2.
Wain KE, Riggs E, Hanson K, et al. The laboratory-clinician team: a professional call to action to improve communication and collaboration for optimal patient care in chromosomal microarray testing. J Genet Couns. 2012;21:631–7.
Bush LW, Beck AE, Biesecker LG, et al. Professional responsibilities regarding the provision, publication and dissemination of patient phenotypes in the context of clinical genetic and genomic testing: points to consider. A statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2018;20:169–71.
Grove ME, White S, Fisk DG, et al. Developing a genomics rotation: practical training around variant interpretation for genetic counseling students. J Genet Couns. 2019;28:466–76.
Harrison SM, Dolinsky JS, Knight Johnson AE, et al. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med. 2017;19:1096–104.
Acknowledgements
The authors would like to thank H. Lester Kirchner for early statistical guidance. We would like to acknowledge and thank the National Society of Genetic Counselors Board of Directors for assistance in disseminating this survey. This work is supported by funding from the National Human Genome Research Institute (U41HG006834, U41HG009649, and U41HG009650). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
A.K.J. is an employee of Invitae. P.K. is an employee of ARUP Laboratories. The other authors declare no conflicts of interest.
Additional information
ANIMAL STUDIES: No nonhuman animal studies were carried out by the authors for this article.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wain, K.E., Azzariti, D.R., Goldstein, J.L. et al. Variant interpretation is a component of clinical practice among genetic counselors in multiple specialties. Genet Med 22, 785–792 (2020). https://doi.org/10.1038/s41436-019-0705-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-019-0705-9
Keywords
This article is cited by
-
Preimplantation Genetic Testing for Inherited Heart Diseases
Current Cardiovascular Risk Reports (2023)