Abstract
Purpose
Moyamoya angiopathy (MMA) is a cerebrovascular disease characterized by occlusion of large arteries, which leads to strokes starting in childhood. Twelve altered genes predispose to MMA but the majority of cases of European descent do not have an identified genetic trigger.
Methods
Exome sequencing from 39 trios were analyzed.
Results
We identified four de novo variants in three genes not previously associated with MMA: CHD4, CNOT3, and SETD5. Identification of additional rare variants in these genes in 158 unrelated MMA probands provided further support that rare pathogenic variants in CHD4 and CNOT3 predispose to MMA. Previous studies identified de novo variants in these genes in children with developmental disorders (DD), intellectual disability, and congenital heart disease.
Conclusion
These genes encode proteins involved in chromatin remodeling, and taken together with previously reported genes leading to MMA-like cerebrovascular occlusive disease (YY1AP1, SMARCAL1), implicate disrupted chromatin remodeling as a molecular pathway predisposing to early onset, large artery occlusive cerebrovascular disease. Furthermore, these data expand the spectrum of phenotypic pleiotropy due to alterations of CHD4, CNOT3, and SETD5 beyond DD to later onset disease in the cerebrovascular arteries and emphasize the need to assess clinical complications into adulthood for genes associated with DD.
Similar content being viewed by others
INTRODUCTION
Moyamoya angiopathy (MMA) is a rare cerebrovascular disease characterized by progressive bilateral stenosis of the distal internal carotid arteries with the development of a compensatory collateral vessel network. MMA leads to strokes starting in childhood through the fourth decade of life resulting in a range of neurologic deficits. Occlusive lesions in the large arteries of MMA patients contain neointimal cells that positively stain for smooth muscle cell (SMC) markers, but lack lipid and inflammatory cell deposition typical of atherosclerosis,1 suggesting that MMA is triggered by pathways distinct from those responsible for atherosclerosis. The risk factors associated with MMA also differ from those for atherosclerosis and include traumatic injury, hypertension, irradiation of the brain, chemotherapy, and hyperthyroidism.2 A genetic predisposition for MMA is supported by the fact that 7–12% MMA patients have similarly affected family members2 and an increased incidence of MMA in Asian countries is due to a founder variant in RNF213.3 Lastly, a number of genetic syndromes are associated with an increased risk for MMA, including neurofibromatosis type I (OMIM 162200, due to NF1 pathogenic variants),4 Alagille syndrome (OMIM 118450, JAG1),5 X-linked moyamoya syndrome (OMIM 300845, BRCC3 and MTCP/MTCP1NB deletions),6 MMA with achalasia and hypertension (OMIM 615750, GUCY1A3),7 Schimke immuno-osseous dysplasia (OMIM 242900, SMARCAL1), and microcephalic osteodysplastic primordial dwarfism type II (OMIM 210720, PCNT).8 Bilateral stenosis of the distal internal carotid arteries also occurs with smooth muscle dysfunction syndrome (OMIM 613834, ACTA2 pathogenic variants)9 and Grange syndrome (OMIM 602531, biallelic YY1AP1 loss-of-function variants).10 These genetic findings highlight potential molecular pathways leading to MMA, including activation of proproliferative pathways (NF1) and loss of nitric oxide signaling (GUCY1A3), and implicate involvement of SMCs (ACTA2).7
MATERIALS AND METHODS
This study was approved by the Institutional Review Board at the University of Texas Health Science Center at Houston. Unrelated sporadic and familial cases of MMA were recruited from the United States, France, and Germany, and informed consent was obtained from all study participants. Diagnosis of MMA was based on magnetic resonance imaging, computed tomography, or cerebral angiography (Supplemental Materials and Methods). Clinical history and demographics were abstracted from medical records, public databases, or obtained by patient report. The cohort includes 197 unrelated probands (172 bilateral, 24 unilateral, and 1 unknown) and 125 relatives (21 affected and 104 clinically healthy) including a total of 39 trios. Individuals diagnosed with established Mendelian diseases predisposing to MMA were excluded from the study. Patients with RNF213 variants were not excluded since RNF213 is considered to be a susceptibility gene for MMA rather than responsible for Mendelian MMA. Exome sequencing assay was performed by the Center for Mendelian Genomics (University of Washington, USA) or Integragen core (Evry, France) on Illumina platforms (Supplemental Materials and Methods). To limit the detection of false de novo variants, we employed stringent quality criteria by restricting our analysis to variants with a high-quality score (Q Phred score ≥30) and a read depth ≥20× in all members of a same trio. We limited our analysis to the following types of variants: nonsense, nonsynonymous, canonical splice site, and indels in coding regions. Variants with a minor allelic frequency exceeding 0.01 in public databases (dbSNP 144, 1000 Genomes Phase 3, Exome Sequencing Project [ESP6500SIV2], ExAC [0.3], gnomAD) were excluded. In silico prediction tools MutationTaster, SIFT, and PolyPhen-2 were used in the variant curation process; missense variants predicted benign by more than one of these tools were excluded from further analysis. Variants with Combined Annotation Dependent Depletion pathogenicity (CADD) scores ≤20 in non–de novo trios cases were excluded. Remaining variants were validated using Sanger sequencing (Supplemental Materials and Methods).
RESULTS
To further identify genes and cellular pathways involved in the molecular pathogenesis of MMA, exome sequencing was pursued for 39 affected probands with MMA of unknown genetic etiology (average age of disease onset 15 years, range: 0.5 months–54 years) and their unaffected parents. After rigorous filtering, 39 genes harboring rare de novo variants were identified in 26 MMA trios (2 trios had 3 de novo variants, 12 trios had 2 de novo variants, 12 trios had 1 de novo variant). Thirteen trios did not have de novo variants that met these criteria (Table S1). Three of these variants were in RNF213, and one was a variant of unknown significance in NF1 (Table S1).
In four MMA patients, we identified de novo variants in three genes, CHD4, CNOT3, and SETD5, previously found to harbor pathogenic de novo variants in children with developmental disorders (DD) (Table 1, Fig. 1). The affected probands had clinical features that overlapped with these previously reported cases (Table S3). CHD4 de novo variants were reported in children with congenital heart disease, developmental delay, and Sifrim–Hitz–Weiss syndrome (OMIM 617159).11,12 We identified a de novo CHD4 variant, p.Ala1178Val (absent in exome databases; CADD score 23.9) in a patient diagnosed with bilateral MMA at 10 years of age with a history of developmental delay, congenital heart defect, abnormal gait, and mildly dysmorphic facies (Tables S3 and S4). Importantly, the fact that MMA was previously reported in a child with intellectual disability due to a de novo CHD4 variant, p.Arg1127Gln, further supports an association of CHD4 variants with MMA (Tables S3 and S4).11 CNOT3 de novo variants have also been identified in children with DD and autism spectrum.12 Two MMA probands in our cohort harbor de novo variants in CNOT3, p.Gln215* and p.Leu522Ile (absent in exome databases; CADD scores 39 and 17.9, respectively). The de novo variant, p.Gln215*, was identified in a woman diagnosed with bilateral MMA at 20 years of age; she had a history of intellectual disability, autism spectrum disorder, and mildly dysmorphic facies (Figure S1B and C, Tables S3 and S4). De novo SETD5 variants, primarily loss-of-function (LoF) variants, have been found in children with intellectual disability, developmental delay, skeletal abnormalities, and variable dysmorphic features (OMIM 615761).12,13 A de novo SETD5 haploinsufficiency variant, p.Glu661Lysfs*5 (Fig. 1), was identified in a patient diagnosed with bilateral MMA at 10 years of age with a history of developmental delay, polydactyly, and mild dysmorphic facial features (Tables S3 and S4). Collectively, the phenotypic overlap between our MMA patients harboring de novo variants and those previously reported with neurodevelopmental disorders indicates that the pleiotropy associated with de novo variants in CHD4, CNOT3, and SETD5 should include MMA.
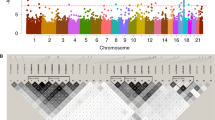
Schematic representation of domains of the proteins encoded by CHD4, CNOT3, and SETD5 and the location of the rare variants identified in individuals with either moyamoya angiopathy (MMA) or developmental disorders. Above the protein schematics are the location of rare de novo variants (red) and variants (black) identified in singleton cases in the MMA cohort. Below the protein schematics are variants identified in children with intellectual disability, congenital heart defects, or developmental disorders (blue).11,12 The red variant in the CHD4 schematic was identified in three unrelated patients, including the patient described with intellectual disability and moyamoya disease11 (CHD4: NP_001264.2, CNOT3: NP_055331.1, SETD5: NP_001073986.1). ATP adenosine triphosphate.
To further validate an association between rare variants in CHD4, CNOT3, and SETD5 and MMA, exome data from 158 unrelated MMA cases and 43 relatives (18 with MMA, and 25 unaffected) were filtered and validated as previously described. Rare CHD4 variants were confirmed in six additional MMA cases, including five missense variants and one splice donor site variant (Table 1). RNA analysis of leukocytes confirmed that the splice site variant is associated with aberrant splicing of exon 11, leading to a frameshift and nonsense-mediated decay. CHD4 is intolerant to both missense (Z-score = 7.05) and LoF variants (probability of being LoF variant intolerant score [pLI] = 1) in the general population. Of the seven rare CHD4 variants in the MMA cases, four were absent in gnomAD and three were reported with very low frequencies (minor allele frequency [MAF] < 1.62e−5) (Table 1). The majority of CHD4 variants associated with DD cluster in the central region of the protein involved in the helicase function of CHD4. Interestingly, three of the variants present in MMA patients (p.Ile1741Val, p.Tyr1758Cys, and p.Pro1880Ser) are located in the C-terminal part of the protein, which is known to interact with pericentrin, encoded by the PCNT gene.14 Homozygous LoF variants in PCTN predispose to MMA (Fig. 1).8 Of the seven individuals with rare CHD4 cases in this study, all had bilateral MMA (Figure S1A; Tables S2, S3, and S4). Intellectual disability was observed in one of the additional cases (Table S4).
A CNOT3 missense variant, p.Gly304Ser, with a MAF of 4.76e−4 (CADD score 27.2) was identified in two unrelated MMA patients (Table 1). CNOT3 is another gene intolerant to both missense (Z-score = 3.89) and LoF variation (pLI = 1). The two patients who harbor the p.Gly304Ser variant, presented with unilateral MMA at 5 and 22 years of age, but these cases lacked developmental delay and other clinical findings previously associated with de novo CNOT3 variants.
Two missense variants, p.Asp339Tyr and p.Arg767Cys, were identified in SETD5 in unrelated MMA cases. The variant, p.Asp339Tyr, is absent in gnomAD, and p.Arg767Cys is present at an extremely low frequency (Table 1). SETD5 is intolerant to LoF variation (pLI = 1) but not missense variation (Z-score = −0.04). All three cases with SETD5 rare variants had bilateral MMA (Tables S2, S3, and S4). In addition to MMA, the patient harboring p.Asp339Tyr had stenosis of other arteries and hypertension (Table S4).
DISCUSSION
These data indicate that rare variants in CHD4 and CNOT3 predispose to MMA in the presence and absence of DD; additional data are required to validate an association between SETD5 rare variants and MMA. CHD4 and CNOT3, along with a previously published gene, YY1AP1,10 are all associated with MMA-like occlusive vascular disease, developmental delay, skeletal changes, and dysmorphic facies. All of these genes encode proteins involved in chromatin remodeling. CHD4 encodes a helicase that is the main component of the nucleosome remodeling and histones deacetylase (NuRD) repressor complex, which pairs adenosine triphosphate (ATP)-dependent chromatin remodeling with histone deacetylase activity. The NuRD complex is involved in the regulation of gene expression during differentiation of pluripotent stem cells.15 CNOT3 encodes the subunit 3 of the CCR4-NOT transcription complex, which executes poly(A)-tail shortening of messenger RNA (mRNA) in the cytoplasm and its conditional deletion results in the formation of autophagic vacuoles and cardiomyocyte death, leading to lethal heart failure accompanied by long QT intervals.16 CNOT3 also controls the acetylation level of histones H3 and H4.17 Inhibition of histone acetylases was shown to reverse the heart defects observed in heterozygous knockout mice suggesting a mechanistic link to epigenetic remodeling.18 Therefore, it remains unclear whether the effect on transcription is direct or indirect through mRNA deadenylation. We previously reported LoF variants in YY1AP1 in individuals with Grange syndrome, which is characterized by MMA-like occlusion in the carotid arteries, along with occlusive lesions in the renal, celiac, and coronary arteries.10 YY1AP1, encoding YY1-associated protein 1, is an enhancer of Yin Yang 1 (YY1) transcriptional activity; both are components of the INO80 chromatin remodeling complex. Interestingly, SETD5 encodes SET domain-containing protein 5 and family of proteins with lysine methyltransferase, but SETD5 has not been shown to have methyltransferase activity. SETD5 interacts closely with the histone deacetylase-containing NCoR complex, and loss of Setd5 increases histone acetylation at transcription start sites.19
The association of MMA or MMA-like with rare variants in CHD4, CNOT3, SETD5, and YY1AP1 indicates that disrupted chromatin remodeling is a predisposing factor for non-atherosclerotic occlusive cerebrovascular disease. The differentiation of progenitor cells into functional cells in organs requires chromatin remodeling for tissue-specific gene expression patterns. Deficiency of Chd4 in mice is known to disrupt neuronal differentiation; neural progenitor cells in Chd4-deficient mice precociously exit the cell cycle, fail to differentiate, and die.15 In contrast, an interaction between retinal ganglion cells and endothelial cells critical for development of the mouse retina vasculature is dependent on Setd5. Setd5 in retinal ganglion cells represses Sema3a, thus allowing expression of miR-126-5p, which regulates angiogenesis by protecting adjacent endothelial cells from apoptosis.20 Deficiency of Setd5 in these ganglion cells decreases production of miR-126-5p and leads to an underdeveloped retinal vasculature. Thus, disruption of CHD4, CNOT3, SETD5, and YY1AP1 potentially predisposes to MMA through altered differentiation of the cells in the arterial wall or altered expression of regulatory factors in the adjacent neuronal cells.
These data expand the phenotypic pleiotropy resulting from CHD4, CNOT3, and SETD5 de novo variants to include childhood to young adult–onset occlusive cerebrovascular disease. Additional studies to assess the penetrance of MMA are required before we can recommend imaging for all the patients carrying de novo variants in CHD4, CNOT3, and SETD5 (Table S4). It is also important to note that individuals with pathogenic variants may present with MMA in adulthood without history of developmental defects. Furthermore, these results suggest that pathogenic variation in other genes that disrupt chromatin remodeling and lead to development defects may be associated with other complications not evident until later in life. Finally, identifying the genetic triggers responsible for MMA large artery occlusive lesions raises the possibility that “atherosclerotic” lesions in ischemic stroke patients in general may result from aberrant cellular chromatin remodeling.
References
Masuda J, Ogata J, Yutani C. Smooth-muscle cell-proliferation and localization of macrophages and T-cells in the occlusive intracranial major arteries in moyamoya disease. Stroke. 1993;24:1960–1967.
Scott RM, Smith ER. Moyamoya disease and moyamoya syndrome. N Engl J Med. 2009;360:1226–1237.
Kamada F, Aoki Y, Narisawa A, et al. A genome-wide association study identifies RNF213 as the first moyamoya disease gene. J Hum Genet. 2011;56:34–40.
Friedman JM, Arbiser J, Epstein JA, et al. Cardiovascular disease in neurofibromatosis 1: report of the NF1 Cardiovascular Task Force. Genet Med. 2002;4:105–111.
Rocha R, Soro I, Leitao A, Silva ML, Leao M. Moyamoya vascular pattern in Alagille syndrome. Pediatr Neurol. 2012;47:125–128.
Miskinyte S, Butler MG, Herve D, et al. Loss of BRCC3 deubiquitinating enzyme leads to abnormal angiogenesis and is associated with syndromic moyamoya. Am J Hum Genet. 2011;88:718–728.
Herve D, Philippi A, Belbouab R, et al. Loss of alpha1beta1 soluble guanylate cyclase, the major nitric oxide receptor, leads to moyamoya and achalasia. Am J Hum Genet. 2014;94:385–394.
Bober MB, Khan N, Kaplan J, et al. Majewski osteodysplastic primordial dwarfism type II (MOPD II): expanding the vascular phenotype. Am J Med Genet A. 2010;152A:960–965.
Milewicz DM, Ostergaard JR, la-Kokko LM, et al. De novo ACTA2 mutation causes a novel syndrome of multisystemic smooth muscle dysfunction. Am J Med Genet A. 2010;152A:2437–2443.
Guo DC, Duan XY, Regalado ES, et al. Loss-of-function mutations in YY1AP1 lead to Grange syndrome and a fibromuscular dysplasia-like vascular disease. Am J Hum Genet. 2017;100:21–30.
Weiss K, Terhal PA, Cohen L, et al. De novo mutations in CHD4, an ATP-dependent chromatin remodeler gene, cause an intellectual disability syndrome with distinctive dysmorphisms. Am J Hum Genet. 2016;99:934–941.
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438.
Powis Z, Farwell Hagman KD, Mroske C, et al. Expansion and further delineation of the SETD5 phenotype leading to global developmental delay, variable dysmorphic features, and reduced penetrance. Clin Genet. 2018;93:752–761.
Sillibourne JE, Delaval B, Redick S, Sinha M, Doxsey SJ. Chromatin remodeling proteins interact with pericentrin to regulate centrosome integrity. Mol Biol Cell. 2007;18:3667–3680.
Nitarska J, Smith JG, Sherlock WT, et al. A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep. 2016;17:1683–1698.
Yamaguchi T, Suzuki T, Sato T, et al. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Sci Signal. 2018;11:eaan3638.
Peng W, Togawa C, Zhang K, Kurdistani SK. Regulators of cellular levels of histone acetylation in Saccharomyces cerevisiae. Genetics. 2008;179:277–289.
Neely GG, Kuba K, Cammarato A, et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153.
Osipovich AB, Gangula R, Vianna PG, Magnuson MA. Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Development. 2016;143:4595–4607.
Villain G, Poissonnier L, Noueihed B, et al. miR-126-5p promotes retinal endothelial cell survival through SetD5 regulation in neurons. Development. 2018;145:dev156232.
Acknowledgements
We are grateful to the patients and their family members for participating in this study. This study was funded by the Henrietta B. and Frederick H. Bugher Foundation (D.M.M.), the Texas Heart Institute Fibromuscular Dysplasia Project (D.M.M.), INSERM (E.T.-L.), the American Heart Association (18POST34020031; A. P.) and the Fondation pour le Recherche Médicale (S.G.). Exome sequencing was funded by the National Human Genome Research Institute and the National Heart, Lung, and Blood Institute grant HG006493 to D.A.N., M.J.B., and Suzanne Leal. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Pinard, A., Guey, S., Guo, D. et al. The pleiotropy associated with de novo variants in CHD4, CNOT3, and SETD5 extends to moyamoya angiopathy. Genet Med 22, 427–431 (2020). https://doi.org/10.1038/s41436-019-0639-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-019-0639-2
Keywords
This article is cited by
-
The genetic landscape and clinical implication of pediatric Moyamoya angiopathy in an international cohort
European Journal of Human Genetics (2023)
-
Updated Stroke Gene Panels: Rapid evolution of knowledge on monogenic causes of stroke
European Journal of Human Genetics (2023)
-
Mutation of key signaling regulators of cerebrovascular development in vein of Galen malformations
Nature Communications (2023)
-
Childhood stroke
Nature Reviews Disease Primers (2022)
-
The angiographic presentation of European Moyamoya angiopathy
Journal of Neurology (2022)