Abstract
Purpose
Preaxial polydactyly (PPD) is a common congenital hand malformation classified into four subtypes (PPD I–IV). Variants in the zone of polarizing activity regulatory sequence (ZRS) within intron 5 of the LMBR1 gene are linked to most PPD types. However, the genes responsible for PPD I and the underlying mechanisms are unknown.
Methods
A rare large four-generation family with isolated PPD I was subjected to genome-wide genotyping and sequence analysis. In vitro and in vivo functional studies were performed in Caco-2 cells, 293T cells, and a knockin transgenic mouse model.
Results
A novel g.101779T>A (reference sequence: NG_009240.2; position 446 of the ZRS) variant segregates with all PPD I–affected individuals. The knockin mouse with this ZRS variant exhibited PPD I phenotype accompanying ectopic and excess expression of Shh. We confirmed that HnRNP K can bind the ZRS and SHH promoters. The ZRS mutant enhanced the binding affinity for HnRNP K and upregulated SHH expression.
Conclusion
Our results identify the first PPD I disease-causing variant. The variant leading to PPD I may be associated with enhancing SHH expression mediated by HnRNP K. This study adds to the ZRS-associated syndromes classification system for PPD and clarifies the underlying molecular mechanisms.
Similar content being viewed by others
INTRODUCTION
Preaxial polydactyly (PPD, OMIM 603596), extra digits occurring on the preaxial side of the hands, is the most common congenital hand malformation, with the prevalence ranging from 5 to 19 per 10,000 live births.1 According to Temtamy–McKusick classification, PPD is generally classified into four subtypes: type I (PPD I, OMIM 174400), thumb/hallux polydactyly; type II (PPD II, OMIM 174500), triphalangeal thumbs (TPT); type III (PPD III, OMIM 174600), polydactyly of an index finger; and type IV (PPD IV, OMIM 174700), polysyndactyly.2,3 Among all four types, PPD I occurs most frequently, but presently, its underlying genetic basis is not well understood.4,5
PPD manifests either by itself or as a part of a syndrome. When it occurs in isolation, autosomal dominant variations in a single gene are typically involved.6 Genetic variants in the zone of polarizing activity regulatory sequence (ZRS, OMIM 605522), a limb-specific cis-regulator located in intron 5 of the LMBR1 on chromosome 7q36, have been linked to most PPD types, including PPD II, PPD II/III, triphalangeal thumb–polysyndactyly syndrome (TPTPS, OMIM 188770), syndactyly type IV (SD4, OMIM 186200), and Werner mesomelic syndrome (WMS, OMIM 188770),7,8,9,10,11,12 indicating that variants at ZRS can lead to different PPD phenotypes. In this region, more than 20 point variants, 10 duplications, 1 triplication, and a base pair insertion have been reported to be associated with PPD.3,13,14 Further study indicated that this highly conserved ~800 bp region is located ~1 Mb upstream from its target gene, sonic hedgehog (SHH, OMIM 600725), and acts as the enhancer of SHH.15 The ZRS normally controls the expression of SHH in the zone of polarizing activity (ZPA) located along the posterior margin of the limb bud.16,17 Point variants and duplications in the ZRS are associated with ectopic expression of SHH in the anterior limb bud and usually result in a PPD phenotype.14,18 However, the potential mechanisms through which specific ZRS variants impact ectopic expression of SHH in the pathophysiology of PPD have only been partially clarified.
In this study, we report the first case of isolated PPD I in a four-generation Chinese family that is associated with a novel pathogenic variation, g.101779T>A in the LMBR1 gene (reference sequence: NG_009240.2; position 446 of the ZRS sequence). We generated a mouse model harboring the g.67973T>A in the Lmbr1 gene (reference sequence: NC_000071.6; position 447 of the mouse ZRS sequence), a variant analogous to the T446A variant associated with PPD I in humans. This novel model exhibited a distinct PPD I phenotype with anterior ectopic expression of Shh in the E11.5 limb buds. In vitro functional studies implicated HnRNP K, which binds both the ZRS region and the SHH promoter. This genetic variant enhanced HnRNP K's binding potential to its target in ZRS and promoted SHH upregulation, which may be associated with PPD I.
MATERIALS AND METHODS
Patient information
A four-generation pedigree with PPD I, including 31 Chinese Han individuals from Guangdong Province, was identified (Fig. 1a). Twenty-six family members (8 affected members, 8 normal members, and 10 undefined members) participated in this study. PPD I was diagnosed based on family history, physical examination, and X-ray examination and was classified according to the criteria.2,19 DNA samples were extracted from peripheral blood using QIAamp DNA blood mini kits (Qiagen, Germany). The study was approved by the First Affiliated Hospital Medical and Animal Ethics Committee of Sun Yat-sen University (MEC-2015-76). Each human participant signed an informed consent form.
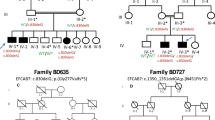
A ZRS point variant is responsible for the pathogenesis of isolated preaxial polydactyly (PPD) I in a four-generation pedigree. (a) Pedigree of isolated PPD I, showing autosomal dominant inheritance with near complete penetrance. Circles denote females and squares denote males. Filled symbols represent affected individuals. Unfilled symbols represent unaffected individuals. Question marks represent undetermined individuals. The arrow indicates the proband. Symbols with slashes represent deceased individuals. Asterisks indicate that a variant is present. WT wild-type. (b) Depicted are normal and X-ray images of both hands in affected, undetermined, and normal individuals. (c) A T/A point variant at position 446 of the the ZRS was identified by Sanger sequencing.
Genotyping and genetic analysis
DNA samples from 11 family members (II-3, II-4, II-7, II-8, II-11, II-12, III-4, III-5, III-8, III-11, and IV-1) (Fig. 1a) were subjected to genome-wide genotyping using the Illumina HumanOmni2.5-Quad Beadchip by the Microarray Core at University of Texas Southwestern Medical Center (UTSW). Data were analyzed using GenomeStudio software (v. 2011.1). We first selected 7037 single-nucleotide polymorphisms (SNPs) across the autosomes from the ~2.5 million SNPs to construct a sparse linkage map. The selected SNPs were spaced ~2 per centiMorgan. A genome-wide affected-only model-free linkage analysis was performed.20 Linked regions were further evaluated by multipoint model-based linkage analysis using both affected and unaffected individuals, assuming a dominant model. Copy-number variation analysis in the linked region was performed using cnvPartition implemented in GenomeStudio. Genome-wide linkage analysis revealed five linked regions on chromosomes 1p13, 4q35, 7q36, 10p15, and 21q21 (Supplementary Fig. 1). Noting that the ZRS locus is directly underneath the linkage peak on chromosome 7, we sequenced the ZRS locus in the LMBR1 using the Big Dye Terminator Cycle Sequence Kit 3.1 (ABI, Foster City, CA, USA). The primers used are listed in Supplementary Table 1. We also sequenced the ZRS locus from 50 healthy unrelated age- and sex-matched Chinese Han control subjects.
Mouse model
A C57BL/6 mouse model with a point variant (T447A) located in introns 5–6 of the mouse Lmbr1 (NM_020295.3; Ensembl: ENSMUSG00000010721) locus was created by Cyagen Biosciences (Suzhou, China) with CRISPR/Cas9-mediated genome engineering. The targeting strategy as well as single guide RNA (sgRNA) and oligo donor designs are shown in Supplementary Fig. 2. After coinjected, the pups were genotyped by polymerase chain reaction (PCR), followed by sequence confirmation (Supplementary Table 2). Skeletal staining of postnatal day (P) 30 mice was conducted as previously reported.21 The PPD incidence of each genotype was calculated according to the phenotype of 368 mice.
E11.5 limb buds were divided into anterior halves and posterior halves. Messenger RNA (mRNA) of each half was extracted and real-time PCR was performed to compare the expression of Shh and Hnrnpk among the three genotypes. The sequences of the primers are shown in Supplementary Table 6.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using a LightShift Chemiluminescent EMSA kit (Pierce, Rockford, IL, USA) with nuclear extracts from Caco-2 cells. The mutant and wild-type 29-bp DNA probes (Supplementary Table 3) were synthesized by Integrated DNA Technologies (Coralville, IA, USA). Biotin-labeled DNA was visualized by chemiluminescence. In the supershift assay, for depletion, the following antibodies (1 µl) were added along with the nuclear extract in a 10-µl reaction system: HnRNP K (ab70492, Abcam), hnRNP A/B (sc98810, Santa Cruz), hnRNP Q (ab10687, Abcam), HnRNP D (ab61193, Abcam), and control IgG (ab46540, Abcam).
DNA pulldown and mass spectrometry (MS)
The biotinylated human SHH promoter probe was generated by PCR. The WT-ZRS and MT-ZRS probes, and negative control probes LacZ were synthesized by Bersin Bio (Guangzhou, China). The primers are shown in Supplementary Table 4. A DNA pulldown assay was performed as described previously22 using Caco-2 cells. For liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses, a Q-Exactive mass spectrometer (Thermo 588 Finnigan, San Jose, CA, USA) coupled with an Easy-nLC1000 HPLC system 589 (Thermo Fisher, San Jose, CA, USA) was used as described previously.23 Sonar tandem MS software was used for protein identification.
Plasmid construct and transfection
Full-length human HnRNP K complementary DNA (cDNA) (GenBank accession number NM_031262) was introduced into the XhoI/KpnI site of the GV219 vector plasmid. GV219 was used as control. The human HnRNP K small interfering RNA (siRNA) and scrambled siRNA (Supplementary Table 5) were synthesized by Bersin Bio (Guangzhou, China). Caco-2 and 293T cells were transfected with plasmid or siRNA (20 nM) using Lipofectamine 3000 (Invitrogen). The efficiency of the transfection was assessed as previously described.24 Primer sequences are described in Supplementary Table 6. The following primary antibodies were used: anti-HnRNP K (ab70492, Abcam), anti-SHH (06–1106, Millipore), and anti-GAPDH (EarthOx, Millbrae, CA, USA). The identification results using the HnRNP K siRNAi are shown in Supplementary Fig. 3. HnRNP K siRNAi3 was used for the EMSA assay and SHH regulation study.
Chromatin immunoprecipitation (ChIP) assays
Mononuclear cells from the peripheral blood of patients with PPD I or normal members were isolated and purified by density gradient centrifugation using Ficoll-Hypaque (1.077 g/ml) (TBD, China). ChIP assays were performed using a Magna ChIP™ Kit (Millipore). Briefly, mononuclear cells (1 × 107) were cross-linked and lysed. The sheared chromatin was incubated with an anti-hnRNP K antibody and magnetic protein A/G beads (Millipore). The IgG (ab46540, Abcam) was used as control. The immune complexes were purified and further analyzed using quantitative PCR for the ZRS and SHH promoter. SHH promoter sequence (−849 ~ +12) was divided into ten different SHH promoter segment sequences, from SHH-1 to SHH-10. The different sequences and the primer sequences are provided in Supplementary Table 6. The MMP12 promoter region (−95 to −20) was used as a positive control.25
Dual-luciferase reporter assay
Vector plasmids bearing the human SHH minimal promoter26 (CS-NEG-GA03-02), ZRS-SHH promoter (CS-ZRS01-GA03), or mutant ZRS-SHH promoter reporter (CS-ZRS02-GA03) were constructed, packaged, and purified by GeneCopoeia (Guangzhou, China), and 293T cells were cotransfected with siRNAs corresponding to HnRNP K and the SHH promoter, ZRS-SHH promoter, or mutant ZRS-SHH promoter reporter. Luciferase activity was measured using a Secrete-Pair Dual Luminescence Assay Kit (GeneCopoeia, China).
Statistical analysis
Statistical analysis was performed using the statistical software SPSS13. Results are expressed as mean ± standard deviation (s.d.) and analyzed for significant differences using analysis of variance (ANOVA) and Student’s t test. P < 0.05 was considered significant (*p < 0.05, **p < 0.01).
RESULTS
PPD I phenotypes in the index family
The disease manifested variable phenotypes in an autosomal dominant pattern of inheritance in the index pedigree. Eight family members (II-3, II-7, II-11, III-4, III-8, III-11, III-12, and IV-1) of this four-generation family were defined as affected individuals (Fig. 1a). The left thumbs of affected members II-11 and IV-1 showed duplication of the distal phalanx, typical of PPD I and Wassel type II according to Wassel’s classification system19 (Fig. 1b). The right thumbs of II-11, IV-1, III-12 (Fig. 1b), and both thumbs of another four affected members (II-7, III-4, III-8 [Fig. 1b], and III-11 [Fig. 1b]) exhibited incomplete duplication of the distal phalanx. The left thumb of III-12 (Fig. 1b) was normal. There were small sesamoids located between two phalanges of the bilateral thumbs of individuals II-2 (Fig. 1b), II-5, II-9 (Fig. 1b), II-13, III-2, III-6, III-7, III-9, III-10, and III-13, who were classified as undefined members. We examined an additional 50 hands of trauma patients with normal X-rays and found 9 with sesamoids similar to those in the members in our pedigree. Members II-4, II-6, II-8, II-12, II-14, III-1, III-5, and III-14 were defined as unaffected individuals (Fig. 1a) with a normal phenotype (Fig. 1b). The affected family members did not have any other apparent congenital anomalies. The clinical features from affected individuals in PPD I pedigree are shown in Supplementary Table 7.
Identification of a point variant in the ZRS
A genome-wide affected-only model-free linkage screen identified five linked regions: 1p13, 4q35, 7q36, 10p15, and 21q21. Model-based linkage analysis under a dominant model with a penetrance of 0.95, sporadic rate of 0.01, and disease-predisposing allele frequency of 0.001 confirmed the results of the model-free analysis and generated a maximum logarithm of the odds (LOD) score of ~1.75 in each linked region (Supplementary Fig. 1). There were no copy-number variations cosegregating with disease in the five regions. Of note, the ZRS locus was directly underneath the linkage peak on chromosome 7.
Sequencing a 1.28-kb region covering the ZRS on chromosome 7 revealed a heterozygous T>A point variant at 446 bp of the ZRS in all affected members and an undefined member, III-9 (Fig. 1a, c and Supplementary Fig. 4). All unaffected members and other undefined members from this family were homozygous T/T (Fig. 1a, c). This variant was not found in ExAC, gnomAD, or in the 50 healthy controls.
Point variant (g.67973T>A) at mouse Lmbr1 leads to a PPD I phenotype and excess Shh expression in the developing limb bud
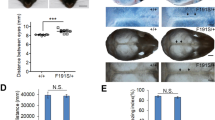
To further explore the functional consequences of this variant, a g.67973T>A knockin (homologous position in mouse to human g.101779T>A) at the mouse Lmbr1 locus was engineered in C57BL/6 mouse (Supplementary Fig. 2). The resulting heterozygous mice exhibited extra preaxial digits only in the hindlimbs and not in the forelimbs (Fig. 2a, ZRS+/−). Most homozygotes (62%) exhibited extra preaxial digits only on the hindlimbs and 38% of those ZRS−/− mice exhibited extra digits on both the hindlimbs and forelimbs (Fig. 2a, ZRS−/−). All wild-type mice exhibited normal preaxial digits (Fig. 2a, ZRS+/+). The penetrance of heterozygous mice was 81% (150/185), compared with 100% in homozygous mice (92/92) (Fig. 2b and Supplementary Table 8). No other apparent abnormalities were observed in the heterozygous or homozygous mice. Analysis of Shh expression in the developing limb buds at E11.5 by quantitative (qPCR) showed that Shh is only expressed in the posterior of developing limb buds in wild-type mouse. Both the heterozygous and homozygous mutant result in ectopic anterior expression and increased posterior expression of Shh compared with wild-type mouse. The Shh expression level in homozygous mutant mouse was the highest among three genotypes (Fig. 2c). Our results show that the g.67973T>A variant in murine Lmbr1 leads to ectopic anterior and increased posterior expression of Shh and a PPD I phenotype in mouse.
Point variant (T447A) in mouse ZRS leads to a preaxial polydactyly (PPD) I phenotype and excess shh expression in the developing limb bud. (a) Whole-mount skeletal specimens from postnatal day (P) 30 of transgenic mouse were stained using Alizarin red and Alcian blue. The extra preaxial digit was identified on the hindlimbs of heterozygous mouse (ZRS+/−). Homozygous mouse (ZRS−/−) showed extra preaxial digits in both hindlimbs and forelimbs or only in hindlimbs. Preaxial digits are indicated by red arrows. (b) The penetrance statistical analysis of polydactyly among wild-type mouse (ZRS+/+), heterozygous mouse (ZRS+/−), and homozygous mouse (ZRS-/-). (c) Real-time polymerase chain reaction (PCR) was performed to compare the expression of Shh in the anterior and posterior halves of E11.5 limb buds among wild-type mouse (ZRS+/+), heterozygous mouse (ZRS+/−), and homozygous mouse (ZRS−/−). Data represent mean ± SD. *p < 0.05, significantly different from control (n = 3). mRNA messenger RNA.
Protein-binding differences between mutant and wild-type ZRS
The protein-binding profile associated with the 29-bp motif encompassing T446A variant on ZRS was analyzed by EMSA. The results showed a specific DNA/protein-binding band for the wild-type probe and mutant probe at the same position, but the mutant probe exhibited enhanced binding intensity compared with the wild-type probe (Fig. 3a, L2 and L5). The binding specificity was demonstrated using unlabeled competitors for either the wild-type or mutant probes (Fig. 3a, L3 and L6).
Identification of HnRNP K binding to the ZRS and the promoter of SHH. (a) Electrophoretic mobility shift assay (EMSA) analysis of the difference in binding of Caco-2 cell nuclear extract (NE) to biotin-labeled mutant (B-MT-DNA) and wild-type ZRS (B-WT-DNA) probes. Lane 2 and lane 5: specific binding bands S of NE to both B-WT-DNA and B-MT-DNA were detected, but the binding quantity of NE to MT-DNA is higher than that of NE to WT-DNA. Lane 3 and Lane 6: 200-fold molar excess unlabeled probes (U-WT-DNA, U-MT-DNA) were added to demonstrate specific binding to both WT-DNA and MT-DNA. (b) EMSA supershift assay to identify the specific binding of the ZRS probe to HnRNP K. Lane 2 shows that the anti-HnRNP K antibody depleted the binding band S to the MT-DNA, but no changes occurred with IgG. (c–d) Caco-2 cells were transfected with HnRNP K small interfering RNA (siRNA) or scrambled siRNA as a control. Knockdown efficiency was measured by real-time polymerase chain reaction (PCR) (c) and western blot (d). (e–f) Caco-2 cells were transfected with HnRNP K overexpression plasmid or control vector plasmid. Real-time PCR (e) and western blot (f) were performed to verify overexpression efficiency. (g) EMSA assay was performed to detect the binding ability of wild-type and mutant ZRS to NEs from HnRNP K–overexpressing or knockdown caco-2 cells. The WT probe and mutant probe bound more protein from HnRNP K–overexpressing cells than from control cells. In contrast, both the WT probe and mutant probe bound less protein from HnRNP K knockdown cells than from control cells. (h) A chromatin immunoprecipitation–quantitative PCR (ChIP-qPCR) assay was performed using NEs from preaxial polydactyly (PPD) I patients’ or normal members’ mononuclear cells with the HnRNP K antibody. The HnRNP K antibody precipitated more ZRS from the patients’ mononuclear cells than from the normal mononuclear cells. (i) A ChIP-qPCR assay was conducted to show that the HnRNP K antibody precipitated the minimal promoter of SHH. SHH-1 to SHH-10 represent different SHH promoter segment sequences. The different sequences' specificities are provided in Supplementary Table 7. The MMP12 promoter region (−95 to −20) was used as a positive control. GAPDH was used as a negative control. Data represent mean ± SD. *p < 0.05, significantly different from control (n = 3). mRNA messenger RNA.
Identification of HnRNP K binding to the ZRS and the promoter of SHH
To identify the specific proteins binding to the ZRS, we employed DNA pulldown and MS assays to analyze the binding protein components. As a result, several protein members from the heterogeneous nuclear ribonucleoprotein (HnRNPs) family, including HnRNP K, HnRNP A/B, HnRNP Q, and HnRNP D, were identified among the specific binding proteins (Supplementary Figs. 5, 6, and Supplementary Table 9), whereas those proteins were not captured with a negative control probe (Supplementary Table 10). To determine the candidate HnRNPs specifically binding to the ZRS sequences, a supershift assay was performed using antibodies specific for HnRNP K, HnRNP A/B, HnRNP Q, and HnRNP D proteins. Only the anti-HnRNP K antibody depleted specific protein binding to the mutant DNA probe (Fig. 3b, Supplementary Fig. 7). To further investigate the binding specificity of HnRNP K to the mutant ZRS, we constructed HnRNP K knockdown (Fig. 3c, d) and overexpression models (Fig. 3e, f) in Caco-2 cells. The EMSA results showed that both the wild-type and mutant probes bound more protein in HnRNP K–overexpressing cells than that in control cells (Fig. 3g, L1–L4). In contrast, both the wild-type and mutant probes bound less protein from HnRNP K knockdown cells (Fig. 3g, L5–L8). However, the mutant probes exhibited higher binding efficiency with HnRNP K in both cells compared with wild-type probes (Fig. 3g)
To test if HnRNP K binds directly to the mutant site on ZRS, we performed ChIP using nuclear extracts isolated from mononuclear cells of patients with PPD I or unaffected family members. We observed that the HnRNP K antibody precipitated more of the ZRS sequence from mutant cells than from normal cells (Fig. 3h). These results suggested that HnRNP K indeed binds the ZRS and has a stronger binding affinity to the mutant ZRS than the wild-type ZRS. Interestingly, the HnRNP K and HnRNP U proteins were also pulled down by the promoter probe of SHH, as shown by the MS result (Supplementary Fig. 8 and Supplementary Table 9). A ChIP-qPCR assay further confirmed that the HnRNP K antibody precipitated the promoter sequence (−668 ~ −596) of SHH (Fig. 3i).
HnRNP K mediates the ZRS variant regulating the transcriptional activity of SHH
To determine whether the ZRS variant regulates the transcriptional activity of SHH and whether HnRNP K mediates the abnormal SHH expression by the ZRS variant, a dual-luciferase reporter assay was performed. The results showed that both wild-type and mutant ZRS significantly increased luciferase activity compared with the promoter-only control; however, the mutant ZRS had increased luciferase activity more dramatically compared with the wild-type ZRS (Fig. 4b). This observation indicated that the wild-type ZRS harbored a potential enhancer of SHH. Furthermore, the mutant ZRS had stronger enhancing activity than the wild-type ZRS. Cotransfection with HnRNP K siRNA and SHH promoter plasmids showed that HnRNP K knockdown not only inhibited the activity of the SHH promoter, but it also completely inhibited the WT- or mutant ZRS-enhanced SHH promoter activity (Fig. 4c). In addition, HnRNP K overexpression or knockdown in 293T cells resulted in a corresponding increase or decrease in SHH both at the mRNA (Fig. 4d, f) and protein levels (Fig. 4e, g). Combined with the results of HnRNP K binding both the ZRS and the promoter of SHH (from the MS studies), we propose that HnRNP K mediates the ability of the variant ZRS to upregulate SHH by directly interacting with the ZRS and the SHH promoter.
ZRS variant regulates SHH hyperexpression via direct mediation by HnRNP K. (a) The scheme of the different constructs and controls used in the dual-luciferase reporter assay. (b) ZRS enhancer activity was analyzed using a dual-luciferase reporter assay in 293T cells. The wild-type ZRS significantly increased luciferase activity compared with the SHH promoter only. The mutant ZRS further enhanced luciferase activity compared with the wild-type ZRS. (c) 293T cells were cotransfected with HnRNP K small interfering RNA (siRNA) or different promoter plasmids and assayed using the dual-luciferase reporter assay. HnRNP K siRNA not only inhibited the activity of the SHH promoter, but also completely inhibited WT-ZRS or MT-ZRS enhancement of SHH promoter activity. (d–e) HnRNP K overexpression in 293T cells resulted in an increase in SHH messenger RNA (mRNA) level as determined by real-time polymerase chain reaction (PCR) (d) and an increase in SHH protein level by western blot (e). (f–g) HnRNP K siRNA-transfected 293T cells exhibited decreased SHH mRNA by real-time PCR (f) and SHH protein by western blot (g). Data represent mean ± SD. *p < 0.05, significantly different from control (n = 3). MT mutant, WT wild-type.
DISCUSSION
PPD I is a common form of polydactyly. PPD I cases are mostly sporadic,27,28 which may be the reason why the genes responsible for it have not yet been identified. In this study, we recruited a four-generation Chinese family with familial isolated PPD I; this family provided us with the opportunity to identify a predisposing variant in individuals with PPD I.
In this family, the affected members show phenotypic variation from typical thumb duplication of the distal phalanx or incomplete thumb duplication to slight sesamoids between the two phalanges of the thumbs. However, III-9 with heterozygous variant manifests small sesamoids, which are found in normal members II-2, II-5, II-9, II-13, III-2, III-6, III-7, III-10, and III-13. Hence, the variant in our PPD family shows incomplete penetrance. The 295C>T variant in ZRS (chr7:156,584,275, hg19) associated with PPD II in multiple English families also shows incomplete penetrance,29 which is consistent with our result. The incomplete penetrance was also observed in our mouse model. The penetrance of heterozygotes was 81% (150/185), compared with 100% in homozygotes (92/92) (Supplementary Table 9). Haploinsufficiency caused by heterozygous variant might account for the variable penetrance.
In this study, we showed that a T/A point variant at 446 bp of ZRS led to PPD I; this is the first report identifying the genetic variation causing isolated PPD I. However, follow-up sequencing in another seven sporadic PPD I patients from different families did not identify a variant in the ZRS. One possible reason is that the sporadic PPD I may be caused by other genetic mechanisms. The generic term “ZRS-associated syndromes” for limb malformations caused by variants of ZRS has been suggested,30 and our study will assist in developing this new genetic classification system for PPD.
The ZRS is a highly conserved region in mammals. To date, variants associated with most PPD types are scattered through the ~800-bp ZRS region. Experiments in animal models showed that these variants can cause ectopic anterior expression of SHH, leading to ectopic digits.31 In our study, the new T>A replacement at position 447 of the ZRS (at position 446 of the human ZRS) also resulted in ectopic anterior expression of Shh in the hindlimbs, which is in agreement with previous reports.
The molecular mechanisms by which ZRS regulates SHH expression remain largely unknown. Studies have suggested that variants might change the affinities of certain transcription factors (TFs) to their binding sites within the ZRS, thereby activating SHH expression at an ectopic site in the limb bud.13,32 For example, there are CDX, ETS factor family, SOX9, and HnRNP U binding sites in the ZRS region and these TFs play distinct roles in the spatial patterning of SHH.33,34 The developing limb buds are preferable for studying the interaction between protein with the ZRS. However, it is difficult to extract E11.5 limb bud nucleoprotein. In this study, we selected the Caco-2 cell line to search for the possible binding protein to the ZRS. Caco-2 cells do express SHH in both differentiated and undifferentiated stages.32 It has been investigated whether the TF-binding properties of the mutated ZRS differs from those of the wild type.32 In contrast to primary fibroblasts or immortalized limb mesenchymal cell lines,35 Caco-2 is more freely available. The results from Caco-2 cells confirmed that HnRNP K binds to the mutant ZRS and that binding differences exist between mutant and wild-type ZRS. HnRNP K, a nucleic acid binding protein, has been identified as a component of hnRNP complexes. HnRNP K play an important role in chromatin organization and transcription.36,37 It possesses sequence-specific binding activity, interacting with both single- and double-stranded DNA, which makes HnRNP K a good candidate for a long-distance transcription factor regulator.38
Here, we provided the first strong evidence confirming that HnRNP K not only preferentially binds the mutant ZRS compared with the wild-type ZRS, but it also binds the promoter of SHH. The dual-luciferase reporter assay further confirmed that HnRNP K knockdown not only inhibited the activity of the SHH promoter but also completely inhibited the enhancement of the SHH promoter by wild-type or mutant ZRS. Kline et al. reported that a syndrome due to loss-of-function variants in HNRNPK includes finger clinodactyly, postaxial polydactyly, and multiple vertebral segmentation defects.39 The current molecular data suggest that the balance between the activation and repression of SHH signaling specifies digit number as well as a polarization of digit identity.40 The abnormal activation or repression of SHH signaling in the limb bud can cause preaxial or postaxial polydactyly respectively.40 Therefore, we speculate that the loss-of-function variants in HNRNPK might repress the activity of SHH and lead to postaxial polydactyly. While in our case, mutant ZRS binds more HnRNP K, which then strengthens SHH transcription and may cause PPD I. Neither postaxial polydactyly nor clinodactyly were observed in our patients.
Nevertheless, our study has some limitations to be addressed in future studies. First, our model does not examine the HnRNP K binding and regulating the ZRS in vivo due to the difficulties of getting enough limb buds from mouse embryos. It would be more convincing to perform EMSA and chip assay with limb bud nuclear extract, as in Lettice et al.33 Second, we did not propose a possible model to explain how the point variant leads to ectopic expression of Shh mediated by HnRNP K. In previous existing models of ETS transcription factors and ZRS action,33 GABPα and ETS1 binding ZRS can activate the transcription of Shh and are expressed mainly in the posterior limb, while ETV4 and ETV5 binding ZRS repress the transcription of Shh and are expressed in both the posterior and anterior limb. When the variant occurs, an increase in the binding sites of the activating ETS factors and a decrease in the binding sites of the repressing ETS factors can lead to Shh ectopic expression, while our study shows that HnRNP K binding mutant ZRS can activate SHH expression. However, there is no difference in the Hnrnpk expression between the posterior and anterior part whatever the genotype (Supplementary Table 9). Therefore the underlying mechanism is not consistent with the reported model and needs further confirmation.
In summary, for the first time, we report a genetic variation that causes PPD I. We further show that the point variant changed the binding affinity of HnRNP K to its target in the ZRS in vitro and also leads to ectopic anterior and increased posterior expression of Shh in developing mouse embryonic limbs. Furthermore, HnRNP K was identified for the first time binding both the promoter of SHH and its long-range cis-regulator ZRS. In vitro functional studies further showed that HnRNP K mediates the regulation of abnormal SHH expression via the ZRS variant by changing its interaction with the ZRS, which may be associated with the PPD I. The identification of additional long-range cis-regulatory elements and their related binding factors, as well as genetic information from noncoding genomic regions, will augment our understanding of how aberrant temporal and spatial expression of SHH may lead to polydactyly.
References
Wu P-F, Guo S, Fan X-F, et al. A novel ZRS mutation in a Chinese patient with preaxial polydactyly and triphalangeal thumb. Cytogenet Genome Res. 2016;149:171–175.
Al-Qattan MM. Preaxial polydactyly of the upper limb viewed as a spectrum of severity of embryonic events. Ann Plast Surg. 2013;71:118–124.
Al-Qattan MM. Zone of polarizing activity regulatory sequence mutations/duplications with preaxial polydactyly and longitudinal preaxial ray deficiency in the phenotype: a review of human cases, animal models, and insights regarding the pathogenesis. Biomed Res Int. 2018;2018:1–9.
Ng ZY, Sau PY, Lim GJ-S, Discordant Type I. Preaxial polydactyly in monozygotic twins on the same hand. Ann Plast Surg. 2015;75:398–400.
Materna-Kiryluk A, Jamsheer A, Wisniewska K, Wieckowska B, Limon J, Borszewska-Kornacka M, et al. Epidemiology of isolated preaxial polydactyly type I: data from the Polish Registry of Congenital Malformations (PRCM). BMC Pediatr. 2013;13:1471–2431.
Lettice LA, Horikoshi T, Heaney SJH, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci U S A. 2002;99:7548–7553.
VanderMeer JE, Lozano R, Sun M, et al. A novel ZRS mutation leads to preaxial polydactyly type 2 in a heterozygous form and Werner mesomelic syndrome in a homozygous form. Hum Mutat. 2014;35:945–948.
Biesecker LG. Polydactyly: how many disorders and how many genes? 2010 update. Dev Dyn. 2011;240:931–942.
Klopocki E, Ott CE, Benatar N, Ullmann R, Mundlos S, Lehmann K. A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J Med Genet. 2008;45:370–375.
Sun M, Ma F, Zeng X, et al. Triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV are caused by genomic duplications involving the long range, limb-specific SHH enhancer. J Med Genet. 2008;45:589–595.
Norbnop P, Srichomthong C, Suphapeetiporn K, Shotelersuk V. ZRS 406A>G mutation in patients with tibial hypoplasia, polydactyly and triphalangeal first fingers. J Hum Genet. 2014;59:467–470.
Wang B, Diao Y, Liu Q, et al. An increased duplication of ZRS region that caused more than one supernumerary digits preaxial polydactyly in a large Chinese family. Sci Rep. 2016;6:38500.
Laurell T, VanderMeer JE, Wenger AM, et al. A novel 13 base pair insertion in the sonic hedgehog ZRS limb enhancer (ZRS/LMBR1) causes preaxial polydactyly with triphalangeal thumb. Hum Mutat. 2012;33:1063–1066.
VanderMeer JE, Ahituv N. Cis-regulatory mutations are a genetic cause of human limb malformations. Dev Dyn. 2011;240:920–930.
Lettice LA, Devenney P, De Angelis C, Hill RE. The conserved sonic hedgehog limb enhancer consists of discrete functional elements that regulate precise spatial expression. Cell Rep. 2017;20:1396–1408.
Anderson E, Hill RE. Long range regulation of the sonic hedgehog gene. Curr Opin Genet Dev. 2014;27:54–59.
Tickle C, Towers M. Sonic hedgehog signaling in limb development. Front Cell Dev Biol. 2017;5:14
Lettice LA, Hill RE. Preaxial polydactyly: a model for defective long-range regulation in congenital abnormalities. Curr Opin Genet Dev. 2005;15:294–300.
Manske MC, Kennedy CD, Huang JI. Classifications in brief: the Wassel Classification for Radial Polydactyly. Clinical Orthop Relat Res. 2016;475:1740–1746.
Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188.
Rigueur D, Lyons KM. Whole-mount skeletal staining. Methods Mol Biol. 2014;1130:113–121.
Zhao Q, Du Q, Wei F, Xie J, Ma X. An infectious disease–associated Il12b polymorphism regulates IL-12/23p40 transcription involving poly(ADP-ribose) polymerase 1. J Immunol. 2017;198:2935–2942.
Gao F, Zhao J, Chen L, et al. The NDH-1L-PSI supercomplex is important for efficient cyclic electron transport in cyanobacteria. Plant Physiol. 2016;172:1451–1464.
Zheng S, Zhou H, Chen Z, et al. Type III transforming growth factor-β receptor RNA interference enhances transforming growth factor β3-induced chondrogenesis signaling in human mesenchymal stem cells. Stem Cell Int. 2018;2018:1–11.
Chung IC, Chen LC, Chung AK, et al. Matrix metalloproteinase 12 is induced by heterogeneous nuclear ribonucleoprotein K and promotes migration and invasion in nasopharyngeal carcinoma. BMC Cancer. 2014;14:348
Petit F, Jourdain A-S, Holder-Espinasse M, et al. The disruption of a novel limb cis-regulatory element of SHH is associated with autosomal dominant preaxial polydactyly-hypertrichosis. Eur J Hum Genet. 2015;24:37–43.
Fujioka H, Ariga T, Horiuchi K, et al. Molecular analysis of non-syndromic preaxial polydactyly: preaxial polydactyly type-IV and preaxial polydactyly type-I. Clin Genet. 2005;67:429–433.
Orioli I, Castilla E. Thumb/hallux duplication and preaxial polydactyly type I. Am J Med Genet. 1999;82:219–224.
Furniss D, Lettice LA, Taylor IB, et al. A variant in the sonic hedgehog regulatory sequence (ZRS) is associated with triphalangeal thumb and deregulates expression in the developing limb. Hum Mol Genet. 2008;17:2417–2423.
Wieczorek D, Pawlik B, Li Y, et al. A specific mutation in the distant sonic hedgehog (SHH)cis-regulator (ZRS) causes Werner mesomelic syndrome (WMS) while complete ZRS duplications underlie Haas type polysyndactyly and preaxial polydactyly (PPD) with or without triphalangeal thumb. Hum Mutat. 2010;31:81–89.
Amano T, Sagai T, Seki R, Shiroishi T. Two types of etiological mutation in the limb-specific enhancer of Shh. G3 (Bethesda). 2017;7:2991–2998.
Farooq M, Troelsen JT, Boyd M, et al. Preaxial polydactyly/triphalangeal thumb is associated with changed transcription factor-binding affinity in a family with a novel point mutation in the long-range cis-regulatory element ZRS. Eur J Hum Genet. 2010;18:733–736.
Lettice Laura A, Williamson I, Wiltshire John H, et al. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev Cell. 2012;22:459–467.
Zhao J, Ding J, Li Y, et al. HnRNP U mediates the long-range regulation of Shh expression during limb development. Hum Mol Genet. 2009;18:3090–3097.
Williamson I, Eskeland R, Lettice LA, et al. Anterior-posterior differences in HoxD chromatin topology in limb development. Development. 2012;139:3157–3167.
Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135:851–867.
Barboro P, Ferrari N, Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 2014;352:152–159.
Saradhi M, Kumari S, Rana M, Mukhopadhyay G, Tyagi RK. Identification and interplay of sequence specific DNA binding proteins involved in regulation of human pregnane and xenobiotic receptor gene. Exp Cell Res. 2015;339:187–196.
Au PYB, You J, Caluseriu O, et al. GeneMatcher aids in the identification of a new malformation syndrome with intellectual disability, unique facial dysmorphisms, and skeletal and connective tissue abnormalities caused by de novo variants in HNRNPK. Hum Mutat. 2015;36:1009–1014.
Malik S. Polydactyly: phenotypes, genetics and classification. Clin Genet. 2014;85:203–212.
Acknowledgements
We thank all participants for their support of this study. This work was supported by the National Natural Science Foundation of China (81472039, 81772302, 81572091, and 81772293) and the Research Fund of Popular Science of Guangzhou City (201803010122).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
The URLs
OMIM, http://www.omim.org/;dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/; Homo sapiens Sonic hedgehog (SHH) promoter, http://genome.ucsc.edu/cgi-bin/hgGene?hgg_gene=uc003wmk.1&hgg_prot=Q15465&hgg_chrom=chr7&hgg_start=155802863&hgg_end=155812273&hgg_type=knownGene&db=hg38&hgsid=419876371_rQNNLqhoMngw09ElHZFiRNkeNNeH; NM 009170, https://www.ncbi.nlm.nih.gov/nuccore/NM_009170.3; NM_031262, https://www.ncbi.nlm.nih.gov/nuccore/NM_031262.3/; CS-NEG-GA03-02, CS-ZRS01-GA03 and CS-ZRS02-GA03, https://www.genecopoeia.co.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, C., Yang, X., Zhou, H. et al. A novel ZRS variant causes preaxial polydactyly type I by increased sonic hedgehog expression in the developing limb bud. Genet Med 22, 189–198 (2020). https://doi.org/10.1038/s41436-019-0626-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-019-0626-7
Keywords
This article is cited by
-
Hnrnpk is essential for embryonic limb bud development as a transcription activator and a collaborator of insulator protein Ctcf
Cell Death & Differentiation (2023)