Abstract
Purpose
Familial exudative vitreoretinopathy (FEVR) is a blindness-causing retinal vascular disease characterized by incomplete vascularization of the peripheral retina and by the absence or abnormality of the second/tertiary capillary layers in the deep retina. Variants in known FEVR disease genes can only explain about 50% of FEVR-affected cases. We aim to identify additional disease genes in patients with FEVR.
Methods
We applied exome sequencing analysis in a cohort of 49 FEVR families without pathogenic variants in known FEVR genes. Functions of the affected proteins were evaluated by reporter assay. Knockout mouse models were generated by endothelial-specific Cre line.
Results
Three novel rare heterozygous variants in Notch ligand JAG1 were identified in FEVR families—c.413C>T p. (A138V), c.1415G>A p. (R472H), and c.2884A>G p. (T962A)—and verified by Sanger sequencing analysis. Notch reporter assay revealed that mutant JAG1 proteins JAG1-A138V and JAG1-T962A lost almost all of their activities, and JAG1-R472H lost approximately 50% of its activity. Deletion of Jag1 in mouse endothelial cells resulted in reduced tip cells at the angiogenic front and retarded vessel growth, reproducing FEVR-like phenotypes.
Conclusion
Our data suggest that JAG1 is a novel candidate gene for FEVR and pinpoints a potential target for therapeutic intervention.
Similar content being viewed by others
INTRODUCTION
Notch signaling is an evolutionarily conserved regulatory system that plays key roles in angiogenesis.1,2,3 Genetic removal of Notch components in mice results in disorganized vessels and embryonic lethality.4,5 The Notch signaling pathway plays an essential role in angiogenesis development, regulating the delicate balance of tip cell and stalk cell specification.6 In mouse models, the Notch ligands Delta like 4 (DLL4, OMIM 605185) and Jagged 1 (JAG1, OMIM 601920) have distinct opposite roles during angiogenesis, with DLL4 serving as an inhibiting protein and JAG1 as a proangiogenic factor.7,8 Consistent with these roles, Dll4 and Jag1 also exhibit distinct spatial expression patterns in the retinal vasculature, with Dll4 highly expressed in tip cells and present at the edge of the growing plexus, while Jag1 expression is low/absent in tips cells but higher in adjacent stalk cells.7 JAG1 is a member of the Jagged/Serrate family and plays important roles in various signaling processes. Pathogenic variants in JAG1 cause Alagille syndrome.9 Additionally, in genetic studies using mouse models, loss of Jag1 in endothelial cells led to reduced sprouting at the angiogenic fronts.7 However, the role of JAG1 in human vascular diseases remains elusive.
Familial exudative vitreoretinopathy (FEVR) is an angiogenesis disease characterized by the incomplete vascularization of the peripheral retina and by the absence or abnormality of the second/tertiary capillary layers in the deep retina (MIM 133780).10 FEVR can lead to neovascularization, vitreous hemorrhaging, traction from the vitreous membranes, and retinal folding and detachment.11 FEVR is a heterogeneous disease, and the presentation of FEVR can be highly variable. Clinical phenotypes of patients harboring the same pathogenic variant might range from no noticeable phenotypes to complete blindness during childhood.12 Unfortunately, no effective intervention is currently available for this disorder.
FEVR can be inherited as an autosomal dominant, recessive, or X-linked disease.13 Variants in nine genes have been associated with FEVR, and another locus has been mapped but remains to be identified; these genes are as follows: NDP (MIM 300658),13 FZD4 (MIM 604579),14,15,16 LRP5 (MIM 603506),17,18,19 TSPAN12 (MIM 613138),20,21 ATOH7 (MIM 609875),22 ZNF408 (MIM 616454),11 KIF11 (MIM 148760),23 RCBTB1 (MIM 607867),24 CTNNB1 (MIM 116806),25 and EVR3 on chromosome 11p12-13 (MIM 605750).26 Variants in these genes can explain only ∼50% of FEVR-affected families.18 In addition, variants in four core components of the Norrin-β-catenin signaling pathway, NDP, FZD4, LRP5, and TSPAN12, account for most FEVR cases. Understanding the genetic causes of the remaining cases might provide insights into the disease mechanisms. Therefore, in this study, we aimed to uncover additional disease genes using exome sequencing (ES), coupled with functional studies. In recent years ES has emerged as a powerful method for identifying retinal disease genes.24,25,27,28,29,30,31,32,33
MATERIALS AND METHODS
Patients and clinical evaluation
This study adhered to the tenets of the Declaration of Helsinki and was approved by the Institutional Research Committee of Sichuan Provincial People’s Hospital and Xinhua Hospital. Written informed consent was obtained from all subjects participating and from guardians for minors. Participants were evaluated by a clinical ophthalmologist based on fundus photographic and angiographic changes by fluorescein angiograph (FFA). Angiographic examinations were assessed by intravenous injection of fluorescein dye. All cases with premature birth, history of oxygen inhalation, and drug abuse were excluded.
Genomic DNA preparation
Peripheral blood from FEVR families and control individuals was collected into EDTA anticoagulant tubes, and genomic DNA was extracted using a blood DNA extraction kit according to the manufacturer’s protocol (Qiagen, Germantown, MD, USA).
Exome sequencing and bioinformatic analysis
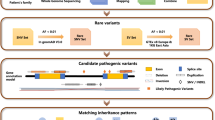
To identify additional genes for FEVR, we applied ES to search for potential genetic variants in 49 FEVR families without pathologic variants in known FEVR genes. ES was performed on the proband from each family. Detailed library construction, sequencing, and data filtering methods are included in the online supplementary methods. Briefly, genomic DNA was fragmented, and exonic regions were captured by SureSelect All Eco V5 Capture Kits (Agilent Technologies, Santa Clara, CA, USA). The libraries were sequenced on a HiSeq2500 sequencer (Illumina, San Diego, CA, USA). Raw reads were aligned to GRCH37 and processed by the Genome Analysis Toolkit (GATK). Variants that presented frequencies greater than 0.1% in the dbSNP database, 1000 Genomes (http://www.internationalgenome.org/), National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP), ExAC Browser (Beta, http://exac.broadinstitute.org/), and Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org/) database were excluded. The remaining variants were annotated with ANNOVAR software (version 2014).
Variants validation
Sanger sequencing was performed to confirm the variants identified by ES. Primers were designed to amplify genomic DNA fragments containing candidate variants (Supplementary table S1). DNA samples from the three FEVR families and 2500 control individuals were subjected to Sanger sequencing.
Site-directed mutagenesis
C-terminal Flag-tagged JAG1 expression vectors were purchased from Origene Inc. (Rockville, MD, USA). Three missense variants were introduced into wild-type (WT) JAG1 complementary DNA (cDNA) by site-directed mutagenesis using a QuikChange® Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). Mutant recombinant plasmids containing JAG1-Flag fusion constructs were verified by DNA sequencing.
Western blotting (WB)
NIH-3T3 cells were seeded onto a 6-well plate (Corning, NY, USA) and transfected with 3000 ng WT plasmid, mutant JAG1 plasmids or empty vectors using Lipofectamine 3000 (Invitrogen, CA, USA) when cell confluence reached approximately 70%. After 48 hours, cells were lysed in sodium dodecyl sulfate (SDS) lysis buffer (2% SDS and 62.5 mM Tris-HCl, pH 6.8, containing protease inhibitor cocktail tablets ordered from Roche Inc.) and sonicated three times for 5 seconds. Equal amounts of protein (20 μg) were loaded onto a 10% polyacrylamide gel and analyzed by immunoblotting. The antibodies used for WB were FLAG (Cat# F1804, Sigma, St. Louis, MO, USA, 1:3000 dilution), JAG1 (Cat# 70109S, Cell Signalling Technology, Danvers, MA, USA, 1:2000 dilution) and GAPDH (Cat# 60004-1-Ig, Proteintech, Wuhan, China, 1:3000 dilution). HRP-conjugated goat antirabbit secondary antibody (Cat# 7074, Cell Signaling Technology, 1:5000 dilution) and goat antimouse secondary antibody (Cat# 7076, Cell Signaling Technology, 1:5000 dilution) were used for immunoblotting.
JAG1 protein activity analysis by luciferase assays
NIH-3T3 cells maintained in Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) were seeded onto a 6-cm dish and a 6-well plate respectively. Cells in the 6-cm dish were transfected with 5 μg plasmid DNA named 4× wt CBF1 Luc134 (the Notch-dependent reporter) using Lipofectamine 3000 (Invitrogen) when they reached 70–80% confluence. NIH-3T3 cells in the 6-well plate were transfected with WT JAG1, JAG1-A138V, JAG1-R472H, JAG1-T962A, and negative control plasmid respectively. After 24 hours, the cells containing 4× wt CBF1 Luc1 plasmid were seeded onto the 6-well plate mentioned above. Luciferase activities were quantified after the cells were cocultured for 36 hours. Briefly, the cells were lysed in 500 μl lysis buffer per well for 10 minutes at room temperature after the plate was washed twice using phosphate buffered saline (PBS). After the cell lysate was centrifuged at 4 °C and 12,000× g for 5 minutes, the supernatant was aspirated into a new tube. Luciferase activity was determined using a Trans Detect Double-Luciferase Reporter Assay Kit (Transgene, Beijing, China). All assays were repeated three times.
Mouse models and genotyping
All animal protocols were approved by the Institutional Animal Care and Use Committee of Sichuan Provincial People’s Hospital. All experimental procedures were performed in accordance with the approved study protocols and relevant regulations. Mice were raised in cyclic lighting conditions with a 12-hour light and 12-hour dark cycle.
Floxed Jag1 mice were obtained from The Jackson Laboratory35(Jag1tm2Grid, https://www.jax.org/strain/031272, named Jag1flox/+). Jag1flox/+ mice were mated to Pdgfb-iCreER transgenic mice36 to generate Jag1flox/flox, Pdgfb-iCreER (hereafter named Jag1 iECKO) mice to inactivate Jag1 in endothelial cells after tamoxifen induction. Tamoxifen stock solution was prepared by dissolving 100 mg tamoxifen salt (Sigma, St. Louis, MO, USA) in 10 ml ethanol. On the day of injection, a 1 mg/ml working solution was prepared by mixing the 10 mg/ml stock solution with corn oil (Sigma). Jag1 iECKO mice and littermate controls of both genders were intraperitoneally injected with a daily dose of 25 mg/kg body weight of the 1 mg/ml tamoxifen solution on postnatal days 1–3.37 Genomic DNA extracted from mouse tails were amplified by polymerase chain reaction (PCR). PCR primer pairs were used to genotype the animals.
Immunostaining of retinal endothelial cells
Retinal dissection was carried out as previously described,37 and whole-mounted retinas were preserved in 0.4% paraformaldehyde (PFA) (Sigma). Enucleated eyes were fixed with 4% PFA and embedded in Tissue-Tek optimal cutting temperature compound (Sakura Finetek, Torrance, CA, USA). Before immunostaining, whole-mounted retinas and cryosections (12 μm, Leica CM1950) were rinsed in PBS (Sigma, St. Louis, MO, USA) three times (5 min/time) and blocked in PBS containing 5% FBS (Invitrogen, Waltham, MA, USA) and 0.2% Triton X-100 for 30 minutes at room temperature, followed by incubation with primary antibodies at 4 °C overnight. The following primary antibodies were diluted in blocking buffer: Isolectin GS-IB4 (1:100; Invitrogen, Waltham, MA; I21411). Then, the sections were washed three times with PBS and labeled for 1–4 hours with Alexa Fluor™-488- or Alexa Fluor™-594-labeled goat antirat or antirabbit IgG or donkey antigoat IgG secondary antibody (1:500; Invitrogen). Images were captured on a LSM 800 confocal scanning microscope (Zeiss, Thornwood, NY, USA). The person who was assigned to cell counting was blind to the genotypes of the samples. No animals were excluded from this study.
Statistical analysis
Statistical significance was determined using Student’s t test or one-way analysis of variance (ANOVA) and was assumed at P < 0.05. Quantitative data are presented as the means ± sem, as indicated in the figure legends. Statistical analyses were performed using GraphPad Prism 6 software. No sample calculation was performed. No randomization was performed for animal studies.
RESULTS
ES unveiled three variants in JAG1 likely to be pathogenic in three FEVR families
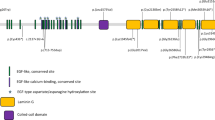
ES was performed to identify pathological variants in 49 FEVR families without pathologic variants in known FEVR genes. Three novel heterozygous variants in Notch ligand JAG1 were identified in three FEVR families: NM_000214.3:c.413C>T p. (A138V), c.1415G>A p. (R472H), and c.2884A>G p. (T962A). Segregation of the three variants was verified by Sanger sequencing analysis in the corresponding families (Fig. 1a–f). The JAG1 gene encodes a 1219 amino acid protein and acts as a ligand in Notch signaling. Previous study reported that variants in JAG1 cause Alagille syndrome.9 The three variants identified in this study have not been reported to cause Alagille syndrome. These variants are unique in our in-house database of 2805 geographically matched individuals analyzed by ES and were not observed in 2500 ethnic-matched control individuals analyzed by Sanger sequencing. For the proband of family 1, subject II:1, 85,685 single-nucleotide polymorphisms (SNPs) and 7410 indels were identified (Supplementary Table S2). For the proband of family 2, subject II:1, 87,186 SNPs and 8309 indels were identified (Supplementary Table S2). For the proband of family 3, subject II:1, 88,320 SNPs and 8368 indels were identified (Supplementary Table S2).
Variants in the JAG1 gene identified in three families with familial exudative vitreoretinopathy (FEVR). (a–f) Three families diagnosed with FEVR and corresponding Sanger sequencing results indicating the genotypes of the family members. Three variants in the JAG1 gene, c.413C>T (p.A138V), c.1451G>A (p.R472H), and c.2884A>G (p.T962A), were verified in families 1, 2, and 3, respectively. Arrows indicate the variant locus. (g) Protein sequence alignment of amino sequences surrounding the JAG1 variants with orthologues from H. sapiens to D. melanogaster. Note that the amino acid sequences surrounding the affected amino acid residues are highly conserved.
In gnomAD (http://gnomad.broadinstitute.org/), the frequency of the p.A138V variant is zero. The frequency of p.R472H is 0.000081 (20/246,230), while the frequency of p.T962A is 0.000085 (21/246,190). FEVR can exhibit incomplete penetrance and variable expressivity. Kashani et al. reported that 58% of the asymptomatic family members demonstrated early stage FEVR and 21% demonstrated clinical or angiographic findings consistent with late stage FEVR, revealed by angiographic screening.12 This can explain the relative high frequency of p.R472H and p.T962A in those presumably unaffected individuals in gnomAD. Those individuals carrying JAG1 p.R472H and p.T962A variants might be asymptomatic. Furthermore, the amino acids affected by these three variants are highly evolutionarily conserved from H. sapiens to D. melanogaster (Fig. 1g), implicating the importance of these amino acid residues.
In family 1 (Fig. 1a), proband II:1 was a 21-year-old girl who developed retinal detachment in her right eye. Fundus photography and fundus fluorescein angiography (FFA) examination of her left eye showed FEVR phenotypes, including temporal tortuous retinal vessels and peripheral ischemia leading to neovascularization and telangiectasis (Fig. 2a, b). Ophthalmic examination of her mother revealed no abnormalities, while for her father, both eyes showed phenotypes similar to those of the proband. Specifically, the peripheral retinal was not fully vascularized with adjacent brush-shaped peripheral retinal vessels and staining of peripheral chorioretinal atrophy (Fig. 2c–f). In family 2 (Fig. 1c), proband II:1 has suffered severe ocular problems since birth, and fundus examination showed partial retinal detachment, falciform folds, and cataracts due to retinal fibrovascular proliferation (Fig. 2g, h). Ocular examination of his mother also showed peripheral avascular retina and dilated peripheral retinal vessels with fluorescein leakage at the late stage of FFA, which are typical FEVR phenotypes (Fig. 2i–l). In family 3 (Fig. 1e), the proband II:1 and his mother were also diagnosed as FEVR by an ophthalmologist.
Fundus photograph and fundus fluorescein angiography (FFA) of patients with familial exudative vitreoretinopathy (FEVR). (a–b) Examination of subject II:1 in family 1 shows temporal tortuous retinal vessels, and peripheral ischemia led to neovascularization. (c–f) FFA examination of subject I:1 in family 1 revealed that the peripheral retina was not fully vascularized with adjacent brush-shaped peripheral retinal vessels and staining of peripheral chorioretinal atrophy. (g–h) Examination of subject II:1 in family 2 revealed partial retinal detachment, falciform folds, and cataract due to retinal fibrovascular proliferation. (i–l) Examination of subject I:2 in family 2 showed peripheral avascular retina and dilated peripheral retinal vessels with fluorescein leakage at the late stage of FFA. OS, oculus sinister (left eye). OD, oculus dexter (right eye).
Three variants identified by ES negatively affect the function of JAG1 protein
The JAG1 gene reportedly plays critical roles in vascular development as a Notch signaling ligand.7 To assess the impacts of these three variants on JAG1 protein, we introduced these variants into the JAG1 coding sequence by site-directed mutagenesis. Western blotting (WB) experiments showed a unique band at approximately the 170 kDa position when probing with JAG1 or FLAG antibodies (Fig. 3a). The expression levels of mutant JAG1 proteins were comparable with those of wild-type JAG1 protein (Fig. 3a). Immunostaining studies on COS7 cells transfected with wild-type or mutant JAG1 expression plasmids showed that both wild-type and mutant JAG1 proteins were localized to the cytoplasm and cell membrane (Fig. S1). The above experiments demonstrated that these three variants identified in patients with FEVR did not impact the expression and localization of JAG1 proteins. To evaluate the effect of these variants on the biological activities of JAG1 protein, we performed a Notch reporter assay in which NIH-3T3 cells containing a Notch-sensitive luciferase construct were cocultured with cells expressing either WT JAG1 or mutant JAG1 proteins, according to a previously reported method.34 NIH-3T3 cells were transfected with a luciferase reporter construct containing four upstream binding sites of wild-type CBF1-binding elements (4× wtCBF1Luc).34 Luciferase assays revealed that the mutant JAG1 proteins JAG1-A138V and JAG1-T962A almost lost all of their activities, and JAG1-R472H lost approximately 50% of its activity (Fig. 3b). These data demonstrate that these three variants negatively affect JAG1 protein activity and are likely pathogenic.
Affected JAG1 proteins failed to activate Notch signaling. (a) Western blot analysis showed that the expression levels of affected JAG1 proteins encoded by three familial exudative vitreoretinopathy (FEVR) variants were comparable with those of wild-type JAG1 protein. A unique immune band under 180 kDa was detected when hybrid with FLAG or JAG1 antibodies, indicating the specificity of the JAG1 antibody. GAPDH was used as a loading control. (b) Luciferase assays indicated that affected JAG1 proteins encoded by three FEVR variants completely or partially lost their activities as Notch ligands. Sample size N = 4, independent biological replicates. The results shown were from one of three independent experiments. ***, p < 0.0001, two-tailed t test. WT wild type.
Deletion of Jag1 in mouse endothelial cells led to FEVR-like phenotypes
To validate the roles of JAG1 in angiogenesis, we generated an endothelial-specific knockout mouse model of Jag1 by crossing the conditional Jag1 allele with an inducible endothelial Cre line carrying Pdgfb-CreER. These Cre transgene mice express a tamoxifen-inducible form of Cre recombinase in endothelial cells under the control of the Pdgfb promoter (Fig. S2). Jag1flox/flox, Pdgfb-iCre-ER (hereafter named Jag1 iECKO), and Jag1flox/flox or Pdgfb-iCre-ER littermate controls were induced by daily intraperitoneal (IP) injections of tamoxifen starting at postnatal day 2 (P2) for three days. Deletion of Jag1 led to delayed angiogenesis (Fig. 4). Flat-mounted retinas obtained from Jag1 iECKO and control mice were stained with Isolectin B4 (IB4) to visualize blood vessel cells. At P7, the horizontal growth of blood vessels was slower in Jag1 iECKO mice than in WT control mice (Fig. 4b, left panel of Fig. 4d). To exclude the potential adverse effects of the Cre transgene in angiogenesis, we also used Pdgfb-iCre-ER mice as controls. Flat mount staining of Pdgfb-iCre-ER and Pdgfb-iCre-ER retinas revealed no abnormalities in blood vessel development. Close examination of retinal wholemounts revealed a sparse blood vessel network in the iECKO retina (Fig. 4c, right panel of Fig. 4d). The vessel density in the iECKO retina was reduced to approximately 50% of that in the control retina. In addition, the removal of Jag1 in ECs led to a reduction in the number of tips and filopodia at the angiogenic front. Immunostaining of retinal frozen sections revealed profound defects in vertical vascular growth into the deeper retinal layers in Jag1 iECKO/iECKO retina, compared with control retinas (Fig. 4e). No secondary and tertiary vessels were observed and the vascular plexus became hyperplasia in Jag1 iECKO/iECKO retinas (Fig. 4e). Therefore, loss of Jag1 in mouse retinal vascular cells leads to angiogenesis defects and certain clinical features seen in FEVR patients, supporting a role of Jag1 in angiogenesis.
Deletion of Jag1 in endothelial cells resulted in delayed radial blood vessel growth. (a) Mice were injected with tamoxifen at P1 for three days and analyzed. (b) Flat-mounted retinas obtained from Jag1 iECKO and control mice were stained with Isolectin B4 (IB4) to visualize blood vessel cells. At P7, the horizontal growth of blood vessels was slower in Jag1 iECKO mice than in wild-type (WT) control mice. Scale bar = 250 μm. (c, d) Close examination of retina wholemounts revealed a sparse blood vessel network in the iECKO retina. d/D (distance of vessel growth/radius length of the retina), was used to measure retinal vessel growth along the retina surface from the optic nerve. The vessel density in the iECKO retina was reduced to approximately 50% of that in the control retina. Sample size N = 4 for each group. Data shown were a typical result from three independent experiments. ***, p < 0.001; **, p < 0.01, two-tailed t test. Error bar, sem. Scale bar = 25 μm. (e) Retinal frozen sections of P9 control and Jag1 iECKO/iECKO mice were costained with IB4 (green) and DAPI (blue). Scale bar = 25 μm. In Jag1 iECKO/iECKO retina, profound defects in vertical vascular growth into the deeper retinal layers were observed and the vascular plexus became hyperplastic. No secondary and tertiary vessels were observed in these retinas.
DISCUSSION
In this study, we reported three heterozygous pathological variants in JAG1 in nonsyndromic FEVR patients and demonstrated that deletion of Jag1 in mice led to angiogenesis defects in the retina. Most previously reported FEVR cases are caused by pathogenic variants in the four core components of the Norrin/β-catenin signaling pathway, NDP, FZD4, LRP5, and TSPAN12. Our study shows that JAG1, an important ligand for the Notch signaling pathway, is a novel FEVR candidate gene and conceptually expands our knowledge regarding pathogenesis of FEVR disease. To the best of our knowledge, this is the first report of possible involvement of Notch ligand JAG1 in the blinding disease FEVR, supported by genetic data, cell biology assays, and animal models. Identification of additional cases of presumably pathologic variants in JAG1 on individuals with FEVR in the future will provide additional evidence for the role of JAG1 in FEVR.
Our data also provide potential targets for therapeutic intervention. Previous studies showed that JAG1 is relevant to proangiogenic activity during angiogenesis. Zeng et al. showed that overexpression of JAG1 in cancer cells promoted neovascularization and the growth of grafted tumors in mice.38 Inhibition of JAG1 in disease conditions might allow the suppression of blood vessel growth, which is important in the treatment of tumor and neovascularization conditions such as age-related macular degeneration or diabetic retinopathy. Moreover, investigating the relationship of Norrin/β-catenin and Jag1/Notch signaling in FEVR pathogenesis is valuable to understand the mechanisms underlying retinal angiogenesis.
References
Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by notch. Dev Cell. 2009;16:196–208.
Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689.
Roca C, Adams RH. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 2007;21:2511–2524.
Hrabe de Angelis M, McIntyre J 2nd, Gossler A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 1997;386:717–721.
Xue Y, Gao X, Lindsell CE, et al. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8:723–730.
Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–784.
Benedito R, Roca C, Sorensen I, et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135.
Pedrosa AR, Trindade A, Fernandes AC, et al. Endothelial Jagged1 antagonizes Dll4 regulation of endothelial branching and promotes vascular maturation downstream of Dll4/Notch1. Arterioscler Thromb Vasc Biol. 2015;35:1134–1146.
Li L, Krantz ID, Deng Y, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251.
Criswick VG, Schepens CL. Familial exudative vitreoretinopathy. Am J Ophthalmol. 1969;68:578–594.
Collin RW, Nikopoulos K, Dona M, et al. ZNF408 is mutated in familial exudative vitreoretinopathy and is crucial for the development of zebrafish retinal vasculature. Proc Natl Acad Sci U S A. 2013;110:9856–9861.
Kashani AH, Learned D, Nudleman E, Drenser KA, Capone A, Trese MT. High prevalence of peripheral retinal vascular anomalies in family members of patients with familial exudative vitreoretinopathy. Ophthalmology. 2014;121:262–268.
Chen ZY, Battinelli EM, Fielder A, et al. A mutation in the Norrie disease gene (NDP) associated with X-linked familial exudative vitreoretinopathy. Nat Genet. 1993;5:180–183.
Robitaille J, MacDonald ML, Kaykas A, et al. Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat Genet. 2002;32:326–330.
Xu Q, Wang Y, Dabdoub A, et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895.
Toomes C, Bottomley HM, Jackson RM, et al. Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet. 2004;74:721–730.
Gong Y, Slee RB, Fukai N, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523.
Salvo J, Lyubasyuk V, Xu M, et al. Next-generation sequencing and novel variant determination in a cohort of 92 familial exudative vitreoretinopathy patients. Invest Ophthalmol Vis Sci. 2015;56:1937–1946.
Jiao X, Ventruto V, Trese MT, Shastry BS, Hejtmancik JF. Autosomal recessive familial exudative vitreoretinopathy is associated with mutations in LRP5. Am J Hum Genet. 2004;75:878–884.
Nikopoulos K, Gilissen C, Hoischen A, et al. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:240–247.
Poulter JA, Ali M, Gilmour DF, et al. Mutations in TSPAN12 cause autosomal-dominant familial exudative vitreoretinopathy. Am J Hum Genet. 2016;98:592.
Khan K, Logan CV, McKibbin M, et al. Next generation sequencing identifies mutations in Atonal homolog 7 (ATOH7) in families with global eye developmental defects. Hum Mol Genet. 2012;21:776–783.
Robitaille JM, Gillett RM, LeBlanc MA, et al. Phenotypic overlap between familial exudative vitreoretinopathy and microcephaly, lymphedema, and chorioretinal dysplasia caused by KIF11 mutations. JAMA Ophthalmol. 2014;132:1393–1399.
Wu JH, Liu JH, Ko YC, et al. Haploinsufficiency of RCBTB1 is associated with Coats disease and familial exudative vitreoretinopathy. Hum Mol Genet. 2016;25:1637–1647.
Panagiotou ES, Sanjurjo Soriano C, Poulter JA, et al. Defects in the cell signaling mediator beta-catenin cause the retinal vascular condition FEVR. Am J Hum Genet. 2017;100:960–968.
Downey LM, Keen TJ, Roberts E, Mansfield DC, Bamashmus M, Inglehearn CF. A new locus for autosomal dominant familial exudative vitreoretinopathy maps to chromosome 11p12-13. Am J Hum Genet. 2001;68:778–781.
Gong B, Zhang H, Huang L, et al. Mutant RAMP2 causes primary open-angle glaucoma via the CRLR-cAMP axis. Genet Med. 2019 Apr 19; 10.1038/s41436-019-0507-0 [Epub ahead of print].
Zhang L, Sun Z, Zhao P, et al. Whole-exome sequencing revealed HKDC1 as a candidate gene associated with autosomal-recessive retinitis pigmentosa. Hum Mol Genet. 2018;27:4157–4168.
Zhou Y, Li S, Huang L, et al. A splicing mutation in aryl hydrocarbon receptor associated with retinitis pigmentosa. Hum Mol Gene. 2018;27:2563–2572.
Xu M, Xie YA, Abouzeid H, et al. Mutations in the spliceosome component CWC27 cause retinal degeneration with or without additional developmental anomalies. Am J Hum Genet. 2017;100:592–604.
El Shamieh S, Neuille M, Terray A, et al. Whole-exome sequencing identifies KIZ as a ciliary gene associated with autosomal-recessive rod-cone dystrophy. Am J Hum Genet. 2014;94:625–633.
Nikopoulos K, Farinelli P, Giangreco B, et al. Mutations in CEP78 cause cone-rod dystrophy and hearing loss associated with primary-cilia defects. Am J Hum Genet. 2016;99:770–776.
Zeitz C, Jacobson SG, Hamel CP, et al. Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2013;92:67–75.
Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959.
Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4.
Claxton S, Kostourou V, Jadeja S, Chambon P, Hodivala-Dilke K, Fruttiger M. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis. 2008;46:74–80.
Pitulescu ME, Schmidt I, Benedito R, Adams RH. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat Protoc. 2010;5:1518–1534.
Zeng Q, Li S, Chepeha DB, et al. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer Cell. 2005;8:13–23.
Acknowledgements
The authors thank all affected individuals and their family members for their participation. This study was supported by grants from the National Key Scientific Research Program (www.most.gov.cn, 2016YFC0905200, 2015CB554100), the National Natural Science Foundation of China (http://www.nsfc.gov.cn/, 81470668, 81700876, 81790643, 81770950), and the Department of Science and Technology of Sichuan Province (www.scst.gov.cn, 2018YSZH0020, 19ZYNLJS, 2016TD0009, 2017TJPT0010, 2018JZ0019, 2014JQ0023, 2015SZ0242, 2015SZ0060, 2019JDZH0029). The funders had no role in study design, data collection and analysis, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, L., Zhang, X., Xu, H. et al. Exome sequencing revealed Notch ligand JAG1 as a novel candidate gene for familial exudative vitreoretinopathy. Genet Med 22, 77–84 (2020). https://doi.org/10.1038/s41436-019-0571-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-019-0571-5
Keywords
This article is cited by
-
Characterization of a novel heterozygous frameshift variant in NDP gene that causes familial exudative vitreoretinopathy in female patients
Molecular Genetics and Genomics (2024)
-
Whole exome sequencing revealed 14 variants in NDP, FZD4, LRP5, and TSPAN12 genes for 20 families with familial exudative vitreoretinopathy
BMC Medical Genomics (2022)
-
Long-term follow-up of a patient with JAG1-associated retinopathy
Documenta Ophthalmologica (2021)